2.1. Description of Data
To demonstrate the efficacy of our analytical approach, data was repurposed from a feed trial assessing the impact of an organic fat supplement on cow health and productivity, through the first 150 days of lactation. All animal handling and experimental protocols were approved by the Colorado State University Institution of Animal Care and Use Committee (Protocol ID: 16-6704AA). The study ran from January through July in 2017, on a USDA Certified Organic dairy in Northern Colorado, enrolling a total of 200 cows over a 1.5-month period into a mixed-parity herd of animals, with predominantly Holstein genetics. Cows were maintained in a closed herd in an open-sided free-stall barn, stocked at roughly half capacity with respect to both feed bunk spaces and stalls. Cows had free access to an adjacent outdoor dry lot while in their home pen, and beginning in April were moved onto pasture at night, to comply with organic grazing standards. Cows were milked three times a day, with free access to TMR between milkings, and were head locked each morning to facilitate data collection and daily health checks. For more details on feed trial protocols, see Manriquez et al. (2018) and Manriquez et al. (2019) [
21,
22].
In addition to standard production and health assessments, behavioral data was also obtained from several PLF data streams [
19]. Milking order, or the sequence in which cows enter the parlor to be milked, is automatically recorded as metadata in all modern RFID-equipped milking systems. Our study cows were milked in a DelPro
TM rotary parlor (DeLaval, Tumba, Sweden). At each morning milking, raw milking logs were exported from the parlor software, and the data were processed in order to extract the single-file order that cows entered the rotary [
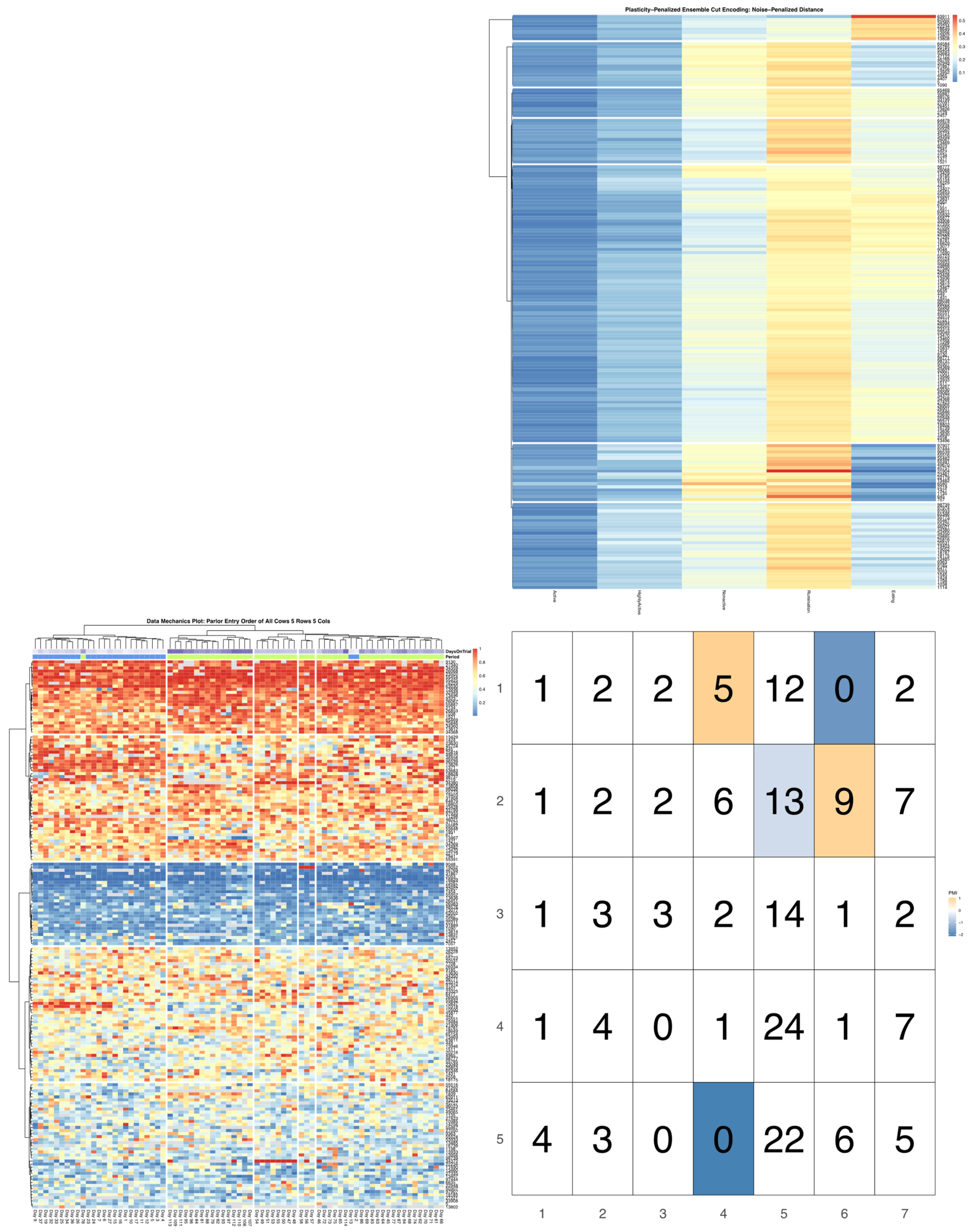
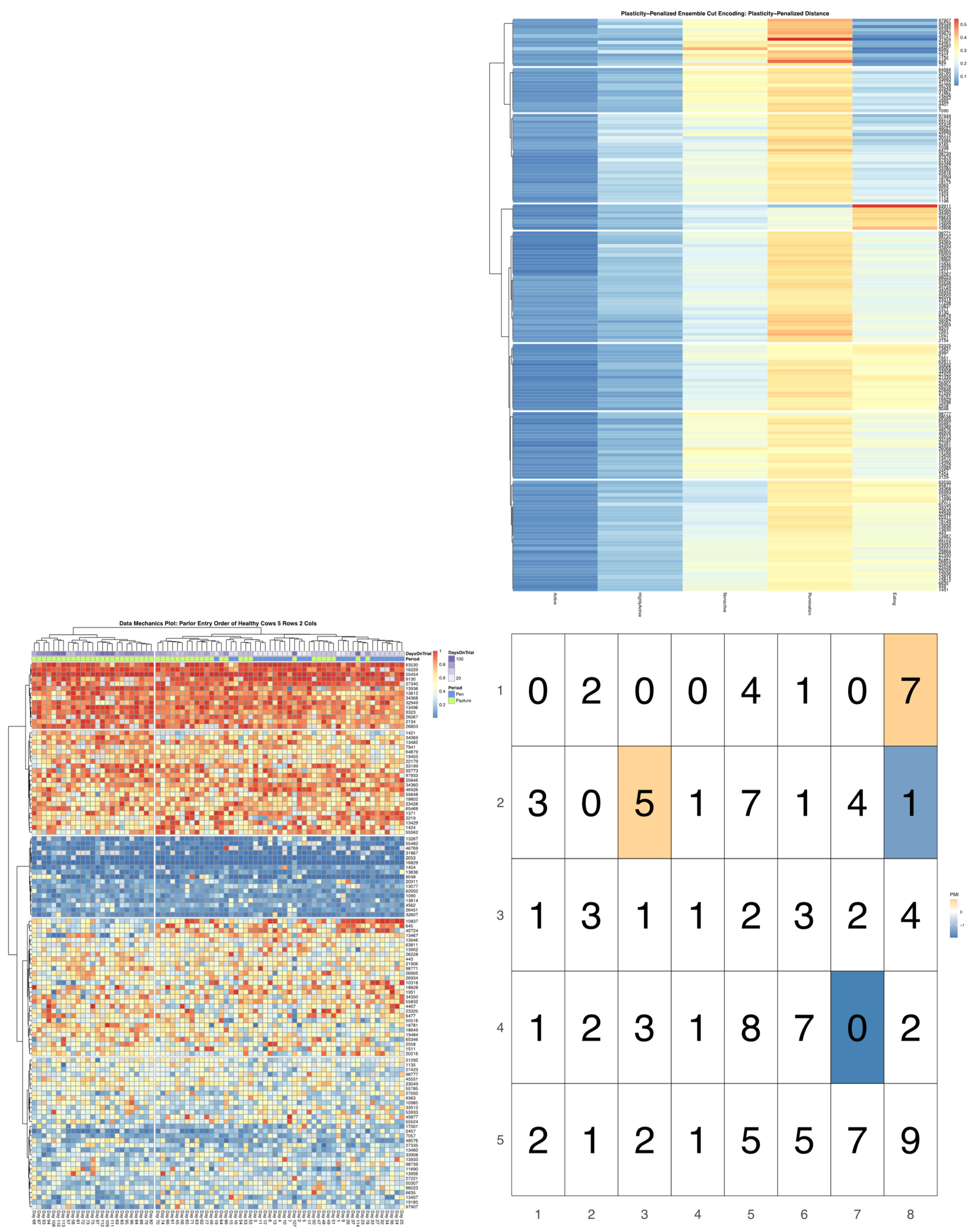
