Sleep Monitoring during Acute Stroke Rehabilitation: Toward Automated Measurement Using Multimodal Wireless Sensors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Equipment
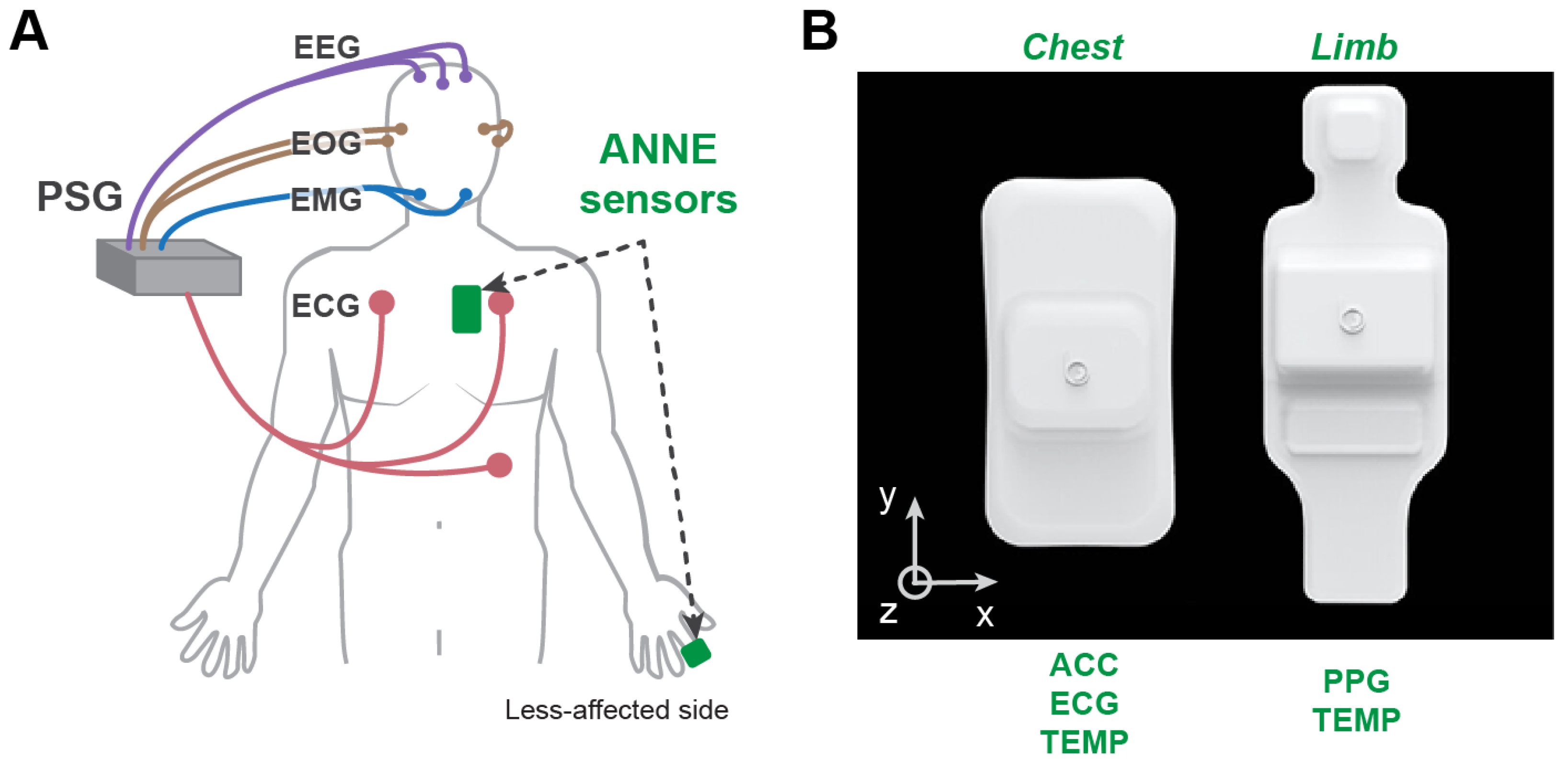
2.2.1. Polysomnography (PSG)
2.2.2. Wearable Sensors
2.3. Procedures
2.4. Data Analysis
2.4.1. PSG for Ground Truth Sleep Staging
2.4.2. Feature Extraction
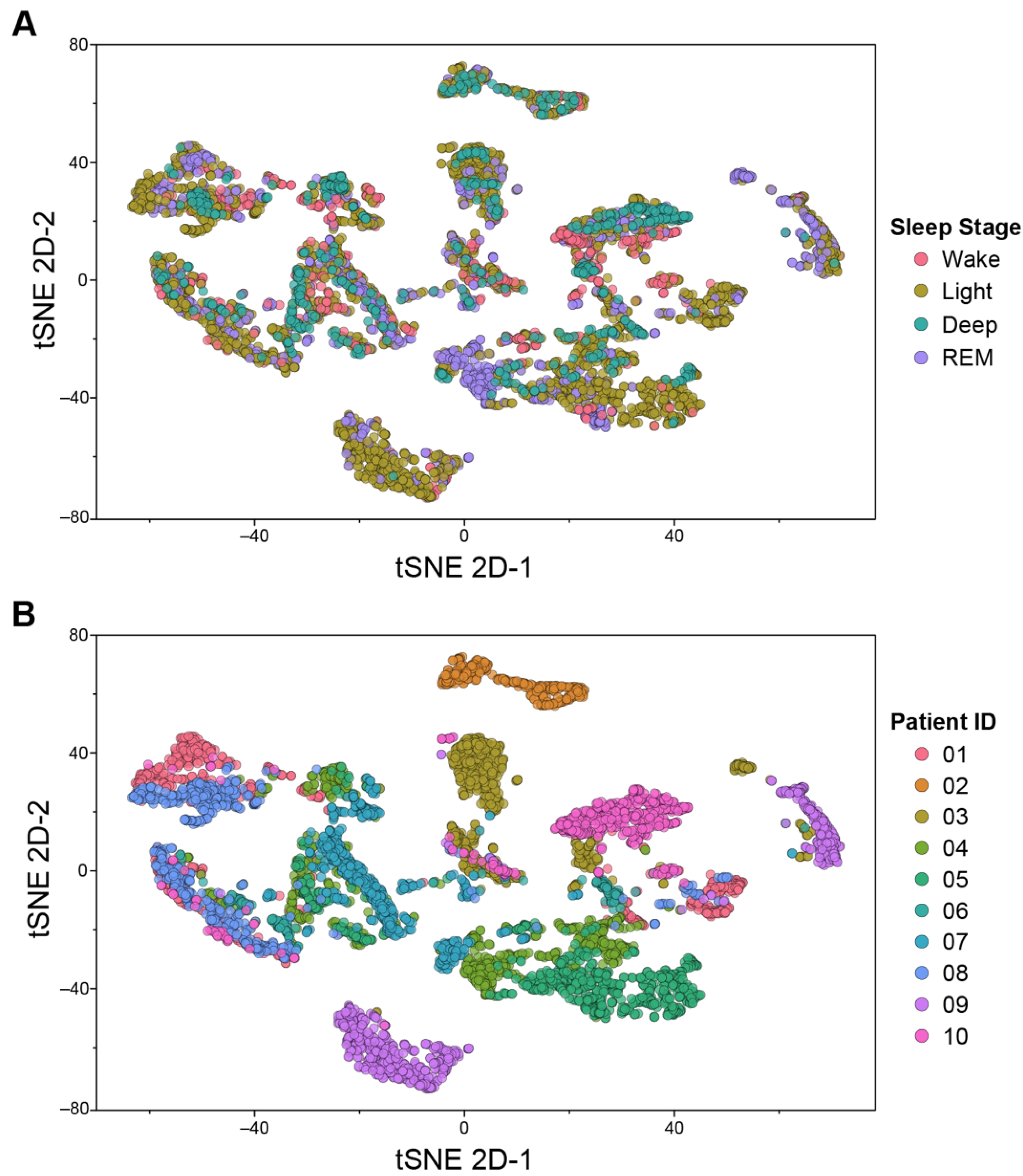
2.4.3. t-Distributed Stochastic Neighbor Embedding (tSNE) Analysis
2.4.4. Class Imbalance
2.4.5. Model Development and Training
2.4.6. Model Evaluation
3. Results
3.1. Sensor Data Visualization
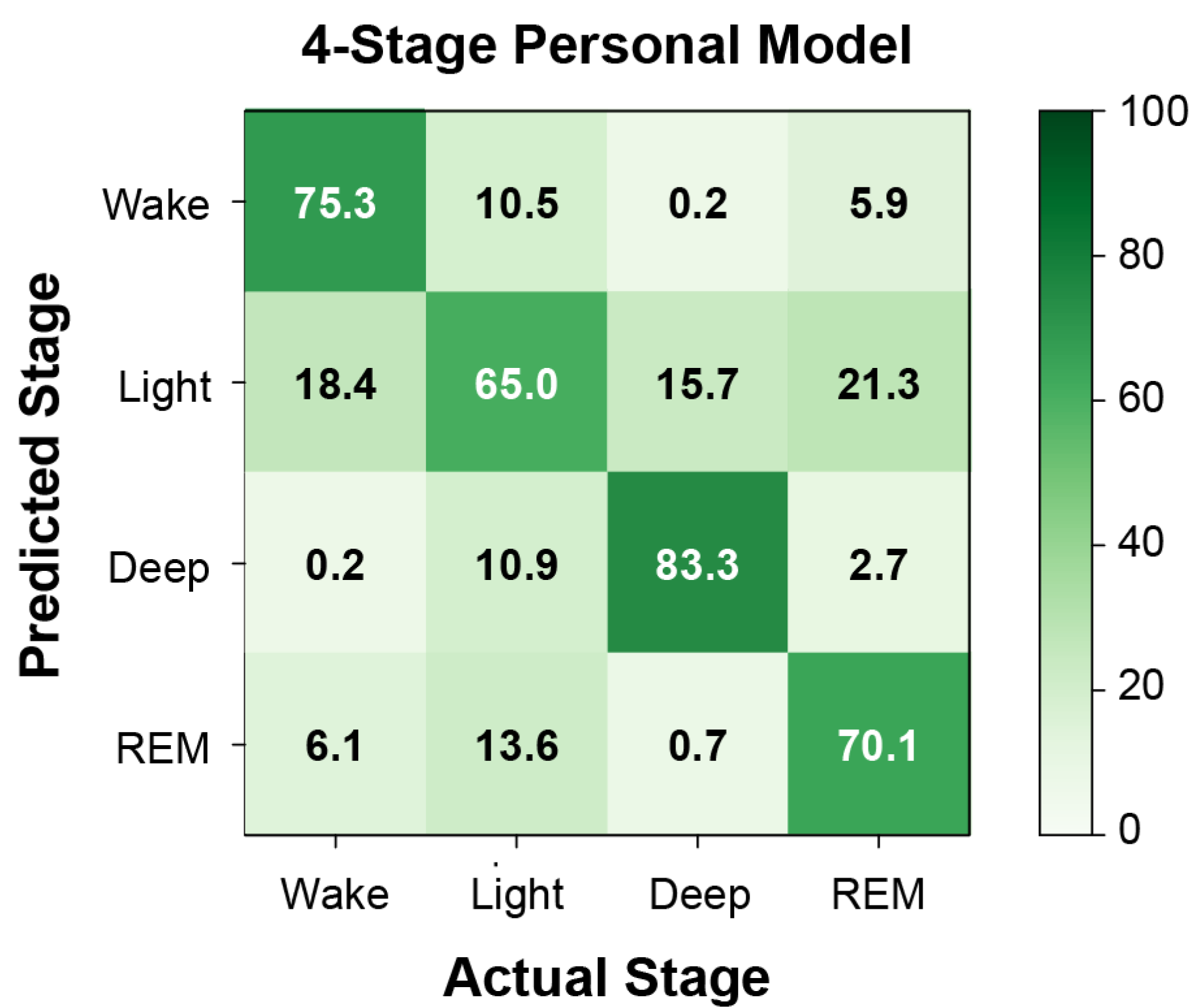
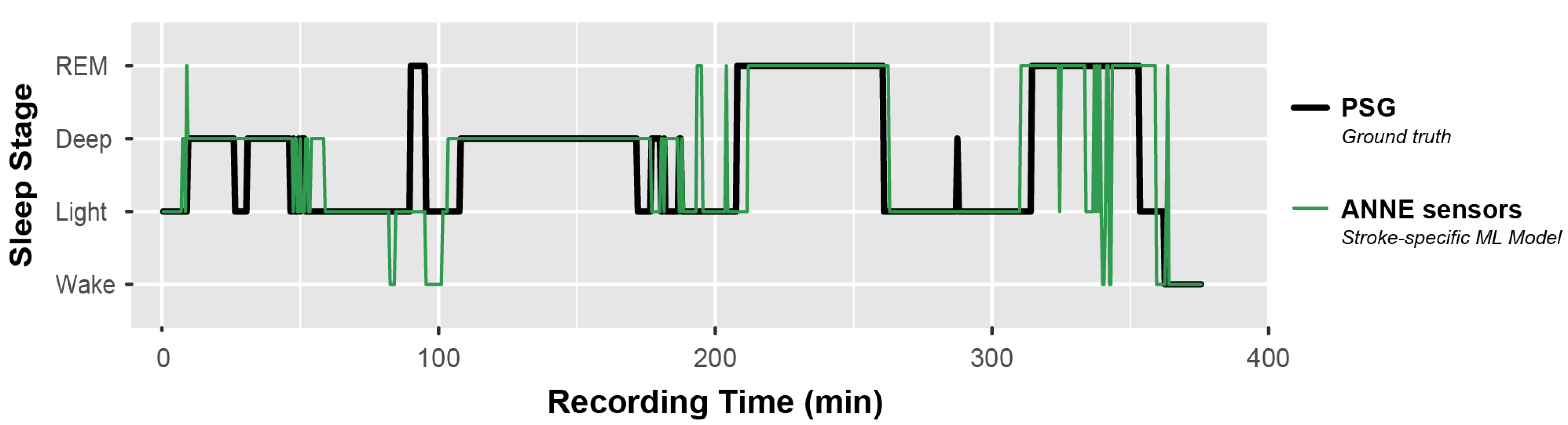
3.2. Machine Learning
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| ID | Stroke Location | Symptom Directed Affect by Stroke | Comorbidities |
|---|---|---|---|
| 1 | Right middle cerebral artery stroke | Reduced balance, coordination, sensation. Hemispatial neglect, inattention, vision deficits. | |
| 2 | Left basal ganglia, caudate, and right parietal-occipital lobes | Impaired ambulation, activities of daily living, eating/swallowing, transfers, bowel and bladder function, cognition, memory, speech. | COPD; deep vein thrombosis and pulmonary embolism |
| 3 | Left subcortical | Reduced strength, endurance, and balance. Dysarthria, spasticity. | |
