Detecting Pest-Infested Forest Damage through Multispectral Satellite Imagery and Improved UNet++
Abstract
1. Introduction
2. Study Area and Data
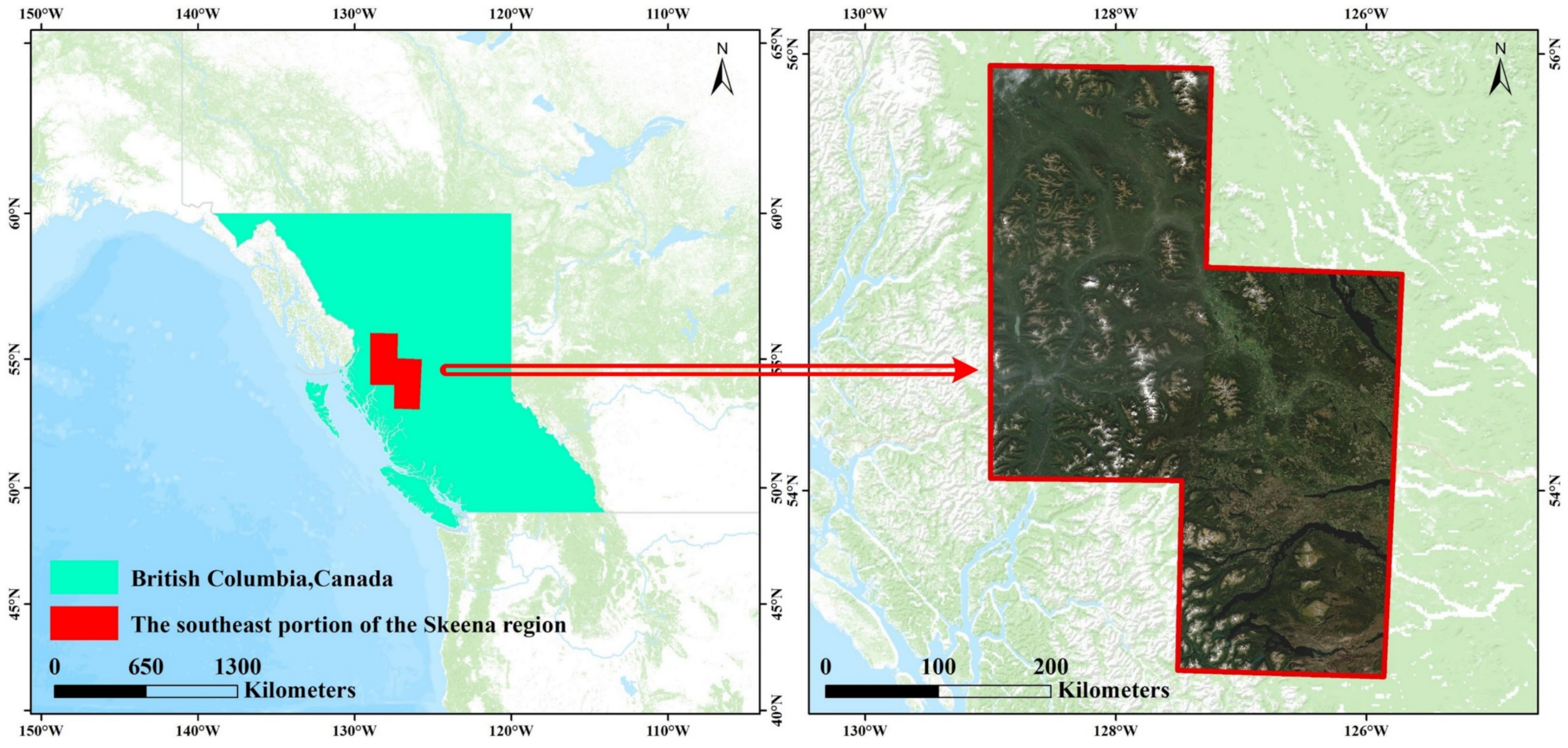
2.1. Study Area
2.2. Sentinel-2 Data and Preprocessing
2.2.1. Sentinel-2 Data
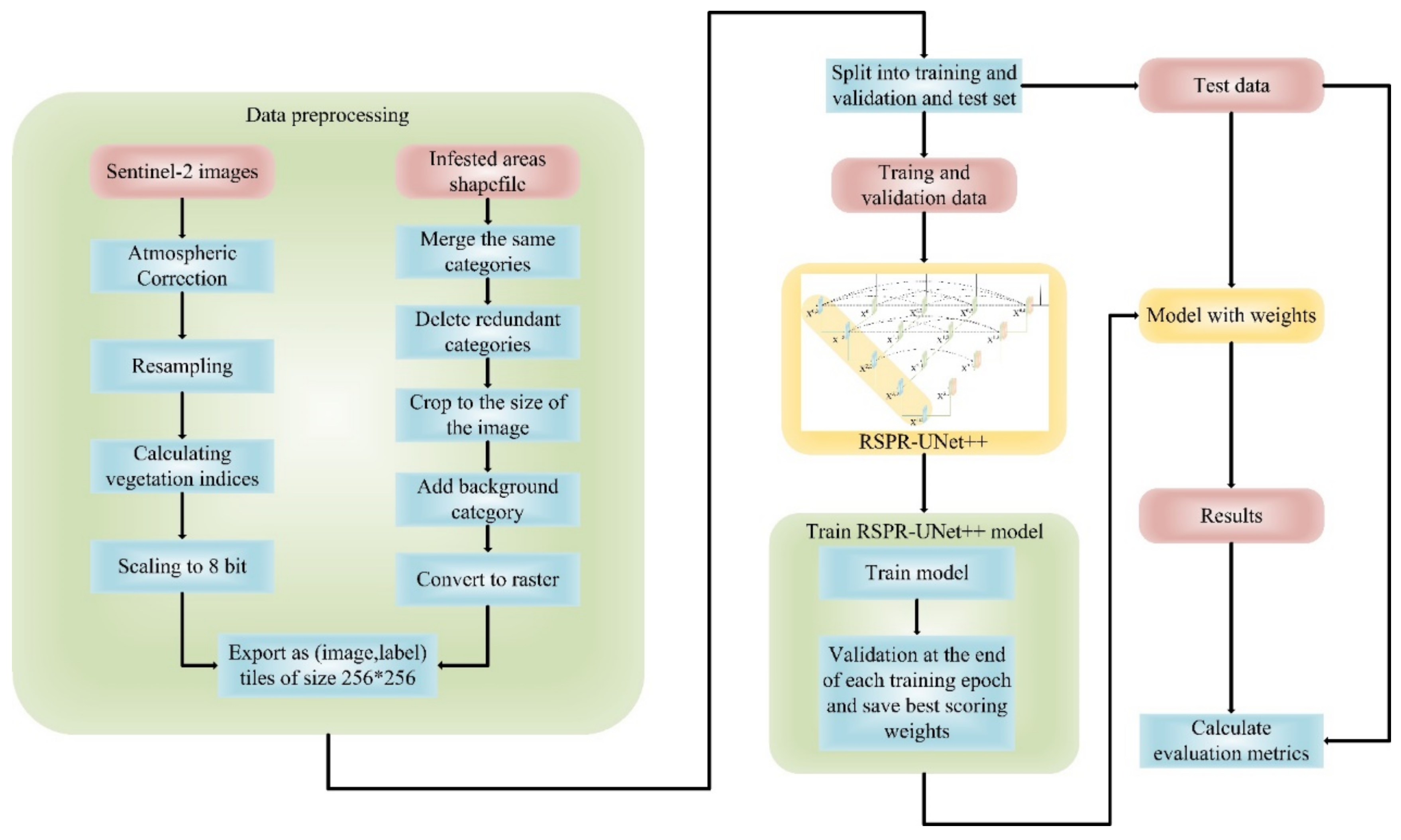
2.2.2. Data Preprocessing
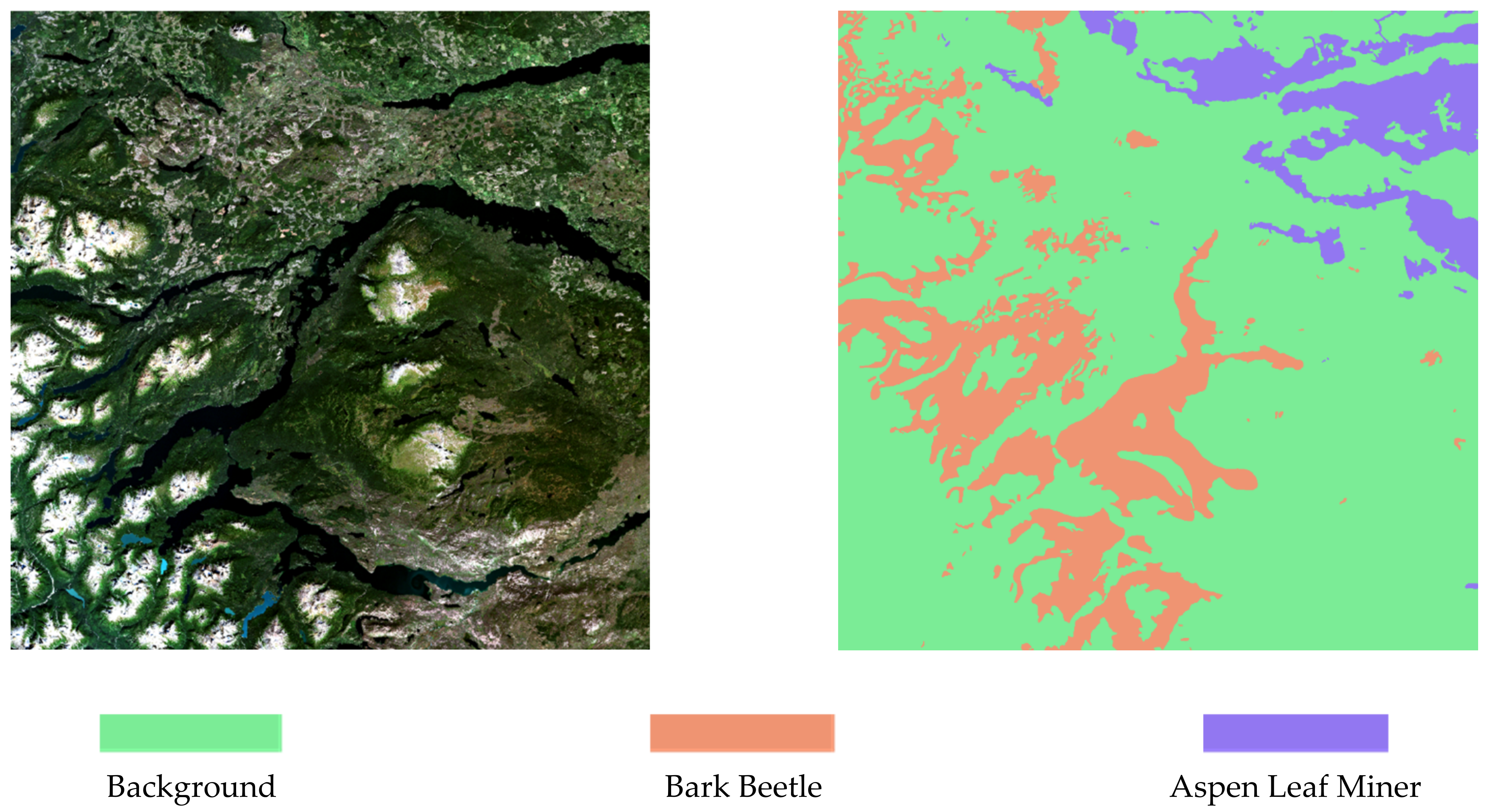
2.3. Dataset
3. Methodology
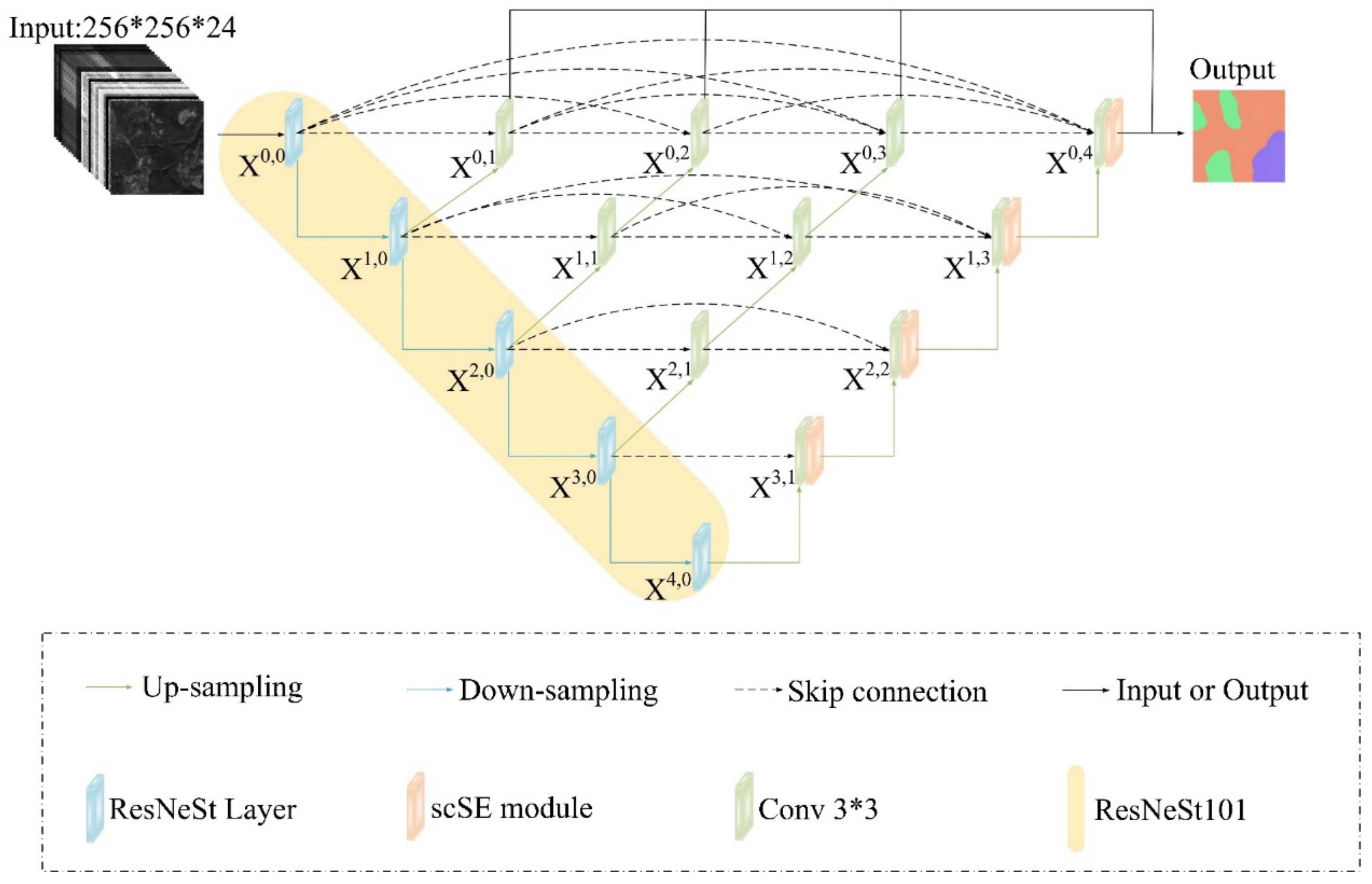
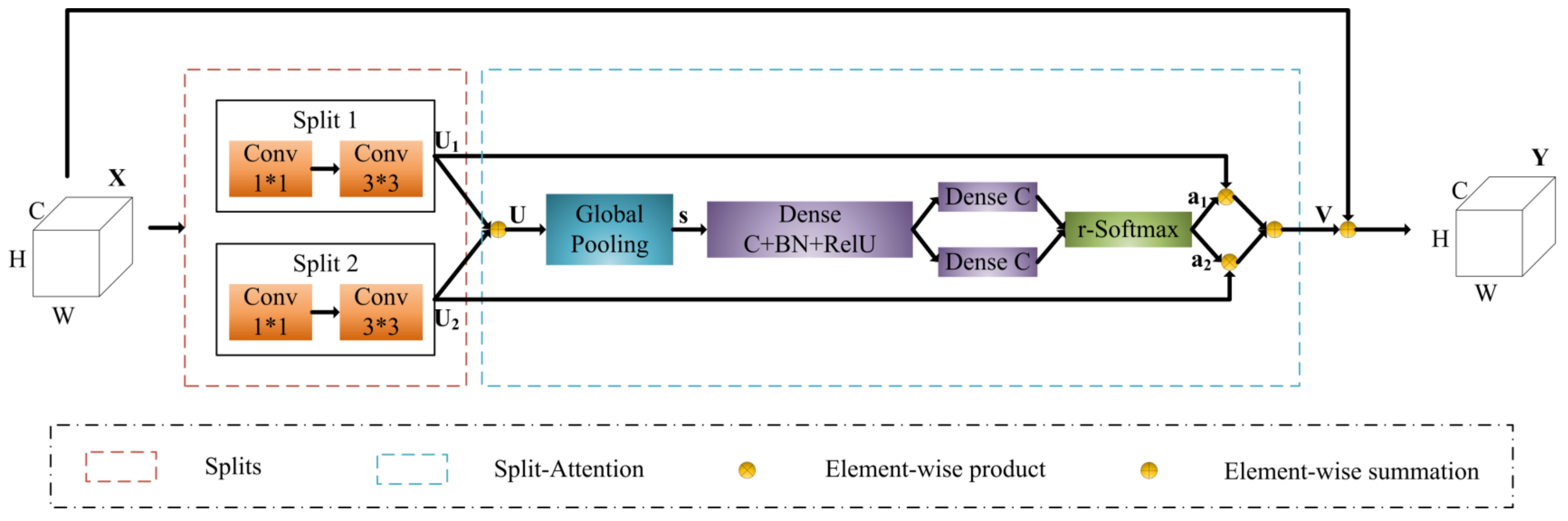
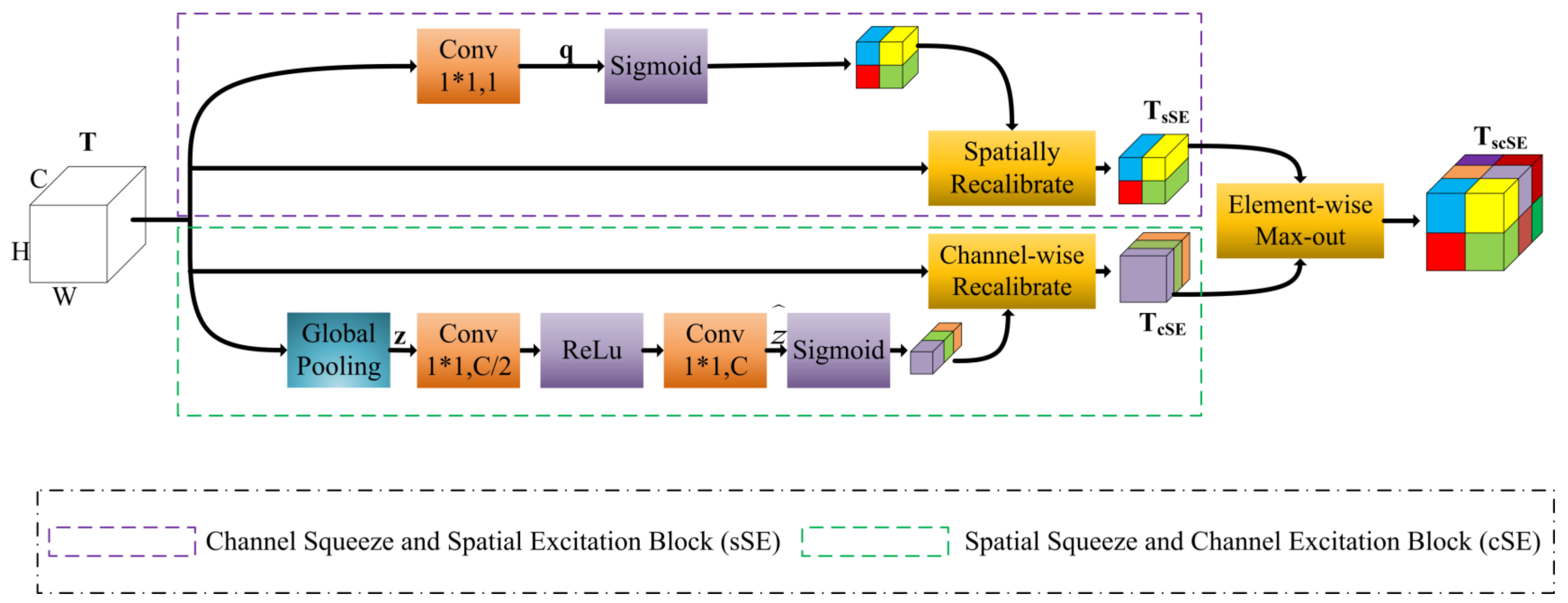
3.1. Model Architecture
3.2. Loss Function
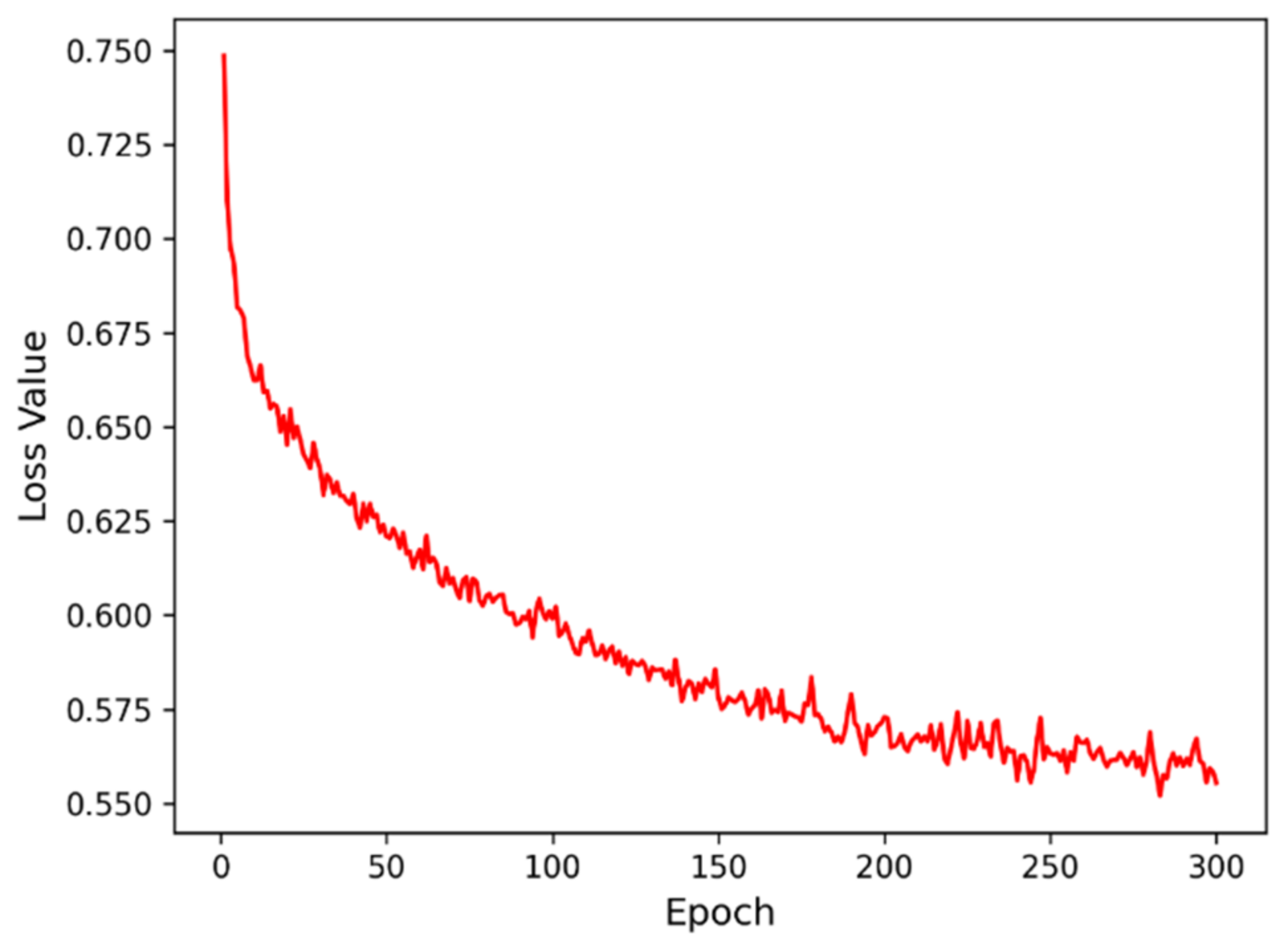
3.3. Model Training
3.4. Evaluation Metrics
3.5. Analyze the Impact of Different Data
4. Results
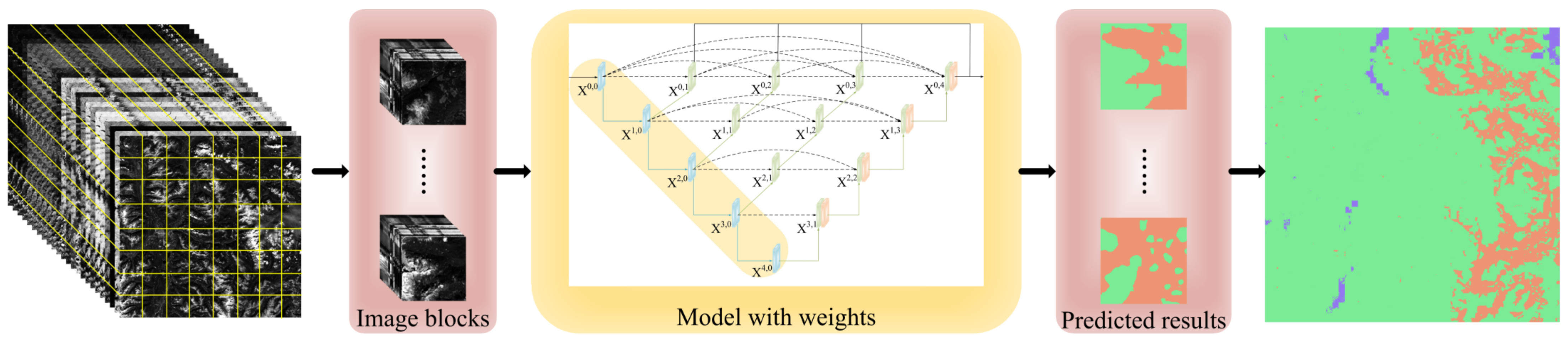
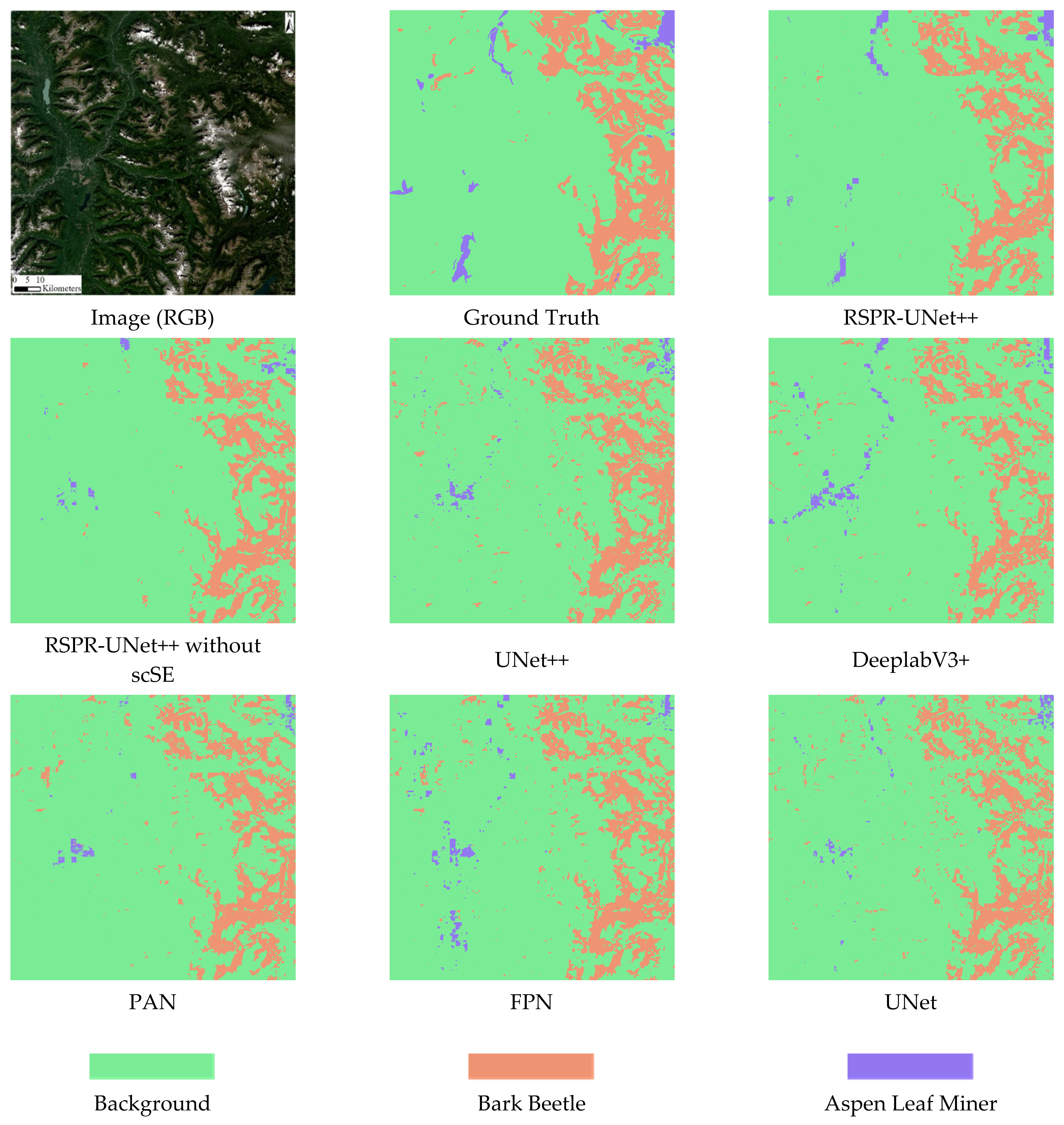
4.1. Comparison of Predicted Results
4.1.1. Comparison between Different Models
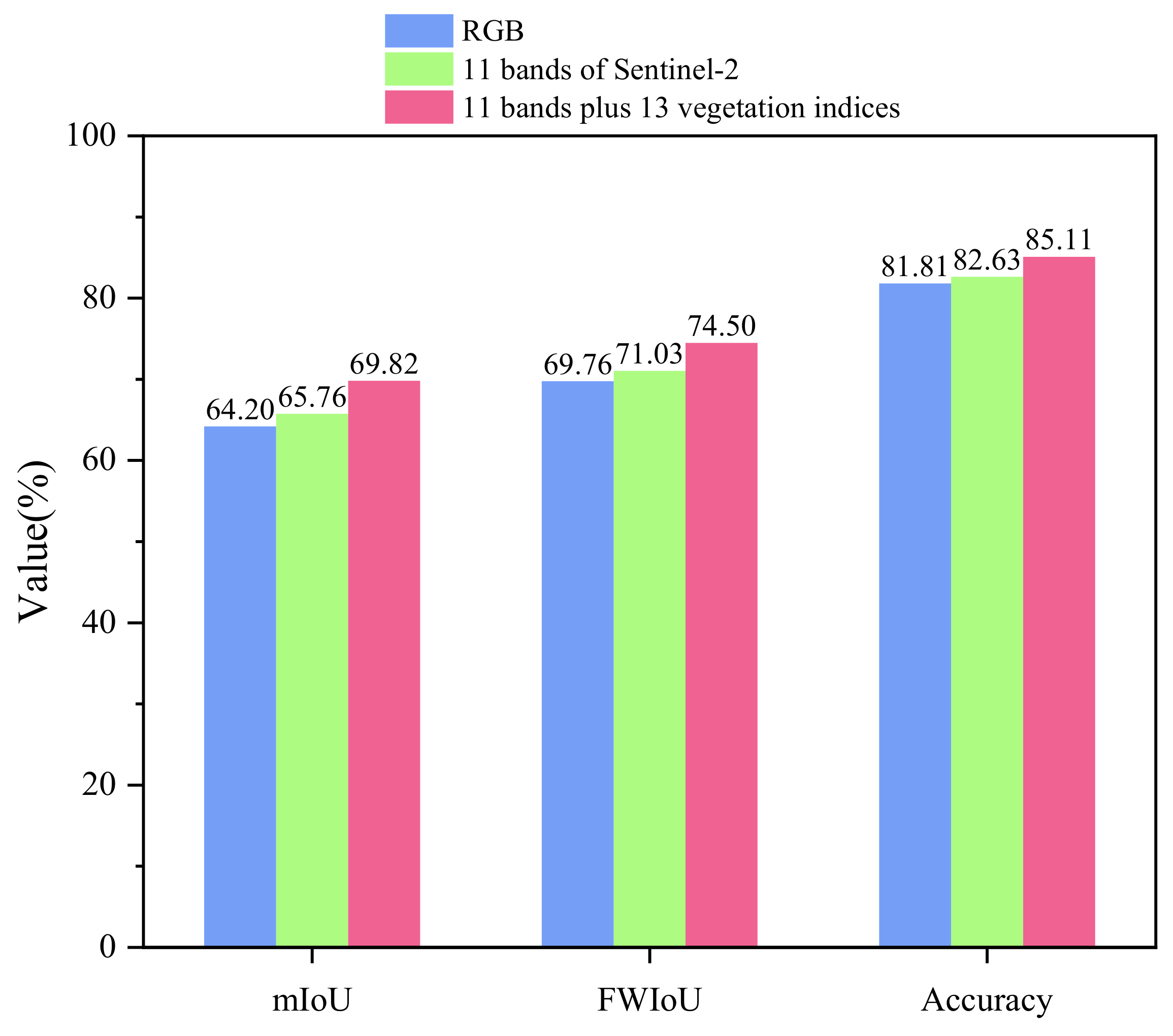
4.1.2. Comparison between RGB Data, Multispectral Data, and Vegetation Indices
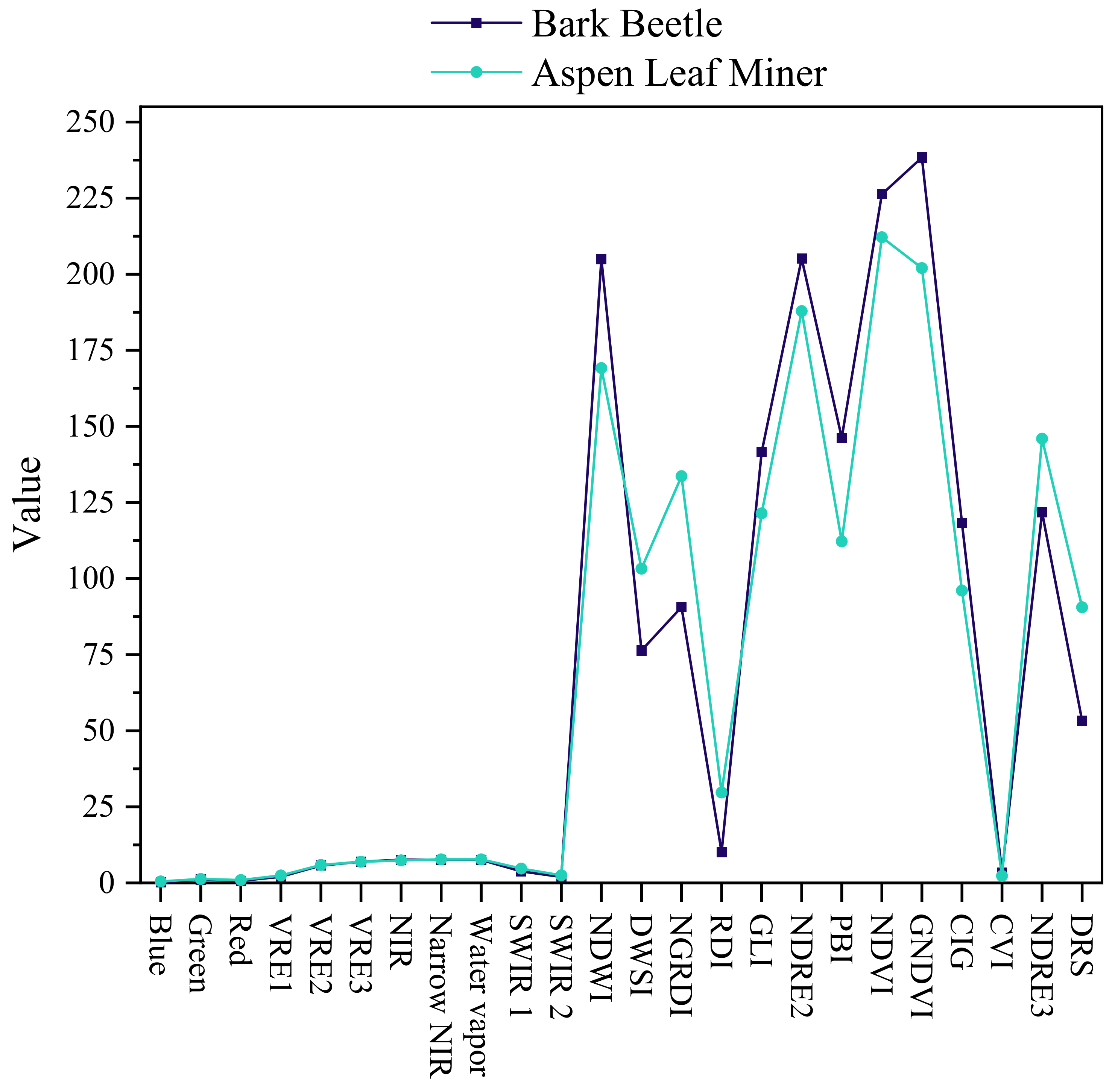
4.2. Comparison of the Spectral Characteristics of Different Infestation Types
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, Z.; Wu, C.; Wang, X. Forest insect-disease monitoring and estimation based on satellite remote sensing data. Geogr. Res. 2019, 38, 831–843. [Google Scholar]
- Hicke, J.A.; Xu, B.; Meddens, A.J.H.; Egan, J.M. Characterizing recent bark beetle-caused tree mortality in the western United States from aerial surveys. For. Ecol. Manag. 2020, 475, 118402. [Google Scholar] [CrossRef]
- Meddens, A.J.H.; Hicke, J.A. Spatial and temporal patterns of Landsat-based detection of tree mortality caused by a mountain pine beetle outbreak in Colorado, USA. For. Ecol. Manag. 2014, 322, 78–88. [Google Scholar] [CrossRef]
- Wagner, D.; DeFoliart, L.; Doak, P.; Schneiderheinze, J. Impact of epidermal leaf mining by the aspen leaf miner (Phyllocnistis populiella) on the growth, physiology, and leaf longevity of quaking aspen. Oecologia 2008, 157, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Doak, P.; Sformo, T.; Steiner, P.M.; Carlson, B. Overwintering physiology and microhabitat use of Phyllocnistis populiella (Lepidoptera: Gracilliariidae) in interior Alaska. Environ. Entomol. 2012, 41, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Young, B.; Wagner, D.; Doak, P.; Clausen, T. Induction of phenolic glycosides by quaking aspen (Populus tremuloides) leaves in relation to extrafloral nectaries and epidermal leaf mining. J. Chem. Ecol. 2010, 36, 369–377. [Google Scholar] [CrossRef]