23]. A total of 80 milk order records—26 recorded while cows remained overnight in a free-stall barn, and 54 following the transition to overnight access to spring pasture—were used to create discrete encodings for parlor entry patterns via data mechanics clustering (see McVey et al. for further analytical details) [
7]. The dendrograms summarizing the distribution of cow entry-order patterns and subsequent heatmap visualizations will be subjected to further analysis, without modifications to the previously reported encodings.
Animals enrolled in this feed trail were also fitted with a CowManager ear tag accelerometer (Agis Automatisering BV, Harmelen, The Netherlands). This commercial sensor platform, while designed and optimized for disease and heat detection, also provides hourly time budget estimates for total time (min) engaged in five mutually exclusive discrete behaviors—eating, rumination, non-activity, activity, and high activity [
24,
25]. Time budget data was collected on all animals for a contiguous period of 65 days (1560 h). The observation window began on 17 February, shortly after trial enrollment was completed, and ended on 23 April, when the grazing season commenced and cows were moved overnight beyond the range of the receiver antennae. After eliminating the data of cows that were removed prematurely from the observation herd due to acute clinical illness, as well as several cows with persistent receiver failure, complete sensor records were available for 179 animals. In order to focus fully on the logistical challenges of encoding and characterizing the complex multivariate dynamics of this system, we have chosen to compress this data over the time axis to consider only the overall time budgets of these cows, and will leave explorations of the longitudinal and cyclical complexity of this dataset for future work.