| 4 | Right pons | Reduced strength, endurance, balance. Impaired cognition. | Anxiety; depression |
| 5 | Right thalamic intracerebral | Fatigue. Reduced strength, endurance, coordination, range of motion. | Urinary incontinence; UTI |
| 6 | Para median pontine (chronic caudate and thalamic infarcts) | Impaired ambulation, activities of daily living, transfers, bowel and bladder function, cognition, memory, speech. Hemiparesis of lower and upper extremities. | Heart murmur; Bordetella infection |
| 7 | Scattered bilateral anterior cerebral artery infarct with left middle cerebral artery distribution | Reduced balance and coordination. Impaired ambulation, activities of daily living, transfers, cognition, and memory. Hemiparesis of lower and upper extremities. | Anxiety; elevated white blood cells |
| 8 | Right lacunar | Reduced balance, coordination, sensation. Spasticity. | Anxiety; spinal stenosis; disc displacement; amnesia; nicotine dependence; GERD; chronic pain |
| 9 | Thalamus and basal ganglia | Impaired ambulation, activities of daily living, transfers, cognition, and speech. | |
| 10 | Perforator of the right basal ganglia and right corona radiata | Reduced balance. Impaired ambulation, activities of daily living, transfers, bladder function, and speech. Hemiparesis of lower and upper extremity weakness. | Hypothyroidism; angina pectoris; UTI; sleep disorder; coronary artery disease |
