An Automatic System for Continuous Pain Intensity Monitoring Based on Analyzing Data from Uni-, Bi-, and Multi-Modality
Abstract
:1. Introduction
1.1. Contributions
2. Related Work
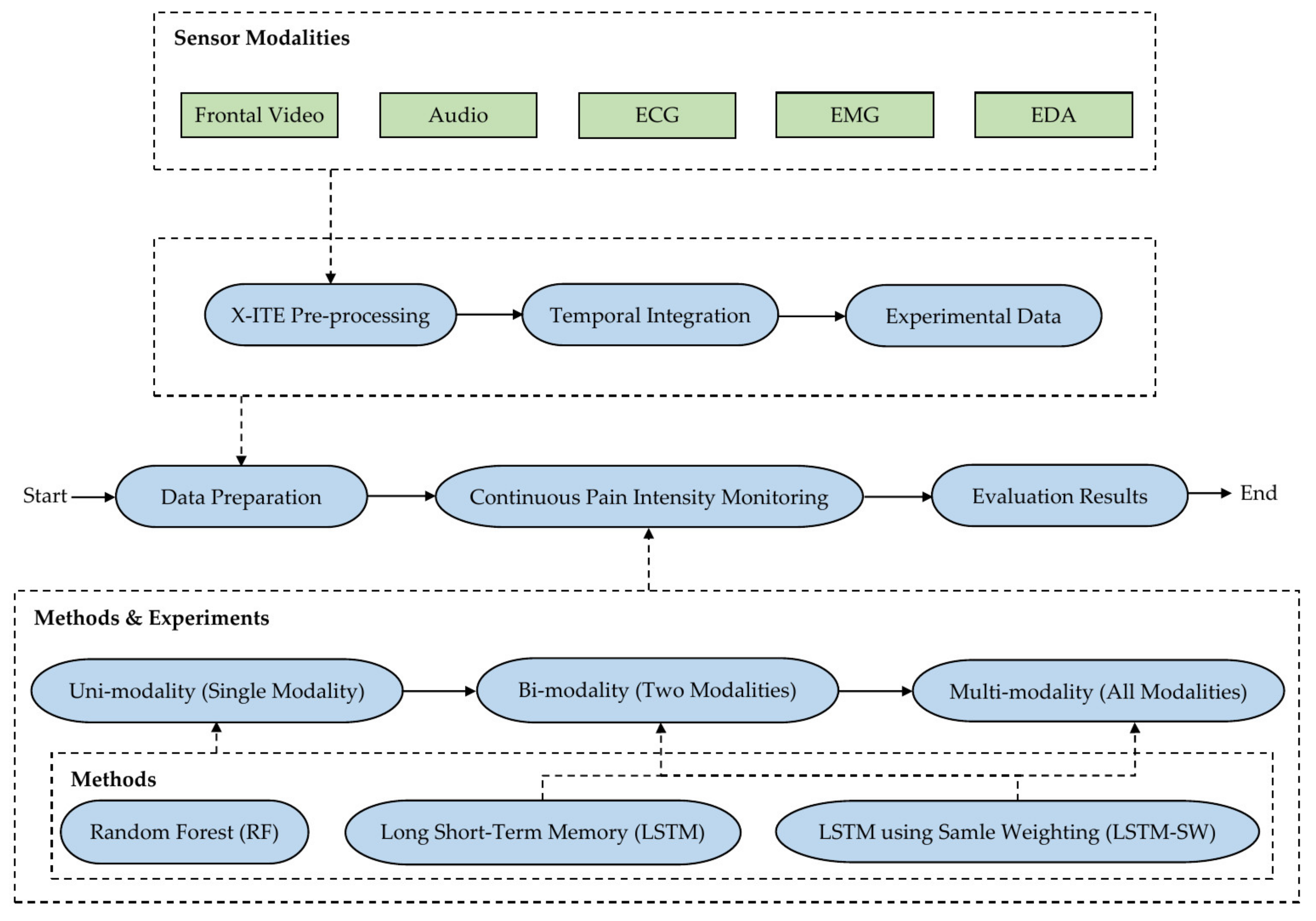
3. Materials and Methods
3.1. X-ITE Pain Database Pre-Processing
3.2. Automatic Pain Intensity Monitoring Methods
3.3. Experiments
4. Results
4.1. Classification vs. Regression
4.2. Classification
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ECG | Electrocardiogram |
| EMG | Electromyogram |
| AUs | Action Units |
| Audio-D | Audio Descriptor |
| two modalities | Bi-modality |
| DDP | delivered duty paid |
| ECG-D | ECG Descriptor |
| EDA | EDA Descriptor |
| EPD | Electrical Phasic Dataset |
| ETD | Electrical Tonic Dataset |
| EDA | Electrodermal Activity |
| EMG | EMG Descriptor |
| FACS | Facial Action Coding System |
| FAD | Facial Activity Descriptor |
| fps | frames per second |
| HRV | Heart Rate Variability |
| HPD | Heat Phasic Dataset |
| HTD | Heat Tonic Dataset |
| ICC | Intraclass Correlation Coefficient |
| HNR | logarithmic Harmonics to Noise Ratio |
| LSTM | Long-Short Term Memory |
| LLD | low-level descriptor |
| LSTM-SW | LSTM using a sample weighting |
| MSE | Mean Squared Error |
| MFCCs | Mel Frequency Cepstral Coefficients |
| Micro avg. F1-score | Micro average F1-score |
| Micro avg. precision | Micro average precision |
| Micro avg. recall | Micro average recall |
| all modalities | Multi-modality |
| PD | Phasic Dataset |
| RF | Random Forest |
| RFc | Random Forest classifier |
| RFr | Random Forest regression |
| REPD | Reduced Electrical Phasic Dataset |
| RETD | Reduced Electrical Tonic Dataset |
| RHPD | Reduced Heat Phasic Dataset |
| RHTD | Reduced Heat Tonic Dataset |
| RPD | Reduced Phasic Dataset |
| RTD | Reduced Tonic Dataset |
| RMS | Root-Mean Square |
| SCL | Skin Conductance Level |
| SCR | Skin Conductance Response |
| STD | Standard Deviation |
| SHS | Subharmonic-Summation |
| SVR | Support Vector Regression |
| TD | Tonic Dataset |
| single modality | Uni-modality |
References
- Williams, A.C.C. Facial Expression of Pain: An Evolutionary Account. Behav. Brain Sci. 2002, 25, 439–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunz, M.; Lautenbacher, S.; LeBlanc, N.; Rainville, P. Are both the sensory and the affective dimensions of pain encoded in the face? Pain 2012, 153, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Thiam, P.; Kestler, H.; Schwenker, F. Multimodal Deep Denoising Convolutional Autoencoders for Pain Intensity Classification based on Physiological Signals. In Proceedings of the 9th International Conference on Pattern Recognition Applications and Methods, Valletta, Malta, 22–24 February 2020; pp. 289–296. [Google Scholar]
- Gruss, S.; Geiger, M.; Werner, P.; Wilhelm, O.; Traue, H.C.; Al-Hamadi, A.; Walter, S. Multi-Modal Signals for Analyzing Pain Responses to Thermal and Electrical Stimuli. J. Vis. Exp. 2019, 146, e59057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekman, P. Facial Expression and Emotion. Am. Psychol. 1993, 48, 384–392. [Google Scholar] [CrossRef]
- Oosterman, J.M.; Zwakhalen, S.; Sampson, E.L.; Kunz, M. The Use of Facial Expressions for Pain Assessment Purposes in Dementia: A Narrative Review. Neurodegener. Dis. Manag. 2016, 6, 119–131. [Google Scholar] [CrossRef]