2.2. Improving Empirical Encodings of Overall Time Budget through Simulation
Regardless of its original distribution, data can always be coarsened into a discrete variable [
26]. For complex or poorly defined systems, where appropriate cutoffs (binning rules) cannot be inferred
a priori, an empirically-determined encoding may provide a more flexible and comprehensive approach to discretizing the underlying behavioral signals. One algorithm that provides a model-free approach to pattern encoding within the larger cannon of UML tools is hierarchical clustering. This approach employs a bottom-up agglomerative strategy to group observational units into discrete clusters of variable sizes, progressively building a coherent picture of herd-level global structures from the similarities in behavioral patterns observed between pairs of individuals [
9,
10]. This series of progressive pairings can be expressed graphically in the form of a dendrogram, which serves as a 2D representation of the data’s geometric distribution in its higher dimensional measurement space, and can subsequently be used in data visualizations to highlight the most prominent structural features of a dataset [
19].
The efficacy of any hierarchical clustering scheme, however, is largely contingent on the adequacy of the estimator used to quantitatively express the pairwise dissimilarity between observational units [
10]. The Euclidean distance (L2 norm) is the default estimator used in most applications of this algorithm [
9,
10,
27], including much of the previous work in precision livestock applications [
13,
14,
18]. The L2 norm is appropriate for many measurement systems where variance is reasonably uniform across a continuous domain of support. Time budget data, however, is distributed multinomially, and as such has significant domain constraints [
26]. Put more simply, we know that the minutes logged for each behavior must sum to an hour. So, if a cow has ruminated for 60 min, then there can be no uncertainty in the remaining axes, because we know these values must be zero. These domain constraints impose co-dependencies between the behavioral axes that become stronger as observations shift towards the boundaries of the distribution’s support, which in turn warp the intrinsic variability of each axis contingent upon their location within the domain.
This statistical tedium also has some intuitive behavioral implications. Suppose we have two cows, Betty and Bessy, who spend 13 and 14 h a day ruminating, respectively. How “different” are these values? Since both cows are exceeding rumination rates needed to sustain a healthy metabolism, we would not anticipate that this difference would have a significant biological impact on these animals, and may ultimately be explained by relatively trivial behavioral fluctuations. Now, suppose instead that we have two other cows, Daisy and Delilah, who spend only 3 and 4 h a day ruminating, respectively. Given that both these cows are now well below the normal threshold for this behavior, this one-hour difference may have significant biological impacts. With a simple L2 norm, however, these two pairs would be given equivalent dissimilarity estimates for this behavioral axis, and so clearly a better estimator is needed.
Relative entropy, also referred to as the Kullback–Leibler divergence, is a classic information theoretic metric specifically designed to contrast discrete probability distributions, and thus a natural candidate for analysis of time budget data [
12]. For any two distributions that utilize the same alphabet of k = 1 … K categorical features (i.e.,—use the same ethogram), relative entropy can be calculated using Equation (1), and converted to a symmetric distance measure using Equation (2). By utilizing the proportion of time that an animal invests in each behavior as both a nominal and relative value, this estimator is able to adjust the relative difference between cows by the absolute position of each observation relative to the boundary of the domain.
Domain constraints are not, however, the only stochastic feature that need be accommodated when working with time budget data. There is also the measurement error attributable to the sensor itself. Returning to the previous example, suppose that we also know that our rumination records are only accurate to ±1 h. Is it then still appropriate to give more weight to the one-hour difference between Daisy and Delilah, than between Betty and Betsy? Since both observations are within the bounds of error, attempting to enhance the underlying biological signal may only succeed in amplifying measurement noise. A closed-form estimator, however, may not be readily generalizable to the wide range of measurement error models encountered with PLF sensors. We therefore propose that a simulation-based approach may offer a more flexible means of accounting for measurement errors in dissimilarity estimates [
11].
The LIT package provides a built-in simulation utility for time budget data that seeks to mimic the stochastic error structure of the original data while still preserving the underlying behavioral signal [
11]. Data is provided as a tensor, with cow indexed on the first axis, time indexed on the second, and the component behaviors on the final axis. The count data at each cow-by-time index is then used to redraw a simulated datapoint from one of three optional distributions [
26]. In the first, the user may sample directly from a multinomial distribution centered around the normalized observed count vector. This model assumes that measurement error should shrink as a cow dedicates larger proportions of an observation window to specific behaviors, and intrinsically prevents estimates from being generated outside the domain of support. Variance can be underestimated at the extremes of the domain, however, if the probability for a behavior is non-negligible, but the observed count is zero due to under-sampling. This issue may be addressed in sampling option two, where samples are redrawn from a multivariate beta distribution (MBD), also known as a Dirichlet distribution, again parameterized using the normalized observed count. While this sampling strategy slightly biases the simulation towards the center of the distribution, it prevents under-sampling at the extremes of the domain. Finally, users may combine these sampling strategies in sampling option three, wherein the probability vector used to parameterize the multinomial is drawn first from the Dirichlet, in order to further increase the uncertainty in the simulated data. After simulation has been completed by redrawing samples at the finest level of temporal granularity supported by the sensor, the data can then be conditionally or fully aggregated along temporal axis as required for downstream analysis as a time budget.
This simulation routine was used to create an ensemble of B = 500 simulated overall time budget matrices that mimicked the stochasticity attributable to a reasonable approximation of the measurement error of the sensor. Stored as a tensor with replication on the last axis, the variance of the ensemble of simulations could then be easily calculated for each combination of cow index and behavioral axis. If the underlying simulation strategy is a reasonable representation of the noise in the sensor, then these variance terms will then serve as a sufficient approximation of the relative uncertainty in each data point. We propose that that this information can then be incorporated into the calculation of dissimilarity estimates by serving as penalty terms in the calculation of an ensemble-weighted distance estimator defined in Equation (3).