| Sleep Stage | Specificity | Precision | Sensitivity | F1 | Balanced Accuracy |
|---|---|---|---|---|---|
| 2-stage | |||||
| Wake | 0.93 (0.03) | 0.45 (0.12) | 0.25 (0.06) | ||
| Sleep | 0.30 (0.07) | 0.93 (0.02) | 0.93 (0.03) | 0.92 (0.02) | |
| 3-stage | |||||
| Wake | 0.87 (0.03) | 0.28 (0.10) | 0.41 (0.07) | 0.28 (0.08) | 0.64 (0.04) |
| NREM | ) | 0.76 (0.03) | 0.58 (0.05) | 0.64 (0.04) | 0.56 (0.02) |
| REM | 0.72 (0.05) | 0.26 (0.06) | 0.31 (0.07) | 0.26 (0.05) | 0.52 (0.04) |
| 4-stage | |||||
| Wake | 0.30 (0.10) | 0.46 (0.09) | 0.29 (0.07) | 0.66 (0.04) | |
| Light | 0.55 (0.05) | 0.59 (0.04) | 0.45 (0.04) | 0.49 (0.03) | 0.50 (0.01) |
| Deep | 0.84 (0.06) | 0.05 (0.03) | 0.09 (0.06) | 0.02 (0.01) | 0.46 (0.03) |
| REM | 0.77 (0.03) | 0.24 (0.06) | 0.22 (0.04) | 0.22 (0.04) | |
References
- Wu, M.P.; Lin, H.J.; Weng, S.F.; Ho, C.H.; Wang, J.J.; Hsu, Y.W. Insomnia Subtypes and the Subsequent Risks of Stroke Report From a Nationally Representative Cohort. Stroke 2014, 45, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, C.L.; Hermann, D.M. Sleep and stroke. Handb. Clin. Neurol. 2011, 99, 1051–1072. [Google Scholar] [PubMed]
- Leppavuori, A.; Pohjasvaara, T.; Vataja, R.; Kaste, M.; Erkinjuntti, T. Insomnia in ischemic stroke patients. Cerebrovasc. Dis. 2002, 14, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Dimyan, M.A.; Cohen, L.G. Neuroplasticity in the context of motor rehabilitation after stroke. Nat. Rev. Neurol. 2011, 7, 76–85. [Google Scholar] [CrossRef]