- Corbett, A.; Achterberg, W.; Husebo, B.; Lobbezoo, F.; de Vet, H.; Kunz, M.; Strand, L.; Constantinou, M.; Tudose, C.; Kappesser, J.; et al. An International Road Map to Improve Pain Assessment in People with Impaired Cognition: The Development of the Pain Assessment in Impaired Cognition (PAIC) Meta-tool. BMC Neurol. 2014, 14, 229. [Google Scholar] [CrossRef] [Green Version]
- Snoek, K.G.; Timmers, M.; Albertyn, R.; van Dijk, M. Pain Indicators for Persisting Pain in Hospitalized Infants in A South African Setting: An Explorative Study. J. Pain Palliat. Care Pharmacother. 2015, 29, 125–132. [Google Scholar] [CrossRef]
- Walter, S.; Gruss, S.; Ehleiter, H.; Tan, J.; Traue, H.C.; Werner, P.; Al-Hamadi, A.; Crawcour, S.; Andrade, A.O.; da Silva, G.M. The BioVid Heat Pain Database: Data for the Advancement and Systematic Validation of an Automated Pain Recognition System. In Proceedings of the IEEE International Conference on Cybernetics (CYBCO), Lausanne, Switzerland, 13–15 June 2013. [Google Scholar]
- Pouromran, F.; Radhakrishnan, S.; Kamarthi, S. Exploration of Physiological Sensors, Features, and Machine Learning Models for Pain Intensity Estimation. PLoS ONE 2021, 16, e0254108. [Google Scholar] [CrossRef]
- Chu, Y.; Zhao, X.; Han, J.; Su, Y. Physiological Signal-Based Method for Measurement of Pain Intensity. Front. Neurosci. 2017, 11, 279. [Google Scholar] [CrossRef]
- Lopez-Martinez, D.; Picard, R. Continuous Pain Intensity Estimation from Autonomic Signals with Recurrent Neural Networks. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 5624–5627. [Google Scholar]
- Thiam, P.; Kessler, V.; Amirian, M.; Bellmann, P.; Layher, G.; Zhang, Y.; Velana, M.; Gruss, S.; Walter, S.; Traue, H.C.; et al. Multi-modal Pain Intensity Recognition based on the SenseEmotion Database. IEEE Trans. Affect. Comput. 2019, 12, 743–760. [Google Scholar] [CrossRef]
- Werner, P.; Al-Hamadi, A.; Gruss, S.; Walter, S. Twofold-Multimodal Pain Recognition with the X-ITE Pain Database. In Proceedings of the 8th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW), Cambridge, UK, 3–6 September 2019. [Google Scholar]
- Walter, S.; Gruss, S.; Limbrecht-Ecklundt, K.; Traue, H.C. Automatic Pain Quantification using Autonomic Parameters. Psychol. Neurosci. 2014, 7, 363–380. [Google Scholar] [CrossRef] [Green Version]
- Walter, S.; Al-Hamadi, A.; Gruss, S.; Frisch, S.; Traue, H.C.; Werner, P. Multimodale Erkennung von Schmerzintensität und-modalität mit maschinellen Lernverfahren. Schmerz 2020, 34, 400–409. [Google Scholar] [CrossRef]
- Susam, B.T.; Riek, N.T.; Akcakaya, M.; Xu, X.; de Sa, V.R.; Nezamfar, H.; Diaz, D.; Craig, K.D.; Goodwin, M.S.; Huang, J.S. Automated Pain Assessment in Children using Electrodermal Activity and Video Data Fusion via Machine Learning. IEEE Trans. Biomed. Eng. 2022, 69, 422–431. [Google Scholar] [CrossRef]
- Subramaniam, S.D.; Dass, B. Automated Nociceptive Pain Assessment using Physiological Signals and a Hybrid Deep Learning Network. IEEE Sens. J. 2021, 21, 3335–3343. [Google Scholar] [CrossRef]
- Odhner, M.; Wegman, D.; Freeland, N.; Steinmetz, A.; Ingersoll, G.L. Assessing Pain Control in Nonverbal Critically ill Adults. Dimens. Crit. Care Nurs. 2003, 22, 260–267. [Google Scholar] [CrossRef]
- Othman, E.; Werner, P.; Saxen, F.; Al-Hamadi, A.; Gruss, S.; Walter, S. Classification Networks for Continuous Automatic Pain Intensity Monitoring in Video using Facial Expression on the X-ITE Pain Database. J. Vis. Commun. Image Represent. 2022, 21, 3273. [Google Scholar]