- Doak, P.; Wagner, D. The role of interference competition in a sustained population outbreak of the aspen leaf miner in Alaska. Basic Appl. Ecol. 2015, 16, 434–442. [Google Scholar] [CrossRef]
- Safonova, A.; Tabik, S.; Alcaraz-Segura, D.; Rubtsov, A.; Maglinets, Y.; Herrera, F. Detection of Fir Trees (Abies sibirica) Damaged by the Bark Beetle in Unmanned Aerial Vehicle Images with Deep Learning. Remote Sens. 2019, 11, 643. [Google Scholar] [CrossRef]
- Yu, R.; Ren, L.L.; Luo, Y.Q. Early detection of pine wilt disease in Pinus tabuliformis in North China using a field portable spectrometer and UAV-based hyperspectral imagery. For. Ecosyst. 2021, 8, 44. [Google Scholar] [CrossRef]
- Dennison, P.E.; Brunelle, A.R.; Carter, V.A. Assessing canopy mortality during a mountain pine beetle outbreak using GeoEye-1 high spatial resolution satellite data. Remote Sens. Environ. 2010, 114, 2431–2435. [Google Scholar] [CrossRef]
- Zhan, Z.; Yu, L.; Li, Z.; Ren, L.; Gao, B.; Wang, L.; Luo, Y. Combining GF-2 and Sentinel-2 Images to Detect Tree Mortality Caused by Red Turpentine Beetle during the Early Outbreak Stage in North China. Forests 2020, 11, 172. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Hong, D.; Gao, L.; Yokoya, N.; Yao, J.; Chanussot, J.; Du, Q.; Zhang, B. More diverse means better: Multimodal deep learning meets remote-sensing imagery classification. IEEE Trans. Geosci. Remote Sens. 2020, 59, 4340–4354. [Google Scholar] [CrossRef]
- Wu, X.; Hong, D.; Chanussot, J. Convolutional neural networks for multimodal remote sensing data classification. IEEE Trans. Geosci. Remote Sens. 2021, 60, 1–10. [Google Scholar] [CrossRef]
- Hong, D.; Gao, L.; Yao, J.; Zhang, B.; Plaza, A.; Chanussot, J. Graph convolutional networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2020, 59, 5966–5978. [Google Scholar] [CrossRef]
- Shi, L.; Li, C.; Li, T.; Peng, Y. A Complementary Spectral–Spatial Method for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60. [Google Scholar] [CrossRef]
- Kotaridis, I.; Lazaridou, M. Remote sensing image segmentation advances: A meta-analysis. ISPRS J. Photogramm. Remote Sens. 2021, 173, 309–322. [Google Scholar] [CrossRef]
- Kislov, D.E.; Korznikov, K.A.; Altman, J.; Vozmishcheva, A.S.; Krestov, P.V.; Disney, M.; Cord, A. Extending deep learning approaches for forest disturbance segmentation on very high-resolution satellite images. Remote Sens. Ecol. Conserv. 2021, 7, 355–368. [Google Scholar] [CrossRef]