- Huang, R.J.; Lai, C.H.; Lee, S.D.; Pai, F.Y.; Chang, S.W.; Chung, A.H.; Chang, Y.F.; Ting, H. Objective Sleep Measures in Inpatients With Subacute Stroke Associated With Levels and Improvements in Activities of Daily Living. Arch. Phys. Med. Rehab. 2018, 99, 699–706. [Google Scholar] [CrossRef]
- Facchin, L.; Schone, C.; Mensen, A.; Bandarabadi, M.; Pilotto, F.; Saxena, S.; Libourel, P.A.; Bassetti, C.L.A.; Adamantidis, A.R. Slow Waves Promote Sleep-Dependent Plasticity and Functional Recovery after Stroke. J. Neurosci. 2020, 40, 8637–8651. [Google Scholar] [CrossRef]
- Krystal, A.D.; Edinger, J.D. Measuring sleep quality. Sleep Med. 2008, 9 (Suppl. S1), S10–S17. [Google Scholar] [CrossRef]
- Good, D.C.; Henkle, J.Q.; Gelber, D.; Welsh, J.; Verhulst, S. Sleep-disordered breathing and poor functional outcome after stroke. Stroke 1996, 27, 252–259. [Google Scholar] [CrossRef]
- Goodwin, G.M.; McCloskey, D.I.; Mitchell, J.H. Cardiovascular and respiratory responses to changes in central command during isometric exercise at constant muscle tension. J. Physiol. 1972, 226, 173–190. [Google Scholar] [CrossRef]