- Othman, E.; Werner, P.; Saxen, F.; Al-Hamadi, A.; Walter, S. Regression Networks for Automatic Pain Intensity Recognition in Video using Facial Expression on the X-ITE Pain Database. In Proceedings of the 25th International Conference on Image Processing, Computer Vision & Pattern Recognition (IPCV’21), Las Vegas, NV, USA, 26–29 July 2021. [Google Scholar]
- Othman, E.; Werner, P.; Saxen, F.; Al-Hamadi, A.; Gruss, S.; Walter, S. Facial Expression and Electrodermal Activity Analysis for Continuous Pain Intensity Monitoringon the X-ITE Pain Database. IEEE Access 2022. submitted. [Google Scholar]
- Craig, K.D. The Facial Expression of Pain Better than a Thousand Words? APS J. 1992, 1, 153–162. [Google Scholar] [CrossRef]
- Prkachin, K.M. The Consistency of Facial Expressions of Pain: A Comparison Across Modalities. Pain 1992, 51, 297–306. [Google Scholar] [CrossRef]
- Schiavenato, M. Facial Expression and Pain Assessment in the Pediatric Patient: The Primal Face of Pain. J. Spec. Pediatric Nurs. 2008, 13, 89–97. [Google Scholar] [CrossRef]
- Tavakolian, M. Efficient Spatiotemporal Representation Learning for Pain Intensity Estimation from Facial Expressions Doctor of Philosophy. Ph.D. Thesis, University of Oulu, Oulu, Finland, 2021. [Google Scholar]
- Feldt, K.S. The Checklist of Nonverbal Pain Indicators (CNPI). Pain Manag. Nurs. 2000, 1, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, S.J.; Riordan, P.A.; Keefe, F.J.; Lefebvre, J.C. Pain Behavior in Rheumatoid Arthritis Patients: Identification of Pain Behavior Subgroups. J. Pain Symptom. Manag. 2008, 36, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Naranjo-Hernández, D.; Reina-Tosina, J.; Roa, L.M. Sensor Technologies to Manage the Physiological Traits of Chronic Pain: A Review. Sensors 2020, 20, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, L. Physiologic Responses to Cutaneous Pain in Neonates. Neonatal Netw. 1987, 6, 18–22. [Google Scholar] [PubMed]
- Greisen, J.; Juhl, C.B.; Grøfte, T.; Vilstrup, H.; Jensen, T.S.; Schmitz, O. Acute Pain Induces Insulin Resistance in Humans. Anesthesiology 2001, 95, 578–584. [Google Scholar] [CrossRef] [Green Version]
- Stevens, B.J.; Johnston, C.C. Physiological Responses of Premature Infants to a Painful Stimulus. Nurs. Res. 1994, 43, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Moscato, S.; Cortelli, P.; Chiari, L. Physiological responses to pain in cancer patients: A systematic review. Comput. Methods Programs Biomed. 2022, 2017, 106682. [Google Scholar] [CrossRef]
- Saccò, M.; Meschi, M.; Regolisti, G.; Detrenis, S.; Bianchi, L.; Bertorelli, M.; Pioli, S.; Magnano, A.; Spagnoli, F.; Giuri, P.G.; et al. The Relationship between Blood Pressure and Pain. J. Clin. Hypertens. 2013, 15, 600–605. [Google Scholar] [CrossRef]
- Littlewort, G.C.; Bartletta, M.S.; Lee, K. Automatic Coding of Facial Expressions Displayed during Posed and Genuine Pain. Image Vis. Comput. 2009, 27, 1797–1803. [Google Scholar] [CrossRef]
- Lucey, P.; Cohn, F.J.; Prkachind, F.K.; Solomon, E.P.; Chewf, S.; Matthews, I. Painful Monitoring: Automatic Pain Monitoring using the UNBC-McMaster Shoulder Pain Expression Archive Database. Image Vis. Comput. 2012, 30, 197–205. [Google Scholar] [CrossRef]