- Zhou, H.; Yuan, X.; Zhou, H.; Shen, H.; Ma, L.; Sun, L.; Fang, G.; Sun, H. Surveillance of pine wilt disease by high resolution satellite. J. For. Res. 2022, 33, 1401–1408. [Google Scholar] [CrossRef]
- Cotrozzi, L. Spectroscopic detection of forest diseases: A review (1970–2020). J. For. Res. 2021, 33, 21–38. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Camino, C.; Beck, P.S.A.; Calderon, R.; Hornero, A.; Hernandez-Clemente, R.; Kattenborn, T.; Montes-Borrego, M.; Susca, L.; Morelli, M.; et al. Previsual symptoms of Xylella fastidiosa infection revealed in spectral plant-trait alterations. Nat. Plants 2018, 4, 432–439. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Hornero, A.; Beck, P.S.A.; Kattenborn, T.; Kempeneers, P.; Hernandez-Clemente, R. Chlorophyll content estimation in an open-canopy conifer forest with Sentinel-2A and hyperspectral imagery in the context of forest decline. Remote Sens. Environ. 2019, 223, 320–335. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Shen, Q.; Deng, J.; Liu, X.; Huang, H. Prediction of bark beetles pests based on temperature vegetation dryness index. Trans. Chin. Soc. Agric. Eng. 2018, 34, 167–174. [Google Scholar]
- Wu, H.; Wang, C.; Miao, Z.; Wang, W.; Wang, X.; Mi, G. Study on Early Warning Technology of Sub-health State of Forest Resources with Spaceborne Remote Sensing. Remote Sens. Technol. Appl. 2021, 36, 1121–1130. [Google Scholar]
- Meng, R.; Gao, R.; Zhao, F.; Huang, C.; Sun, R.; Lv, Z.; Huang, Z. Landsat-based monitoring of southern pine beetle infestation severity and severity change in a temperate mixed forest. Remote Sens. Environ. 2022, 269, 112847. [Google Scholar] [CrossRef]
- Goodwin, N.R.; Coops, N.C.; Wulder, M.A.; Gillanders, S.; Schroeder, T.A.; Nelson, T. Estimation of insect infestation dynamics using a temporal sequence of Landsat data. Remote Sens. Environ. 2008, 112, 3680–3689. [Google Scholar] [CrossRef]
- Zhu, C.; Qu, S.; Zhang, X. Dendrolimus tabulaeformis disaster monitoring and analysis of its influencing factors through remote sensing technology. J. Remote Sens. 2016, 20, 653–664. [Google Scholar]
- Hais, M.; Jonášová, M.; Langhammer, J.; Kučera, T. Comparison of two types of forest disturbance using multitemporal Landsat TM/ETM+ imagery and field vegetation data. Remote Sens. Environ. 2009, 113, 835–845. [Google Scholar] [CrossRef]