- de Zambotti, M.; Trinder, J.; Silvani, A.; Colrain, I.M.; Baker, F.C. Dynamic coupling between the central and autonomic nervous systems during sleep: A review. Neurosci. Biobehav. Rev. 2018, 90, 84–103. [Google Scholar] [CrossRef]
- Swanson, L.W. Brain Architecture: Understanding the Basic Plan; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Boe, A.J.; McGee Koch, L.L.; O’Brien, M.K.; Shawen, N.; Rogers, J.A.; Lieber, R.L.; Reid, K.J.; Zee, P.C.; Jayaraman, A. Automating sleep stage classification using wireless, wearable sensors. NPJ Digit. Med. 2019, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Korpelainen, J.T.; Sotaniemi, K.A.; Myllyla, V.V. Autonomic nervous system disorders in stroke. Clin. Auton. Res. 1999, 9, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Ni, X.Y.; Lee, J.Y.; Arafa, H.; Pe, D.J.; Xu, S.; Avila, R.; Irie, M.; Lee, J.H.; Easterlin, R.L.; et al. Mechano-acoustic sensing of physiological processes and body motions via a soft wireless device placed at the suprasternal notch. Nat. Biomed. Eng. 2020, 4, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, D.; Blake, S.; Kalluri, L.; Walter, J.; Davies, C.; Zee, P.; Xu, S.; Power, T. 403 Comparative Study of Wireless Sensors Versus Type III Home Sleep Apnea Test for Home-based Diagnosis of Obstructive Sleep Apnea. Sleep 2021, 44, A160. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn Res. 2011, 12, 2825–2830. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 12 July 2022).