- Borsook, D.; Moulton, E.A.; Schmidt, K.F.; Becerra, L.R. Neuroimaging Revolutionizes Therapeutic Approaches to Chronic Pain. Mol. Pain 2007, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Aslaksen, P.M.; Myrbakk, I.N.; Høifødt, R.S.; Flaten, M.A. The Effect of Experimenter Gender on Autonomic and Subjective Responses to Pain Stimuli. Pain 2006, 129, 260–268. [Google Scholar] [CrossRef]
- Koenig, J.; Jarczok, M.N.; Ellis, R.J.; Hillecke, T.K.; Thayer, J.F. Heart Rate Variability and Experimentally Induced Pain in Healthy Adults: A systematic Review. Eur. J. Pain 2014, 18, 301–314. [Google Scholar] [CrossRef]
- Werner, P.; Al-Hamadi, A.; Niese, R.; Walter, S.; Gruss, S.; Traue, H.C. Automatic Pain Recognition from Video and Biomedical Signals. In Proceedings of the 22nd International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014. [Google Scholar]
- Storm, H. Changes in Skin Conductance as a Tool to Monitor Nociceptive Stimulation and Pain. Curr. Opin. Anaesthesiol. 2008, 12, 796–804. [Google Scholar] [CrossRef]
- Ledowski, T.; Bromilow, J.; Paech, M.J.; Storm, H.; Hacking, R.; Schug, S.A. Monitoring of Skin Conductance to Assess Postoperative Pain Intensity. Br. J. Anaesth. 2006, 97, 862–865. [Google Scholar] [CrossRef] [Green Version]
- Loggia, M.L.; Juneau, M.; Bushnell, C.M. Autonomic Responses to Heat Pain: Heart Rate, Skin Conductance, and their Relation to Verbal Ratings and Stimulus Intensity . Pain 2011, 152, 592–598. [Google Scholar] [CrossRef]
- Werner, P.; Al-Hamadi, A.; Limbrecht-Ecklundt, K.; Walter, S.; Gruss, S.; Traue, H.C. Automatic Pain Assessment with Facial Activity Descriptors. IEEE Trans. Affect. Comput. 2017, 8, 286–299. [Google Scholar] [CrossRef]
- Kächele, M.; Thiam, P.; Amirian, M.; Werner, P.; Walter, S.; Schwenker, F.; Palm, G. Multimodal Data Fusion for Person-Independent, Continuous Estimation of Pain Intensity. In Proceedings of the Engineering Applications of Neural Networks: 16th International Conference, Rhodes, Greece, 25–28 September 2015. [Google Scholar]
- Erekat, D.; Hammal, Z.; Siddiqui, M.; Dibeklioğlu, H. Enforcing Multilabel Consistency for Automatic Spatio-Temporal Assessment of Shoulder Pain Intensity. In Proceedings of the International Conference on Multimodal Interaction (ICMI’20 Companion), Utrecht, The Netherlands, 25–29 October 2020; pp. 156–164. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Othman, E.; Werner, P.; Saxen, F.; Al-Hamadi, A.; Gruss, S.; Walter, S. Automatic vs. Human Recognition of Pain Intensity from Facial Expression on the X-ITE Pain Database. Sensors 2021, 21, 3273. [Google Scholar] [CrossRef]
- Othman, E.; Werner, P.; Saxen, F.; Al-Hamadi, A.; Walter, S. Cross-Database Evaluation of Pain Recognition from Facial Video. In Proceedings of the 11th International Symposium on Image and Signal Processing and Analysis (ISPA), Dubrovnik, Croatia, 23–25 September 2019. [Google Scholar]
- Thiam, P.; Kessler, V.; Schwenker, F. Hierarchical Combination of Video Features for Personalised Pain Level Recognition. In Proceedings of the European Symposium on Artificial Neural Networks (ESANN), Bruges, Belgium, 26–28 April 2017; pp. 465–470. [Google Scholar]
- Thiam, P.; Schwenker, F. Combining Deep and Hand-Crafted Features for Audio-Based Pain Intensity Classification. In Proceedings of the IAPR Workshop on Multimodal Pattern Recognition of Social Signals in Human-Computer Interaction, Bejing, China, 20 August 2018; pp. 49–58. [Google Scholar]