- Bárta, V.; Lukeš, P.; Homolová, L. Early detection of bark beetle infestation in Norway spruce forests of Central Europe using Sentinel-2. Int. J. Appl. Earth Obs. Geoinf. 2021, 100, 102335. [Google Scholar] [CrossRef]
- Choi, W.-I.L.; Kim, E.-S.; Yun, S.-J.; Lim, J.-H.; Kim, Y.-E. Quantification of One-Year Gypsy Moth Defoliation Extent in Wonju, Korea, Using Landsat Satellite Images. Forests 2021, 12, 545. [Google Scholar] [CrossRef]
- Kern, A.; Marjanović, H.; Csóka, G.; Móricz, N.; Pernek, M.; Hirka, A.; Matošević, D.; Paulin, M.; Kovač, G. Detecting the oak lace bug infestation in oak forests using MODIS and meteorological data. Agric. For. Meteorol. 2021, 306, 108436. [Google Scholar] [CrossRef]
- Xu, Z.H.; Huang, X.Y.; Lin, L.; Wang, Q.F.; Liu, J.; Chen, C.C.; Yu, K.Y.; Zhou, H.K.; Zhang, H.F. Dendrolimus Punctatus Walker Damage Detection Based on Fisher Discriminant Analysis and Random Forest. Spectrosc. Spectr. Anal. 2018, 38, 2888–2896. [Google Scholar] [CrossRef]
- Zou, H. The Effect and Method of Quantitative Analysis of “Red Edge”of Vegetation. Remote Sens. Inf. 2010, 4, 112–116. [Google Scholar]
- Boochs, F.; Kupfer, G.; Dockter, K.; Kühbauch, W. Shape of the red edge as vitality indicator for plants. Remote Sens. 1990, 11, 1741–1753. [Google Scholar] [CrossRef]
- Filella, I.; Penuelas, J. The red edge position and shape as indicators of plant chlorophyll content, biomass and hydric status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Abdullah, H.; Skidmore, A.K.; Darvishzadeh, R.; Heurich, M.; Pettorelli, N.; Disney, M. Sentinel-2 accurately maps green-attack stage of European spruce bark beetle (Ips typographus, L.) compared with Landsat-8. Remote Sens. Ecol. Conserv. 2018, 5, 87–106. [Google Scholar] [CrossRef]
- Eitel, J.U.; Vierling, L.A.; Litvak, M.E.; Long, D.S.; Schulthess, U.; Ager, A.A.; Krofcheck, D.J.; Stoscheck, L. Broadband, red-edge information from satellites improves early stress detection in a New Mexico conifer woodland. Remote Sens. Environ. 2011, 115, 3640–3646. [Google Scholar] [CrossRef]
- Carter, G.A.; Miller, R.L. Early detection of plant stress by digital imaging within narrow stress-sensitive wavebands. Remote Sens. Environ. 1994, 50, 295–302. [Google Scholar] [CrossRef]