The rescaling strategy employed in our proposed dissimilarity estimator is strongly inspired by traditional analysis of variance (ANOVA) techniques, thereby providing several insights into its anticipated behavior. First, because the simulations were generated using the multinomial or one of its analogs, we can infer that these penalty terms will not be homogenous across the domain of support, but should shrink as observations approach the boundary. This will allow the ensemble-weighted distance estimator to emulate the rescaling dynamic achieved with the KL distance; however, rescaling at the extremes of the domain will ultimately be bounded by our simulated measurement error, so as not to exceed the precision of the sensor. Second, because we have here emulated measurement error in our simulation using sampling uncertainty, the central limit theorem will apply [
9]. Thus, we can anticipate that as the number of observations per animal increases, the impact of measurement error on our inferences will shrink, allowing progressively more subtle differences between animals to come into resolution. Taking this property to its limit, however, can it be said that with enough observation minutes the differences between cows can be inferred with near certainty? That intuition, of course, is at odds with our characterization of a dairy herd as a complex system, and highlights an additional stochastic element that must be accommodated—the behavioral plasticity of the cows themselves in response to changes in the production environment [
4].
Given the extended observation window of this particular data set, it would be possible to recalculate time budget conditional on the day of observation, and then use the variance in daily time budget along each behavioral axis as a penalty term. Such estimates would collectively reflect heterogeneity in variance attributed to domain constraints, measurement error, and behavioral plasticity. Such an approach would not, however, be feasible for datasets collected over shorter time intervals with fewer replications, or in applications with behavioral responses where there is no clear hierarchy in the temporal structure of the same. We therefore propose that our stochastic simulation model can be extended to also provide a generalizable means to approximate the uncertainty of the underlying behavioral signal.
As before, the measurement error was simulated by redrawing samples at the finest temporal granularity provided by the sensor. Prior to compression along the temporal axis, however, a random subsample of observations days was selected across all cows, and only these values were used to calculate the simulated overall time budget. If all cows demonstrated comparable levels of consistency in their daily time budgets, then reducing the effective sample size of our simulated data sets through a subsampling routine would increase the ensemble variance estimates. This, in turn, would make our approximation of measurement error hyper-conservative, but this increase would be uniform