- Tsai, F.-S.; Hsu, Y.-L.; Chen, W.-C.; Weng, Y.-M.; Ng, C.-J.; Lee, C.-C. Toward Development and Evaluation of Pain Level-Rating Scale for Emergency Triage based on Vocal Characteristics and Facial Expressions. In Proceedings of the 17th Annual Conference of the International Speech Communication Association, San Francisco, CA, USA, 8–12 September 2016. [Google Scholar]
- Salekin, M.S.; Zamzmi, G.; Hausmann, J.; Goldgof, D.; Kasturi, R.; Kneusel, M.; Ashmeade, T.; Ho, T.; Suna, Y. Multimodal Neonatal Procedural and Postoperative Pain Assessment Dataset. Data Brief 2021, 35, 106796. [Google Scholar] [CrossRef]
- Thiam, P.; Hihn, H.; Braun, D.A.; Kestler, H.A.; Schwenker, F. Multi-Modal Pain Intensity Assessment Based on Physiological Signals: A Deep Learning Perspective. Front. Physiol. 2021, 1, 720464. [Google Scholar] [CrossRef]
- Hinduja, S.; Canavan, S.; Kaur, G. Multimodal Fusion of Physiological Signals and Facial Action Units for Pain Recognition. In Proceedings of the 15th IEEE International Conference on Automatic Face and Gesture Recognition (FG 2020), Buenos Aires, Argentina, 16–20 November 2020. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Baltrušaitis, T.; Robinson, P.; Morency, L.-P. OpenFace: An Open Source Facial Behavior Analysis Toolkit. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Placid, NY, USA, 7–10 March 2016. [Google Scholar]
- Eyben, F.; Wöllmer, M.; Schuller, B. OpenSMILE—The Munich Versatile and Fast Open-Source Audio Feature Extractor. In Proceedings of the 18th ACM International Conference on Multimedia (MM ‘10), New York, NY, USA, 25–29 October 2010; pp. 1459–1462. [Google Scholar]
- Hamilton, P.S.; Tompkins, W.J. Quantitative Investigation of QRS Detection Rules Using the MIT/BIH Arrhythmia Database. IEEE Trans. Biomed. Eng. 1986, 33, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Othman, E.; Saxen, F.; Bershadskyy, D.; Werner, P.; Al-Hamadi, A.; Weimann, J. Predicting Group Contribution Behaviour in a Public Goods Game from Face-to-Face Communication. Sensors 2019, 19, 2786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrout, P.E.; Fleiss, J.L. Intraclass Correlations: Uses in Assessing Rater Reliability. Psychol. Bull. 1979, 86, 420–428. [Google Scholar] [CrossRef]
- Thiam, P.; Schwenker, F. Multi-modal Data Fusion for Pain Intensity Assessment and Classification. In Proceedings of the 7th International Conference on Image Processing Theory, Tools and Applications (IPTA), Montreal, QC, Canada, 28 November–1 December 2017. [Google Scholar]
- Kächele, M.; Amirian, M.; Thiam, P.; Werner, P.; Walter, S.; Palm, G.; Schwenker, F. Adaptive Confidence Learning for The Personalization of Pain Intensity Estimation Systems. Evol. Syst. 2017, 8, 71–83. [Google Scholar] [CrossRef]


| Type | Modality | Intensities | |||
|---|---|---|---|---|---|
| Severe | Moderate | Low | No Pain (77%) | ||
| Phasic | H | PH3 = 3 (2%) | PH2 = 2 (2.1%) | PH1 = 1 (2.1%) | BL = 0 |
| E | PE3 = −3 (2.6%) | PE2 = −2 (2.6%) | PE1 = −1 (2.6%) | ||
| Tonic | H | TH3 = 6 (1%) | TH2 = 5 (1%) | TH1 = 4 (1%) | BL = 0 |
| E | TE3 = −6 (1%) | TE2 = −5 (1%) | TE1 = −4 (1%) | ||
| Subsets (Experimental Data) | No Pain | Pain Intensities | ||
|---|---|---|---|---|