- Pan, J.; Tompkins, W.J. A Real-Time Qrs Detection Algorithm. Ieee T Bio-Med. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef]
- Iber, C.; American Academy of Sleep Medicine. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology, and Technical Specifications; American Academy of Sleep Medicine: Westchester, IL, USA, 2007. [Google Scholar]
- Kräuchi, K.; Cajochen, C.; Werth, E.; Wirz-Justice, A. Functional link between distal vasodilation and sleep-onset latency? Am. J. Physiol. -Regul. Integr. Comp. Physiol. 2000, 278, R741–R748. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach Learn Res. 2008, 9, 2579–2605. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Korkalainen, H.; Aakko, J.; Duce, B.; Kainulainen, S.; Leino, A.; Nikkonen, S.; Afara, I.O.; Myllymaa, S.; Toyras, J.; Leppanen, T. Deep learning enables sleep staging from photoplethysmogram for patients with suspected sleep apnea. Sleep 2020, 43, zsaa098. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- De Vries, H.; Elliott, M.N.; Kanouse, D.E.; Teleki, S.S. Using Pooled Kappa to Summarize Interrater Agreement across Many Items. Field Methods 2008, 20, 272–282. [Google Scholar] [CrossRef]
- Zhang, X.; Kou, W.; Chang, E.I.; Gao, H.; Fan, Y.; Xu, Y. Sleep stage classification based on multi-level feature learning and recurrent neural networks via wearable device. Comput. Biol. Med. 2018, 103, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Z.; Lan, K.; Liu, X.; Zhang, Z.; Li, P.; Cao, D.; Zheng, J.; Pan, J. Sleep stage classification using bidirectional lstm in wearable multi-sensor systems. In Proceedings of the IEEE INFOCOM 2019-IEEE Conference on Computer Communications Workshops (INFOCOM WKSHPS), Paris, France, 29 April–2 May 2019; pp. 443–448. [Google Scholar]
- Beattie, Z.; Oyang, Y.; Statan, A.; Ghoreyshi, A.; Pantelopoulos, A.; Russell, A.; Heneghan, C. Estimation of sleep stages in a healthy adult population from optical plethysmography and accelerometer signals. Physiol. Meas. 2017, 38, 1968–1979. [Google Scholar] [CrossRef]
- Hussain, Z.; Sheng, Q.Z.; Zhang, W.E.; Ortiz, J.; Pouriyeh, S. Non-invasive techniques for monitoring different aspects of sleep: A comprehensive review. ACM Trans. Comput. Healthc. (HEALTH) 2022, 3, 1–26. [Google Scholar] [CrossRef]
- Lees, T.; Shad-Kaneez, F.; Simpson, A.M.; Nassif, N.T.; Lin, Y.G.; Lal, S. Heart Rate Variability as a Biomarker for Predicting Stroke, Post-stroke Complications and Functionality. Biomark Insights 2018, 13, 1177271918786931. [Google Scholar] [CrossRef]
- Oppenheimer, S.M.; Hachinski, V.C. The cardiac consequences of stroke. Neurol. Clin. 1992, 10, 167–176. [Google Scholar] [CrossRef]
- Perslev, M.; Jensen, M.H.; Darkner, S.; Jennum, P.J.; Igel, C. U-Time: A Fully Convolutional Network for Time Series Segmentation Applied to Sleep Staging. Adv. Neur. In. 2019, 32. [Google Scholar] [CrossRef]