across all cows. If, instead, some cows were less consistent in their time budgets across days, then the sampling error imposed by the subsampling routine would be greater, resulting in a larger ensemble variance estimate. Thus, we would expect a stronger penalty to be applied to cows who demonstrated greater plasticity in their behavioral responses to both transient and persistent changes in the production environment. For small datasets with a limited number of replications, the number of subsamples could be set quite close to the size of the complete sample, and would thus emulate a jackknife approach to variance estimation [
9,
10,
28]. For larger datasets, however, the subsample size could be set smaller, to make the resulting ensemble variance estimates progressively more sensitive to the uncertainty in the underlying behavioral signal.
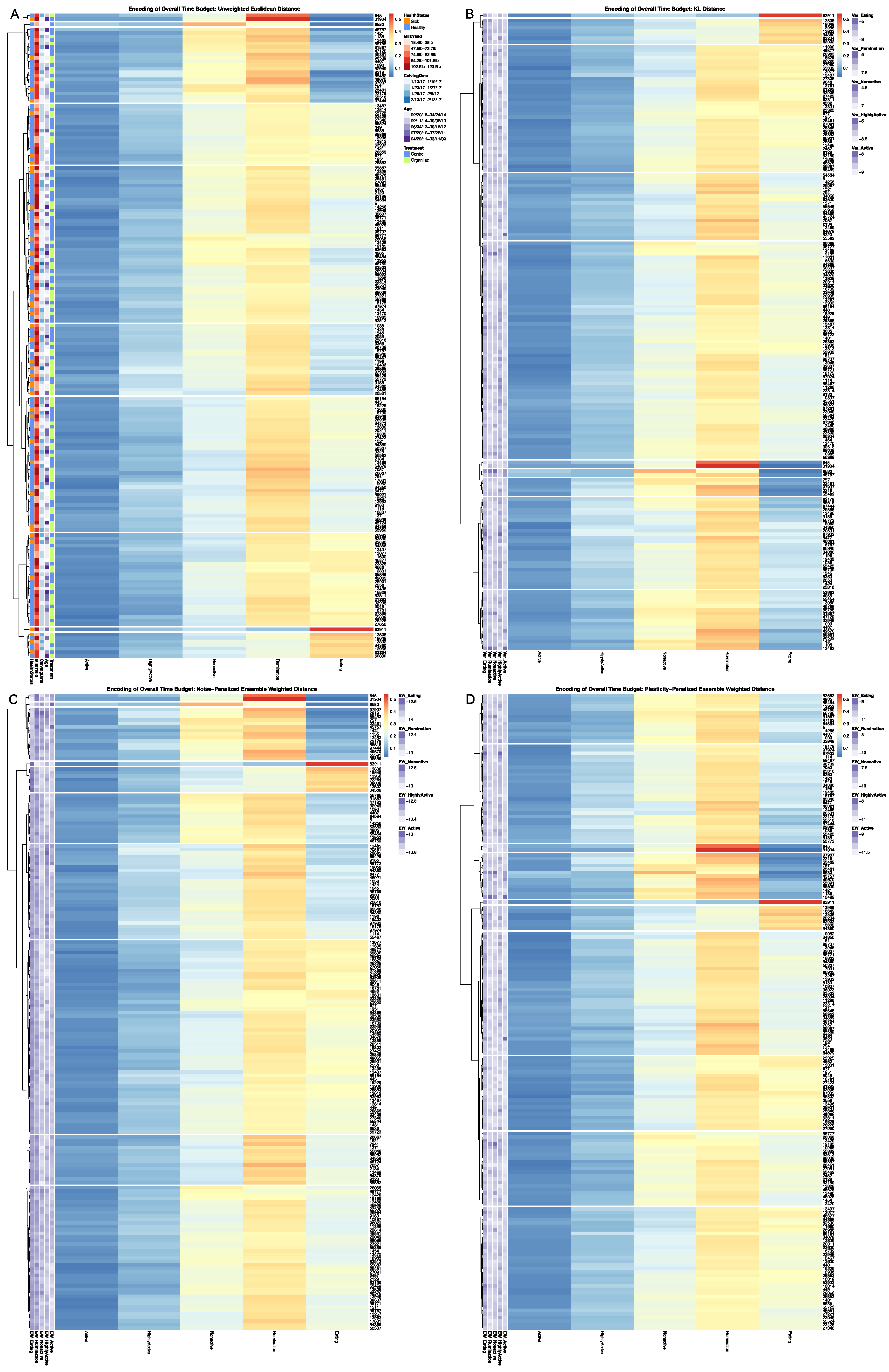
To evaluate the empirical performance of these dissimilarity estimators, distance matrices were calculated for the 177 cows with complete CowManager time budget records. Euclidean distance and KL Distance were calculated using base R utilities, with speed up options utilizing the
Rfast package [
23,
29]. An ensemble-weighted dissimilarity matrix was first calculated using simulated values accounting only for measurement error using the most conservative joint Dirichlet-multinomial sampling scheme, hereafter referred to as noise-penalized distance. A second ensemble-weighted dissimilarity matrix was then calculated using the same sampling scheme for measurement noise but aggregated over a 14-day subsample to account for behavioral plasticity in daily time budgets, hereafter referred to as plasticity-penalized distance. The LIT package provides users a clustering visualization utility, which converts dissimilarity matrices into a dendrogram using the
hclust utility in base R with default Ward D2 linkage [
23], and the generates heatmap visualizations of the resulting clustering results using the
pheatmap package [
30]. Heatmaps were generated on a grid of cluster values from k = 1 … 10 for each of the four dissimilarity estimators, with complete results provided in
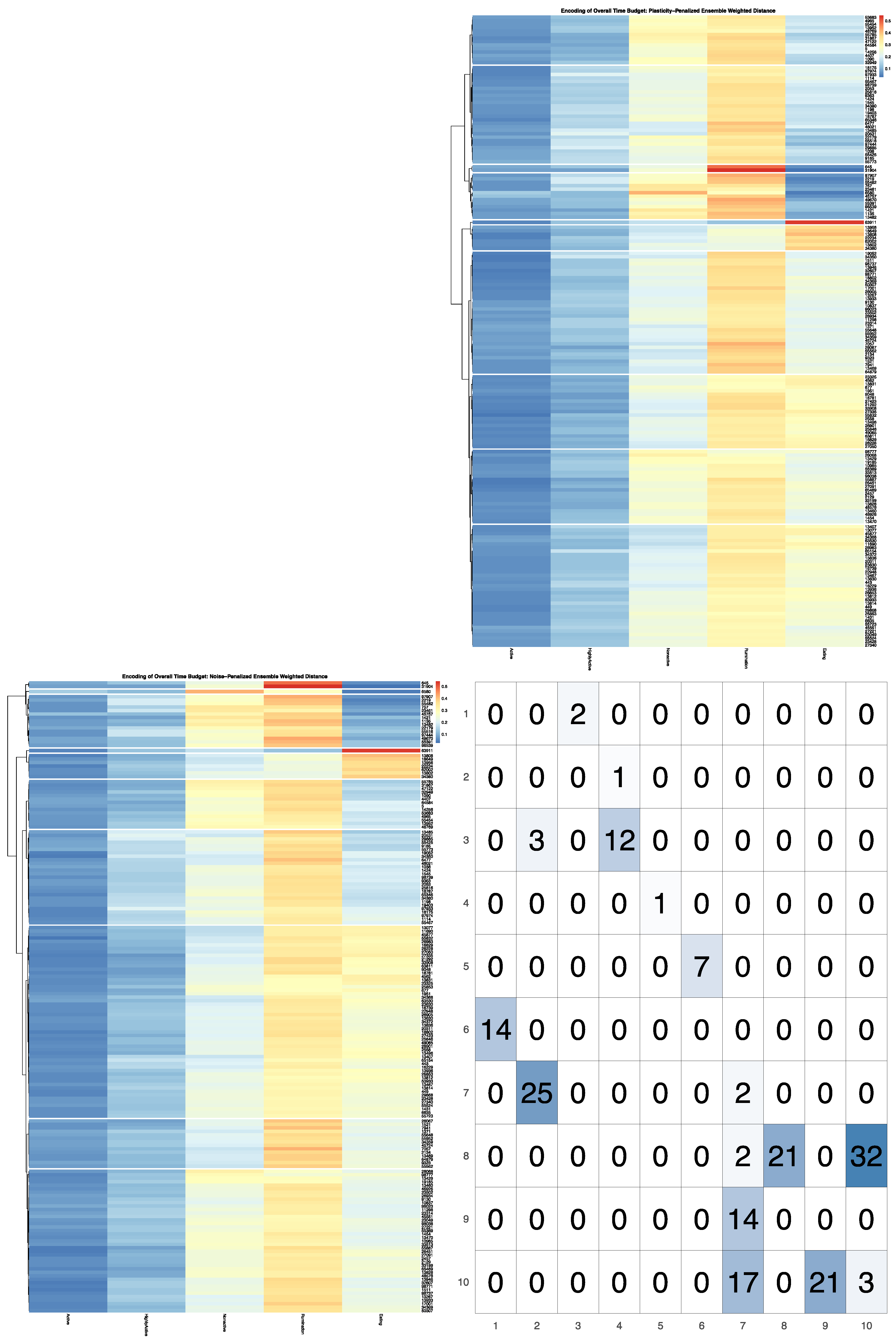
Supplementary Materials, the results for k = 10 clusters are provided. The LIT package also provides users with a plotting utility to visually contrast the broader patterns between behavioral encodings. Outputs from the clustering utility are passed in to create a contingency matrix generated using
ggplot2 with cells colored by their corresponding cell count [
31]. The heatmap visualizations for each encoding are then added to the row and column margins of the contingency matrix using the
ggpubr package [
32], and arranged such that each row cluster in either heatmap matches the order of the contingency matrix reading either up-down or left-to-right, allowing for direct and detailed visual comparison of the discretized behavioral patterns. Comparisons between the noise-penalized and plasticity-penalized encodings are provided.