| PD | Phasic Dataset | Exclude tonic samples and no pain samples before these samples and also after samples with −10, −11 labeled. | 77.7% | 22.23% |
| HPD | Heat Phasic Dataset | Exclude electrical samples from PD and no-pain samples before these frames. | 87.5% | 21.5% |
| EPD | Electrical Phasic Dataset | Exclude heat samples from PD and no pain frames before these frames. | 86.1% | 13.9% |
| TD | Tonic Dataset | Exclude phasic samples and no pain samples before these samples and also after samples with −10, −11 labeled. | 70.3% | 29.7% |
| HTD | Heat Tonic Dataset | Exclude electrical samples from TD and no pain frames before these frames. | 20.0% | 80.0% |
| ETD | Electrical Tonic Dataset | Exclude heat samples from TD and no pain frames before these frames. | 82.0% | 18.0% |
| Reduced Subsets (Experimental Data) | No Pain | Pain Intensities | ||
| RPD | Reduced Phasic Dataset | Reduce the no pain frames in PD to about 50%. | 50.0% | 50.0% |
| RHPD | Reduced Heat Phasic Dataset | Reduce the no pain frames in HPD to about 50%. | 50.1% | 49.9% |
| REPD | Reduced Electrical Phasic Dataset | Reduce the no pain frames in EPD to about 50%. | 50.0% | 50.0% |
| RTD | Reduced Tonic Dataset | Reduce the no pain frames in TD to about 38%. | 38.1% | 61.9% |
| RETD | Reduced Electrical Tonic Dataset | Reduce the no pain frames in ETD to about 49%. | 49.0% | 51.0% |
| Layer Type | Attribute | Classification | Regression | ||||
|---|---|---|---|---|---|---|---|
| A(c) | B(c) | C(c) | D(c) | A(r) | B(r) | ||
| Input | Size: | 10 × 252 | 10 × 252 | 10 × 252 | 10 × 252 | 10 × 252 | 10 × 252 |
| Timestep: | 10 | 10 | 10 | 10 | 10 | 10 | |
| Features: | 252 | 252 | 252 | 252 | 252 | 252 | |
| LSTM | Activation: | ReLU | ReLU | ReLU | ReLU | ReLU | ReLU |
| No. of units: | 4 | 8 | 4 | 8 | 4 | 8 | |
| Dropout | with p: | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Flatten | Output: | 80 | 40 | 80 | 40 | 80 | 40 |
| Dense1 | Activation: | ReLU | ReLU | ReLU | ReLU | ReLU | ReLU |
| No. of units: | 128 | 64 | 128 | 64 | 128 | 64 | |
| Dense2 | Activation: | Softmax | Softmax | Softmax | Softmax | Linear/Sigmoid | |
| No. of units: | 7 | 7 | 4 | 4 | 1 | 1 | |
| Output | Continuous | - | - | - | - | √ | √ |
| Discrete | √ | √ | √ | √ | - | - | |
| 7 levels | 7 levels | 4 levels | 4 levels | ||||
| Layer Type | Attribute | Architectures Configurations (Bi-Modality) | |||||
|---|---|---|---|---|---|---|---|
| Classification | Regression | ||||||
| A-Bi(c) | B-Bi(c) | C-Bi(c) | D-Bi(c) | A-Bi(r) | B-Bi(r) | ||
| Concatenate (after dense1) | Modality X | A(c) | B(c) | C(c) | D(c) | A(r) | B(r) |
| + | + | + | + | + | + | + | |
| Modality Y | A(c) | B(c) | C(c) | D(c) | A(r) | B(r) | |
| Dense2 | Activation: | Softmax | Sigmoid | ||||
| No. of units: | 7 | 7 | 4 | 4 | 1 | 1 | |
| Output | Continuous | - | - | - | - | √ | √ |
| Discrete | √ | √ | √ | √ | - | - | |
| 7 levels | 7 levels | 4 levels | 4 levels | ||||
| Layer Type | Attribute | Architectures Configurations (Multi-Modality) | |||||
|---|---|---|---|---|---|---|---|
| Classification | Regression | ||||||
| A-Mu(c) | B-Mu(c) | C-Mu(c) | D-Mu(c) | A-Mu(r) | B-Mu(r) | ||
| Concatenate (after dense1) | Modality 1 | A(c) | B(c) | C(c) | D(c) | A(r) | B(r) |
| + | + | + | + | + | + | + | |
| Modality 2 | A(c) | B(c) | C(c) | D(c) | A(r) | B(r) | |
| + | + | + | + | + | + | + | |
| Modality 3 | A(c) | B(c) | C(c) | D(c) | A(r) | B(r) | |
| + | + | + | + | + | + | + | |