- Sridhar, N.; Shoeb, A.; Stephens, P.; Kharbouch, A.; Ben Shimol, D.; Burkart, J.; Ghoreyshi, A.; Myers, L. Deep learning for automated sleep staging using instantaneous heart rate. Npj Digital. Med. 2020, 3, 106. [Google Scholar] [CrossRef]
- Yang, X.D.; Shah, S.A.; Ren, A.F.; Zhao, N.; Zhao, J.X.; Hu, F.M.; Zhang, Z.Y.; Zhao, W.; Rehman, M.U.; Alomainy, A. Monitoring of Patients Suffering From REM Sleep Behavior Disorder. Ieee J. Electromagn. Rf Microw. Med. Biol. 2018, 2, 138–143. [Google Scholar] [CrossRef]
- Penzel, T.; Bunde, A.; Heitmann, J.; Kantelhardt, J.W.; Peter, J.H.; Voigt, K. Sleep stage-dependent heart rate variability in patients with obstructive sleep apnea. In Proceedings of the Computers in Cardiology 1999. Vol.26 (Cat. No.99CH37004), Hannover, Germany, 26–29 September 1999; pp. 249–252. [Google Scholar]
- Tobaldini, E.; Nobili, L.; Strada, S.; Casali, K.R.; Braghiroli, A.; Montano, N. Heart rate variability in normal and pathological sleep. Front. Physiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Hermann, D.M.; Bassetti, C.L. Sleep-related breathing and sleep-wake disturbances in ischemic stroke. Neurology 2009, 73, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
| ID | Age | Sex | BMI | Race | Stroke Type | Affected Side (Left/Right) | Experiencing Pain (Yes/No, Self-Report) | No. of Medications with Sleep-Related Side Effects (Drowsiness, Insomnia) |
|---|---|---|---|---|---|---|---|---|
| 1 | 53 | M | 23.6 | C | Isc+Hem | L | Y | 2 D, 2 I |
| 2 | 52 | F | 40.3 | AA | Isc | R | N | 0 D, 3 I |
| 3 | 56 | M | 38.6 | C | Isc | R | N | 2 D, 2 I |
| 4 | 48 | F | 30.4 | C | Isc | L | N | 1 D, 1 I |
| 5 | 64 | F | 27.5 | AA | Hem | L | Y | 1 D, 4 I |
| 6 | 70 | F | 21.0 | AA | Isc | L | N | 1 D, 5 I |
| 7 | 37 | F | 32.6 | PI | Isc | R | Y | 2 D, 3 I |
| 8 | 56 | M | 39.1 | NA | Isc | L | Y | 2 D, 2 I |
| 9 | 65 | M | 25.8 | A | Hem | L | N | 0 D, 1 I |
| 10 | 80 | M | 23.1 | C | Isc | L | N | 0 D, 2 I |
| Mean (SD)or Count | 58.1 (12.1) | 5 F, 5 M | 30.2 (7.2) | 4 C, 3 AA, 1 A, 1 PI, 1 NA | 7 Isc, 2 Hem, 1 Isc+Hem | 7 L, 3 R | 4 Y, 6 N | 1.1 D, 2.5 I |
| Sensor Modality | Sampling Freq (Hz) | No. of Features | Features | ||
|---|---|---|---|---|---|
| ACC (Chest) | 52 | 33 | Mean (x, y, z) Min (x, y, z) Max (x, y, z) Range (x, y, z) | IQR (x, y, z) SD (x, y, z) Kurtosis (x, y, z) RMS (x, y, z) | Variance (x, y, z) rho (x, y, z) p (x, y, z) |
| ECG | 512 | 19 | HR mean HR min HR max SDNN RMSSD NN50 NN20 | PNN50 PNN20 VLF power VLF peak LF power LF peak | HF power HF peak LFHF ratio R-R mean R-R min R-R max |
| TEMP | 5 | 6 | DPG mean DPG min | DPG max DPG range | Chest (proximal) mean Limb (distal) mean |
| PPG | 256 | 15 | SpO2 mean SpO2 min SpO2 variance SpO2 rho SpO2 ZC | SpO2 DI TSA95 TSA90 TSA85 TSA80 | TSA70 ODI2 ODI3 ODI4 ODI5 |