2.3. Improving Tree Pruning Decisions through Simulation
An optimal encoding strategy seeks to minimize the loss of relevant information by retaining as much of the underlying deterministic signal as possible, while hemorrhaging only noise [
26]. In a hierarchical clustering framework, this is achieved by pruning the dendrogram built from the dissimilarity matrix at the point where the branches cease to represent differences in the underlying signal. Standard pruning strategies allow users to either: (1) provide a dissimilarity cutoff, below which value all further branches are grouped into the same bin, or (2) extract the first K branches of the tree [
9,
10]. As with the default Euclidean distance dissimilarity estimator, this approach may be appropriate for datasets with relatively homogenous variance structures. For data drawn from intrinsically heterogenous distributions, however, the branch lengths cannot be directly compared across the domain of support, making globally-defined pruning rules a suboptimal strategy for analysis of time budget data.
More fundamentally, a homogenous pruning strategy may be too simplistic for many PLF sensor datasets, for which the underlying signal often represents a complex composite of behavioral mechanisms that operate at multiple scales. Although some environmental factors might be expected to have an impact on cattle behaviors that are uniform across the herd, other factors might elicit responses that differ in magnitude for different subgroups within the larger population, or even become isolated within smaller social cliques. For example, we might expect the number of times cows are moved each day for milking will place similar constraints on the time left to lie down across all animals, but overstocking with respect to stall spaces might have a much larger magnitude of impact on the lying patterns of subordinate heifers than the more dominant older cows [
33]. In such a complex system, we would expect the heterogeneity imposed by the underlying biological signal to differ in scale across the dataset. Subsequently, in attempting to employ a global cutoff decision to encode information for such a dataset, we would always be faced with the difficult decision to either ignore the subtler behavioral patterns present in some branches of the tree, or else allow noise to contaminate our encoding of other branches with intrinsically coarser behavioral patterns.
Although all the components that contribute to the signal in a complex livestock system might be difficult to anticipate a priori, we propose that a more dynamic pruning algorithm might still be achieved, by again employing flexible simulation-based approaches to emulate the comparably simpler sources of uncertainty. If each branch of the dendrogram is viewed as a pairwise contrast between two groups of animals, then we need only to determine whether the bifurcation under inspection represents a difference in the underlying signal that can be reliably distinguished from noise. If it can, then the two groups should be split in the final encoding to capture this feature of the data’s distribution. If a branch falls below the intrinsic resolution of the data, however, then the branch may be pruned so that all animals are placed into the same cluster, with no loss of meaningful information. By implementing such a branch-level test recursively, we can gradually work our way down the tree with adaptive locally-defined pruning decisions.
To evaluate the reliability of the behavioral signal encoded at each bifurcation of the tree [
34], our branch test utility utilizes two mimicries. The first set of simulations are generated under the alternative hypothesis that assumes a branch contains an underlying deterministic signal that is only partially obscured by stochastic noise. Thus, we can simply repurpose the ensemble of simulated data sets used previously to calculate the ensemble weighted dissimilarity metrics by mimicking the uncertainty in the observed data. The second set of simulations are generated under the null hypothesis that a given branch contains only noise. As the null implies that animals demonstrate equivalent patterns of behavior within the resolution of the sample, this mimicry can be generated quite efficiently using a standard bootstrapping routine [
28], wherein time budgets simulated under the alternative are unconditionally resampled from amongst all animals in a given branch. HClustering is then performed independently on each data mimicry in either ensemble, and the first k branches are extracted to create an ensemble of discrete encodings.