| Modality 4 | A(c) | B(c) | C(c) | D(c) | A(r) | B(r) | |
| + | + | + | + | + | + | + | |
| Modality 5 | A(c) | B(c) | C(c) | D(c) | A(r) | B(r) | |
| Dense2 | Activation: | Softmax | Softmax | Softmax | Softmax | Sigmoid | Sigmoid |
| No. of units: | 7 | 7 | 4 | 4 | 1 | 1 | |
| Output | Continuous | - | - | - | - | √ | √ |
| Discrete | √ | √ | √ | √ | - | - | |
| 7 levels | 7 levels | 4 levels | 4 levels | ||||
| Meas. | Task | Classification | Regression | |||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Uni-Modality | Bi-Modality | Multi-Modality | Uni-Modality | Bi-Modality | Multi-Modality | ||
| MSE | Subsets | PD | 0.09 EDA-D | 0.08 EMG-D EDA-D | 0.08 | 0.06 EDA-D | 0.06 EMG-D EDA-D | 0.06 |
| HPD | 0.10 EDA-D | 0.09 EMG-D EDA-D | 0.10 | 0.08 EDA-D | 0.07 EMG-D EDA-D | 0.09 | ||
| EPD | 0.06 EDA-D | 0.06 EMG-D EDA-D | 0.05 | 0.05 EDA-D | 0.04 EMG-D EDA-D | 0.04 | ||
| TD | 0.11 EDA-D | 0.11 EMG-D EDA-D | 0.11 | 0.09 EDA-D | 0.10 EMG-D EDA-D | 0.08 | ||
| HTD | 0.15 EDA-D (LSTM-SW) EMG-D (LSTM) | 0.13 EMG-D EDA-D | 0.15 | 0.11 EDA-D | 0.10 EMG-D EDA-D | 0.10 | ||
| ETD | 0.11 (RFc) | 0.08 EMG-D EDA-D | 0.08 | 0.07 EDA-D | 0.06 EMG-D EDA-D | 0.06 | ||
| STD | 0.03 | 0.02 | 0.03 | 0.02 | 0.02 | 0.02 | ||
| Mean | 0.10 | 0.09 | 0.10 | 0.08 | 0.07 | 0.07 | ||
| Reduced Subsets | RPD | 0.05 EDA-D (both LSTM) | 0.05 EMG-D EDA-D | 0.05 | 0.04 EDA-D | 0.04 EMG-D EDA-D | 0.04 | |
| RHPD | 0.07 EDA-D | 0.07 EMG-D EDA-D | 0.08 | 0.05 EDA-D (both LSTM) | 0.05 EMG-D EDA-D | 0.08 | ||
| REPD | 0.05 EDA-D (both LSTM) | 0.05 EMG-D EDA-D (both LSTM) | 0.05 (both LSTM) | 0.03 EDA-D | 0.04 EMG-D EDA-D | 0.06 | ||
| RTD | 0.19 EDA-D | 0.19 EMG-D EDA-D | 0.19 | 0.11 EDA-D | 0.11 EMG-D EDA-D | 0.04 | ||
| RETD | 0.16 EDA-D | 0.15 EMG-D EDA-D | 0.16 | 0.10 EDA-D | 0.09 EMG-D EDA-D (both LSTM) | 0.09 | ||
| STD | 0.07 | 0.06 | 0.07 | 0.04 | 0.03 | 0.02 | ||
| Mean | 0.10 | 0.10 | 0.11 | 0.07 | 0.07 | 0.05 | ||
| Meas. | Task | Classification | Regression | |||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Uni-Modality | Bi-Modality | Multi-Modality | Uni-Modality | Bi-Modality | Multi-Modality | ||
| ICC | Subsets | PD | 0.40 EDA-D | 0.45 EMG-D EDA-D | 0.46 | 0.43 EDA-D | 0.51 EMG-D EDA-D | 0.49 |
| HPD | 0.30 EDA-D | 0.41 EMG-D EDA-D | 0.39 | 0.32 EDA-D | 0.41 EMG-D EDA-D | 0.40 | ||
| EPD | 0.50 EDA-D | 0.53 EMG-D EDA-D | 0.57 | 0.53 EDA-D | 0.58 EMG-D EDA-D | 0.58 | ||
| TD | 0.15 EDA-D | 0.18 EMG-D EDA-D | 0.23 | 0.17 EDA-D | 0.26 EMG-D EDA-D | 0.30 | ||
| HTD | 0.33 EDA-D (LSTM-SW) EMG-D (LSTM) | 0.42 EMG-D EDA-D | 0.35 | 0.30 EDA-D | 0.32 EMG-D EDA-D | 0.38 | ||
| ETD | 0.14 (RFc) | 0.22 EMG-D EDA-D | 0.26 | 0.21 EDA-D | 0.31 EMG-D EDA-D | 0.33 | ||
| STD | 0.14 | 0.14 | 0.13 | 0.13 | 0.13 | 0.10 | ||
| Mean | 0.30 | 0.39 | 0.38 | 0.33 | 0.40 | 0.41 | ||
| Reduced Subsets | RPD | 0.83 EDA-D (both LSTM) | 0.83 EMG-D EDA-D | 0.82 | 0.84 EDA-D | 0.85 EMG-D EDA-D | 0.82 | |
| RHPD | 0.76 EDA-D | 0.79 EMG-D EDA-D | 0.74 | 0.81 EDA-D (both LSTM) | 0.83 EMG-D EDA-D | 0.73 | ||
| REPD | 0.84 EDA-D (both LSTM) | 0.85 EMG-D EDA-D (both LSTM) | 0.81 (both LSTM) | 0.88 EDA-D | 0.87 EMG-D EDA-D | 0.80 | ||
| RTD | 0.31 EDA-D | 0.32 EMG-D EDA-D | 0.28 | 0.24 (EDA-D) | 0.33 EMG-D EDA-D | 0.29 | ||
| RETD | 0.47 EDA-D | 0.52 EMG-D EDA-D | 0.44 | 0.49 EDA-D | 0.52 EMG-D EDA-D (both LSTM) | 0.56 | ||