- Ahern, F. The effects of bark beetle stress on the foliar spectral reflectance of lodgepole pine. Int. J. Remote Sens. 1988, 9, 1451–1468. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf optical properties in higher plants: Linking spectral characteristics to stress and chlorophyll concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. Isprs J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Fernandez-Manso, A.; Fernandez-Manso, O.; Quintano, C. SENTINEL-2A red-edge spectral indices suitability for discriminating burn severity. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 170–175. [Google Scholar] [CrossRef]
- Bhattarai, R.; Rahimzadeh-Bajgiran, P.; Weiskittel, A.; MacLean, D.A. Sentinel-2 based prediction of spruce budworm defoliation using red-edge spectral vegetation indices. Remote Sens. Lett. 2020, 11, 777–786. [Google Scholar] [CrossRef]
- Hojas-Gascon, L.; Belward, A.; Eva, H.; Ceccherini, G.; Hagolle, O.; Garcia, J.; Cerutti, P. Potential improvement for forest cover and forest degradation mapping with the forthcoming Sentinel-2 program. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 40, 417–423. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Westfall, J.; Ebata, T.; Bains, B. Summary of Forest Health Conditions in British Columbia; BC Ministry of Forests and Range, Forest Practices Branch: Victoria, BC, Canada, 2020.
- Zabihi, K.; Surovy, P.; Trubin, A.; Singh, V.V.; Jakuš, R. A review of major factors influencing the accuracy of mapping green-attack stage of bark beetle infestations using satellite imagery: Prospects to avoid data redundancy. Remote Sens. Appl. Soc. Environ. 2021, 24, 100638. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Lin, H.; Zhang, Z.; Sun, Y.; He, T.; Mueller, J.; Manmatha, R. Resnest: Split-attention networks. arXiv 2020, arXiv:2004.08955. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Roy, A.G.; Navab, N.; Wachinger, C. Recalibrating fully convolutional networks with spatial and channel “squeeze and excitation” blocks. IEEE Trans. Med. Imaging 2018, 38, 540–549. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Lin, T.-Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Li, H.; Xiong, P.; An, J.; Wang, L. Pyramid attention network for semantic segmentation. arXiv 2018, arXiv:1805.10180. [Google Scholar]
- Wang, Z.; Wang, J.; Yang, K.; Wang, L.; Su, F.; Chen, X. Semantic segmentation of high-resolution remote sensing images based on a class feature attention mechanism fused with Deeplabv3+. Comput. Geosci. 2022, 158, 104969. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, C.; Han, M. MLFC-net: A multi-level feature combination attention model for remote sensing scene classification. Comput. Geosci. 2022, 160, 105042. [Google Scholar] [CrossRef]