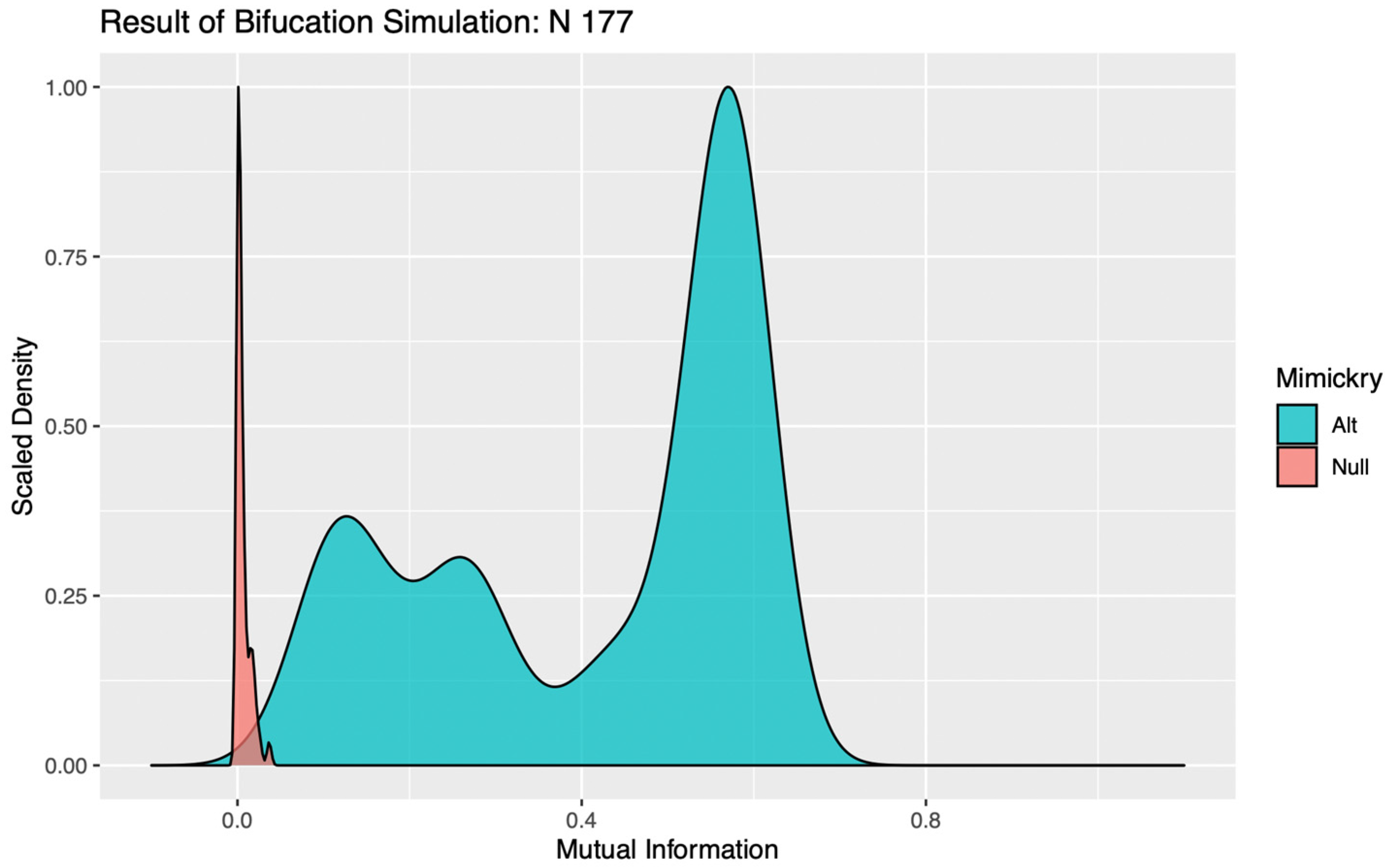
Under the alternative hypothesis, a strong signal should produce a robust tree structure such that, even after the addition of simulated noise, the resulting encoding would still closely mirror that of the original observed data. As the stochastic component of a dataset becomes stronger relative to the signal, these bifurcation points will become progressively less stable, and the subsequent encodings less reliably aligned with the original data. When the signal falls below the resolution of the data, the tree structures of the simulated data would then seldom match that of the original data, and so would become poorly distinguished from encodings generated under the null, with no signal component. We propose that mutual information, which can be calculated without any additional distributional assumptions, can be used to quantify the similarity between the observed data and each mimicked dataset, and subsequently used to determine if simulations under the alternative are distinguishable from the null [
12]. In our study, a bifurcation was determined to be significant if less than 5% of the MI values calculated for data simulated under the alternative hypothesis fell below the 95th quantile of MI values calculated for data simulated under the null. If a bifurcation was instead deemed insignificant, the branch was pruned and all cows within it assigned to the same cluster in subsequent encodings.
In evaluating the significance of a bifurcation, it seems intuitive that a k = 2 binary encoding should be utilized. For complex systems subject to the influence of multiple competing drivers of behavioral responses, however, a false negative result can occur with this parameterization if the addition of stochastic noise perturbs the order in which two significant mechanisms with similar magnitudes of impact are bifurcated. Such trivial destabilizations of the tree structures can be readily identified in visualizations of the distributions of MI values calculated against simulations under the alternative, as the “flip flopping” between bifurcation points produces clear evidence of multimodality (see
Figure 1). To circumvent this issue, the LIT package provides users the option to re-test any bifurcations deemed insignificant, using a binary encoding with a more granular discretization (k > 2). This effectively allows the algorithm to “look down the branch” to absorb any irrelevant flip-flopping between competing signals, thereby preventing spurious over-pruning that would hemorrhage information on significant behavioral patterns from the final encoding.