| STD | 0.24 | 0.23 | 0.24 | 0.28 | 0.24 | 0.22 | ||
| Mean | 0.64 | 0.66 | 0.62 | 0.65 | 0.75 | 0.62 | ||
| Measure | Datasets | HPD | HTD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Triv. | RFc | LSTM | LSTM-SW | Triv. | RFc | LSTM | LSTM-SW | |
| Accuracy % | EDA-D (Uni-modality) | 78.5 | 78.1 | 79.8 * | 79 * | 20 | 41.0 | 48.4 | 47.7 |
| FAD and EDA-D (Bi-modality) | 78.5 | - | 80.5 * | 80.2 * | 20 | - | 47.4 * | 49.8 * | |
| Multi-modality | 78.5 | - | 79.3 | 77.6 | 20 | - | 41.6 | 42.2 | |
| Micro avg. precision% | EDA-D (Uni-modality) | 0 | 24.6 | 36.6 * | 32.2 * | 0 | 42.7 | 48.2 | 47.7 |
| FAD and EDA-D (Bi-modality) | 0 | - | 42.8 | 40.1 * | 0 | - | 47.2 | 48.7 | |
| Multi-modality | 0 | - | 34.9 * | 29.2 | 0 | - | 41.8 | 42.0 | |
| Micro avg. recall% | EDA-D (Uni-modality) | 0 | 3.4 | 9.9 * | 10.9 * | 0 | 71.0 | 94.6 * | 100 * |
| FAD and EDA-D (Bi-modality) | 0 | - | 16.3 * | 21.4 * | 0 | - | 92.9 * | 97 * | |
| Multi-modality | 0 | - | 19.8 * | 22.3 * | 0 | - | 90.8 * | 99.9 * | |
| Micro avg. F1-Score% | EDA-D (Uni-modality) | 0 | 5.9 | 15.2 * | 15.5 * | 0 | 52.9 | 62.3 * | 62.5 * |
| FAD and EDA-D (Bi-modality) | 0 | - | 22.3 * | 26.3 * | 0 | - | 60.7 * | 63.3 * | |
| Multi-modality | 0 | - | 23.6 * | 24 * | 0 | - | 56 | 57.9 * | |
| Model | Dataset | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HPD | HPD | ||||||||||
| BL | PH1 | PH2 | PH3 | Mean | BL | TH1 | TH2 | TH3 | Mean | ||
| EDA-D (Uni-modality) | Trivial | 100 | 0 | 0 | 0 | 25 | 100 | 0 | 0 | 0 | 25 |
| RFc | 98.7 | 1.5 | 2 | 6.3 | 27.1 | 34.4 | 27.3 | 47 | 54.5 | 40.8 | |
| LSTM | 99.3 | 5.2 | 5.4 | 15.2 | 31.3 | 9.4 | 73 | 35.1 | 67.8 | 46.3 | |
| LSTM-SW | 98 | 3.1 | 1.7 | 23.3 | 31.5 | 0.30 | 72 | 39.8 | 68 | 45 | |
| FAD and EDA-D (Bi-modality) | Trivial | 100 | 0 | 0 | 0 | 25 | 100 | 0 | 0 | 0 | 25 |
| RFc | - | - | - | - | - | - | - | - | - | - | |
| LSTM | 98.7 | 6.5 | 5.2 | 29.5 | 35 | 20.3 | 45.4 | 56.1 | 62.5 | 46.1 | |
| LSTM-SW | 97.4 | 7.4 | 7.2 | 37.9 | 37.5 | 18.6 | 45.8 | 62.7 | 65.5 | 48.2 | |
| Multi-modality | Trivial | 100 | 0 | 0 | 0 | 25 | 100 | 0 | 0 | 0 | 25 |
| RFc | - | - | - | - | - | - | - | - | - | - | |
| LSTM | 96.8 | 6.2 | 4.6 | 36.2 | 36 | 11.2 | 56.3 | 28.9 | 63.5 | 40 | |
| LSTM-SW | 94.1 | 5.9 | 6.3 | 39.5 | 36.5 | 2.7 | 40.4 | 55.2 | 61.8 | 40 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Othman, E.; Werner, P.; Saxen, F.; Fiedler, M.-A.; Al-Hamadi, A. An Automatic System for Continuous Pain Intensity Monitoring Based on Analyzing Data from Uni-, Bi-, and Multi-Modality. Sensors 2022, 22, 4992. https://doi.org/10.3390/s22134992
Othman E, Werner P, Saxen F, Fiedler M-A, Al-Hamadi A. An Automatic System for Continuous Pain Intensity Monitoring Based on Analyzing Data from Uni-, Bi-, and Multi-Modality. Sensors. 2022; 22(13):4992. https://doi.org/10.3390/s22134992
Chicago/Turabian StyleOthman, Ehsan, Philipp Werner, Frerk Saxen, Marc-André Fiedler, and Ayoub Al-Hamadi. 2022. "An Automatic System for Continuous Pain Intensity Monitoring Based on Analyzing Data from Uni-, Bi-, and Multi-Modality" Sensors 22, no. 13: 4992. https://doi.org/10.3390/s22134992
APA StyleOthman, E., Werner, P., Saxen, F., Fiedler, M.-A., & Al-Hamadi, A. (2022). An Automatic System for Continuous Pain Intensity Monitoring Based on Analyzing Data from Uni-, Bi-, and Multi-Modality. Sensors, 22(13), 4992. https://doi.org/10.3390/s22134992