- Wang, D.; Chen, X.; Jiang, M.; Du, S.; Xu, B.; Wang, J. ADS-Net: An Attention-Based deeply supervised network for remote sensing image change detection. Int. J. Appl. Earth Obs. Geoinf. 2021, 101, 102348. [Google Scholar]
- Bright, B.C.; Hudak, A.T.; Egan, J.M.; Jorgensen, C.L.; Rex, F.E.; Hicke, J.A.; Meddens, A.J.H. Using Satellite Imagery to Evaluate Bark Beetle-Caused Tree Mortality Reported in Aerial Surveys in a Mixed Conifer Forest in Northern Idaho, USA. Forests 2020, 11, 529. [Google Scholar] [CrossRef]
- Meigs, G.W.; Kennedy, R.E.; Gray, A.N.; Gregory, M.J. Spatiotemporal dynamics of recent mountain pine beetle and western spruce budworm outbreaks across the Pacific Northwest Region, USA. For. Ecol. Manag. 2015, 339, 71–86. [Google Scholar] [CrossRef]
- Ye, S.; Rogan, J.; Zhu, Z.; Hawbaker, T.J.; Hart, S.J.; Andrus, R.A.; Meddens, A.J.H.; Hicke, J.A.; Eastman, J.R.; Kulakowski, D. Detecting subtle change from dense Landsat time series: Case studies of mountain pine beetle and spruce beetle disturbance. Remote Sens. Environ. 2021, 263, 112560. [Google Scholar] [CrossRef]
- Hart, S.J.; Veblen, T.T. Detection of spruce beetle-induced tree mortality using high- and medium-resolution remotely sensed imagery. Remote Sens. Environ. 2015, 168, 134–145. [Google Scholar] [CrossRef]
- Zhang, B.; Ye, H.; Lu, W.; Huang, W.; Wu, B.; Hao, Z.; Sun, H. A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 2083. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Chanussot, J.; Zhu, X.X. An augmented linear mixing model to address spectral variability for hyperspectral unmixing. IEEE Trans. Image Process. 2018, 28, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
| Band | Band Name | Resolution (m) |
|---|---|---|
| B1 | Coastal aerosol | 60 |
| B2 | Blue | 10 |
| B3 | Green | 10 |
| B4 | Red | 10 |
| B5 | Vegetation Red Edge 1 | 20 |
| B6 | Vegetation Red Edge 2 | 20 |
| B7 | Vegetation Red Edge 3 | 20 |
| B8 | NIR | 10 |
| B8A | Narrow NIR | 20 |
| B9 | Water vapor | 60 |
| B10 | SWIR-Cirrus | 60 |
| B11 | SWIR 1 | 20 |
| B12 | SWIR 2 | 20 |
| Vegetation Indices | Calculation Method | Calculation Details in Sentinel-2 |
|---|---|---|
| NDWI | ||
| DWSI | ||
| NGRDI | ||
| RDI | ||
| GLI | ||
| NDRE2 | ||
| PBI | ||
| NDVI | ||
| GNDVI | ||
| CIG | ||
| CVI | ||
| NDRE3 | ||
| DRS |
| Model | Characteristics | Reference |
|---|---|---|
| UNet | The architecture contains 2 paths (contraction path and symmetric expanding path). It is an end-to-end fully convolutional network (FCN). | [53] |