| Algorithm | Sleep Stage Resolution (No. Classes) | Population Model | Personalized Model |
|---|---|---|---|
| Bagging Classifier | 2 | 0.249 | 0.483 |
| 3 | 0.132 | 0.473 | |
| 4 | 0.003 | 0.527 | |
| Random Forest | 2 | 0.248 | 0.577 |
| 3 | 0.171 | 0.532 | |
| 4 | 0.061 | 0.517 | |
| Gradient Boosting | 2 | 0.268 | 0.549 |
| 3 | 0.110 | 0.602 | |
| 4 | 0.037 | 0.617 | |
| XGBoost * | 2 | 0.249 | 0.660 |
| 3 | 0.128 | 0.600 | |
| 4 | 0.014 | 0.531 | |
| ActiWatch Autoscore | 2 | 0.477 | |
| Sleep Stage | Specificity | Precision | Sensitivity | F1 | Balanced Accuracy |
|---|---|---|---|---|---|
| 2-stage | |||||
| Wake | 0.97 (0.01) | 0.81 (0.05) | 0.68 (0.06) | 0.68 (0.04) | 0.83 (0.03) |
| Sleep | 0.68 (0.06) | 0.96 (0.01) | 0.97 (0.01) | 0.97 (0.01) | 0.83 (0.03) |
| 3-stage | |||||
| Wake | 0.94 (0.01) | 0.78 (0.03) | 0.81 (0.04) | 0.74 (0.03) | 0.88 (0.02) |
| NREM | 0.80 (0.03) | 0.90 (0.02) | 0.81 (0.03) | 0.83 (0.04) | 0.81 (0.03) |
| REM | 0.89 (0.03) | 0.75 (0.05) | 0.76 (0.04) | 0.71 (0.05) | 0.82 (0.03) |
| 4-stage | |||||
| Wake | 0.96 (0.01) | 0.78 (0.04) | 0.77 (0.04) | 0.72 (0.03) | 0.87 (0.02) |
| Light | 0.79 (0.03) | 0.73 (0.09) | 0.63 (0.07) | 0.66 (0.08) | 0.71 (0.05) |
| Deep | 0.91 (0.02) | 0.57 (0.10) | 0.70 (0.11) | 0.58 (0.10) | 0.76 (0.10) |
| REM | 0.86 (0.04) | 0.69 (0.05) | 0.74 (0.05) | 0.67 (0.05) | 0.80 (0.04) |
| ActiWatch Autoscore Algorithm (2-stage) | |||||
| Wake | 0.92 (0.02) | 0.56 (0.05) | 0.51 (0.08) | 0.50 (0.07) | 0.72 (0.04) |
| Sleep | 0.52 (0.09) | 0.90 (0.02) | 0.93 (0.02) | 0.91 (0.02) | 0.72 (0.04) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.-W.; O’Brien, M.K.; Horin, A.P.; McGee Koch, L.L.; Lee, J.Y.; Xu, S.; Zee, P.C.; Arora, V.M.; Jayaraman, A. Sleep Monitoring during Acute Stroke Rehabilitation: Toward Automated Measurement Using Multimodal Wireless Sensors. Sensors 2022, 22, 6190. https://doi.org/10.3390/s22166190
Chen P-W, O’Brien MK, Horin AP, McGee Koch LL, Lee JY, Xu S, Zee PC, Arora VM, Jayaraman A. Sleep Monitoring during Acute Stroke Rehabilitation: Toward Automated Measurement Using Multimodal Wireless Sensors. Sensors. 2022; 22(16):6190. https://doi.org/10.3390/s22166190
Chicago/Turabian StyleChen, Pin-Wei, Megan K. O’Brien, Adam P. Horin, Lori L. McGee Koch, Jong Yoon Lee, Shuai Xu, Phyllis C. Zee, Vineet M. Arora, and Arun Jayaraman. 2022. "Sleep Monitoring during Acute Stroke Rehabilitation: Toward Automated Measurement Using Multimodal Wireless Sensors" Sensors 22, no. 16: 6190. https://doi.org/10.3390/s22166190
APA StyleChen, P.-W., O’Brien, M. K., Horin, A. P., McGee Koch, L. L., Lee, J. Y., Xu, S., Zee, P. C., Arora, V. M., & Jayaraman, A. (2022). Sleep Monitoring during Acute Stroke Rehabilitation: Toward Automated Measurement Using Multimodal Wireless Sensors. Sensors, 22(16), 6190. https://doi.org/10.3390/s22166190