| DeeplabV3+ | The spatial pyramid pooling module and the encoder–decoder structure were combined. The depthwise separable convolution was applied to both the Atrous Spatial Pyramid Pooling and decoder modules. | [54] |
| Feature Pyramid Networks (FPN) | Developed a top-down architecture with lateral connections for building high-level semantic feature maps at all scales. | [55] |
| Pyramid Attention Network (PAN) | Exploited the impact of global contextual information in semantic segmentation. | [56] |
| UNet++ | The architecture is an encoder–decoder network where the encoder and decoder sub-networks are connected through a series of nested, dense skip pathways. It optimizes the topology of UNet and is an improved version of the UNet network structure. | [47] |
| Vegetation Indices | Calculation Method | Calculation Details in Sentinel-2 |
|---|---|---|
| ND790/670 | ||
| NDVI690-710 | ||
| NDRE | ||
| NDVI65 | ||
| GNDVIhyper | ||
| RENDVI1 | ||
| RENDVI2 | ||
| RI |
| Model | Category | Precision (%) | Recall (%) | F1 (%) | IoU (%) | mIoU (%) | FWIoU (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|
| UNet | BG | 85.44 | 88.63 | 87.00 | 77.00 | 63.84 | 70.09 | 82.21 |
| BB | 76.64 | 69.74 | 73.03 | 57.52 | ||||
| ALM | 74.03 | 71.26 | 72.62 | 57.01 | ||||
| FPN | BG | 86.33 | 87.93 | 87.12 | 77.18 | 64.51 | 70.60 | 82.52 |
| BB | 75.09 | 74.12 | 74.60 | 59.49 | ||||
| ALM | 75.08 | 70.11 | 72.51 | 56.87 | ||||
| PAN | BG | 86.39 | 88.34 | 87.36 | 77.55 | 64.84 | 70.87 | 82.71 |
| BB | 75.00 | 73.05 | 74.01 | 58.75 | ||||
| ALM | 76.15 | 71.21 | 73.59 | 58.22 | ||||
| DeeplabV3+ | BG | 87.63 | 86.22 | 86.92 | 76.86 | 65.09 | 70.75 | 82.51 |
| BB | 74.85 | 75.81 | 75.33 | 60.42 | ||||
| ALM | 71.49 | 75.44 | 73.41 | 57.99 | ||||
| UNet++ | BG | 86.81 | 87.81 | 87.31 | 77.47 | 65.18 | 71.06 | 82.82 |
| BB | 75.34 | 74.71 | 75.03 | 60.03 | ||||
| ALM | 75.00 | 71.93 | 73.44 | 58.02 | ||||
| RSPR-UNet++ without scSE | BG | 89.61 | 87.06 | 88.32 | 79.08 | 68.83 | 73.76 | 84.61 |
| BB | 75.60 | 82.15 | 78.74 | 64.94 | ||||
| ALM | 76.83 | 76.98 | 76.90 | 62.47 | ||||
| RSPR-UNet++ | BG | 89.92 | 87.53 | 88.71 | 79.70 | 69.82 | 74.50 | 85.11 |
| BB | 78.10 | 79.52 | 78.81 | 65.02 | ||||
| ALM | 75.16 | 82.33 | 78.58 | 64.72 |
| Model | Accuracy (%) |
|---|---|
| UNet | 86.79 |
| FPN | 86.31 |
| PAN | 87.40 |
| DeeplabV3+ | 85.87 |
| UNet++ | 87.49 |
| RSPR-UNet++ without scSE | 88.29 |
| RSPR-UNet++ | 89.10 |
| Data | Category | Precision (%) | Recall (%) | F1 (%) | IoU (%) | mIoU (%) | FWIoU (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|
| RGB | BG | 87.06 | 85.67 | 86.36 | 75.99 | 64.20 | 69.76 | 81.81 |
| BB | 73.45 | 73.32 | 73.38 | 57.96 | ||||
| ALM | 71.32 | 76.76 | 73.94 | 58.66 | ||||
| 11 bands | BG | 88.96 | 84.98 | 86.93 | 76.87 | 65.76 | 71.03 | 82.63 |
| BB | 70.04 | 81.40 | 75.29 | 60.38 | ||||
| ALM | 76.43 | 73.65 | 75.02 | 60.02 | ||||
| RGB plus 13 vegetation indices | BG | 89.79 | 84.79 | 87.22 | 77.34 | 67.06 | 72.01 | 83.31 |
| BB | 76.23 | 78.65 | 77.42 | 63.16 | ||||
| ALM | 68.99 | 83.43 | 75.52 | 60.67 | ||||
| 11 bands plus 13 vegetation indices | BG | 89.92 | 87.53 | 88.71 | 79.70 | 69.82 | 74.50 | 85.11 |
| BB | 78.10 | 79.52 | 78.81 | 65.02 | ||||
| ALM | 75.16 | 82.33 | 78.58 | 64.72 | ||||
| 8 bands plus 10 vegetation indices | BG | 86.43 | 89.63 | 88.00 | 78.57 | 68.17 | 72.76 | 84.13 |
| BB | 80.66 | 74.73 | 77.58 | 63.37 | ||||
| ALM | 78.76 | 75.28 | 76.98 | 62.57 | ||||
| 11 bands plus 21 vegetation indices | BG | 84.68 | 90.62 | 87.55 | 77.85 | 68.12 | 72.17 | 83.85 |
| BB | 81.21 | 75.63 | 78.32 | 64.37 | ||||
| ALM | 84.00 | 70.48 | 76.65 | 62.14 |
| Attention Module | Accuracy (%) |
|---|---|
| scSE | 85.11 |
| cSE | 84.93 |
| sSE | 84.85 |
| None | 84.61 |
| Parameters | Accuracy (%) |
|---|---|
| 16, 32, 64, 128 and 256 | 85.11 |
| 32, 64, 128, 256 and 512 | 85.03 |
| 64, 128, 256, 512 and 1024 | 83.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Cong, S.; Zhang, G.; Ma, Y.; Zhang, Y.; Huang, J. Detecting Pest-Infested Forest Damage through Multispectral Satellite Imagery and Improved UNet++. Sensors 2022, 22, 7440. https://doi.org/10.3390/s22197440
Zhang J, Cong S, Zhang G, Ma Y, Zhang Y, Huang J. Detecting Pest-Infested Forest Damage through Multispectral Satellite Imagery and Improved UNet++. Sensors. 2022; 22(19):7440. https://doi.org/10.3390/s22197440
Chicago/Turabian StyleZhang, Jingzong, Shijie Cong, Gen Zhang, Yongjun Ma, Yi Zhang, and Jianping Huang. 2022. "Detecting Pest-Infested Forest Damage through Multispectral Satellite Imagery and Improved UNet++" Sensors 22, no. 19: 7440. https://doi.org/10.3390/s22197440
APA StyleZhang, J., Cong, S., Zhang, G., Ma, Y., Zhang, Y., & Huang, J. (2022). Detecting Pest-Infested Forest Damage through Multispectral Satellite Imagery and Improved UNet++. Sensors, 22(19), 7440. https://doi.org/10.3390/s22197440