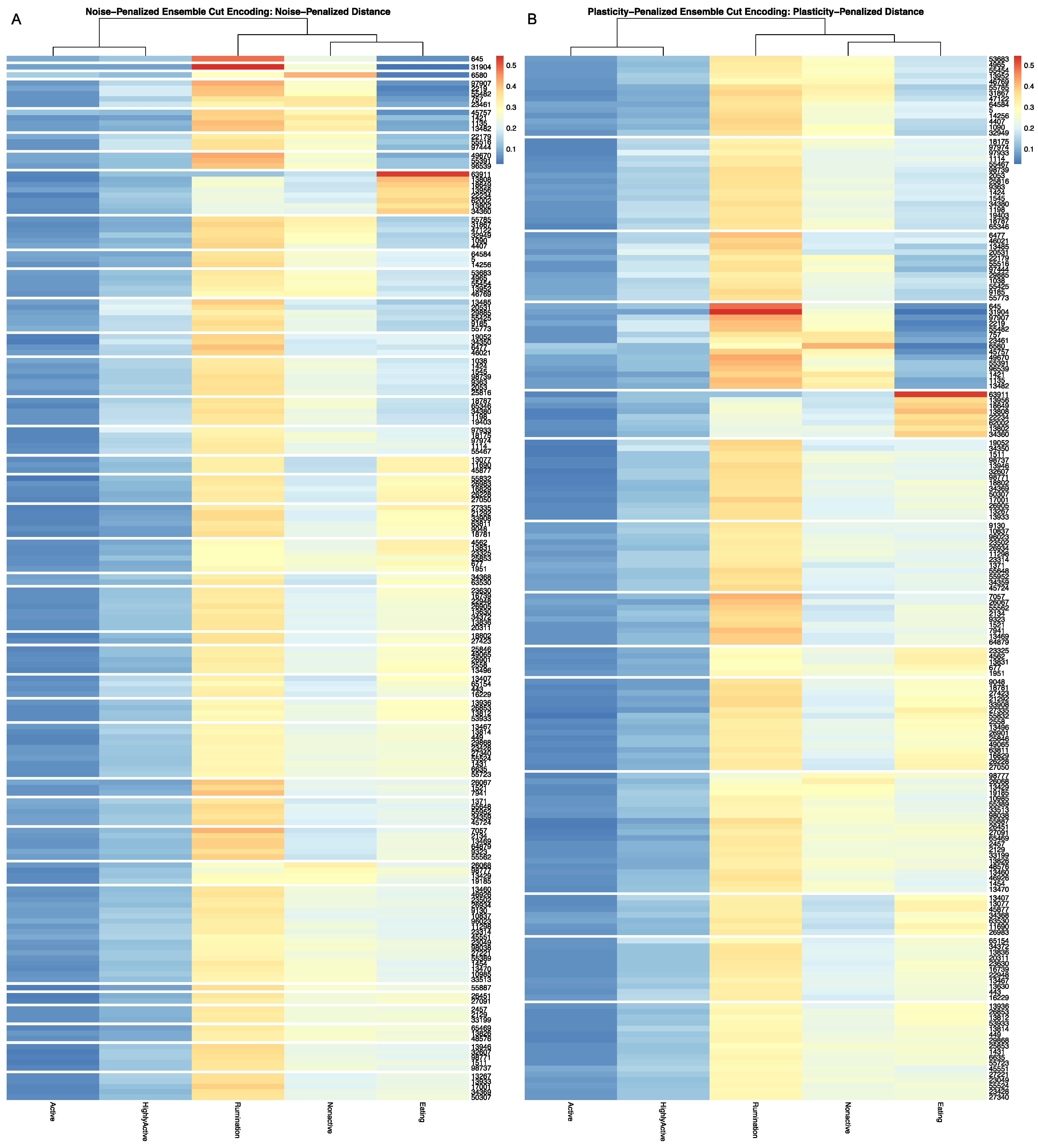
Full results for the application of our ensemble-cut algorithm to dendrograms generated using each of the four dissimilarity estimators discussed in the previous section, and using both the noise-penalized and plasticity-penalized encodings, are provided in the
Supplementary Materials. A summary of results for the application of the ensemble-cut algorithm applied to dendrograms generated from the noise- and plasticity-penalized ensemble-weighted distance metrics, the noise-penalized and plasticity-penalized encodings, respectively, are provided.
2.4. An Information Theoretic Framework for Cross-Sensor Inferences
Equipped with an appropriate encoding to discretely represent the heterogeneity in overall time budgets within this herd, and provided with the encoding of longitudinal patterns in parlor entry position from previous work with this data set, a potential question to ask would be: how does a cow’s time budget, which is largely determined by her behaviors in the home pen, relate to her behavior in the milking queue? There are a number of nonparametric and parametric techniques available to evaluate the overall strength of association between two discrete variables, by evaluating the distribution of animals in the joint encoding [
26]. There is, however, perhaps greater practical utility in characterizing low and high points within the joint encodings, which would provide more detailed insights into the tradeoffs between specific behavioral patterns recovered from the data streams in these distinct farm contexts. Towards this end, information theory offers a more comprehensive approach to decomposing the stochasticity within discretely encoded variables, and thus may provide a more holistic approach to evaluating both the global and local features of a joint encoding, while employing few structural assumptions [
12].
First, to evaluate the strength of the overall relationship between two discretized behavioral responses, the LIT package provides users a permutation-based bivariate testing utility that uses the mutual information estimator to quantify the amount of information entropy that is redundant between the two encodings [
7,
12]. We can anticipate, however, that the efficacy of this test in recovering significant relationships between the underlying biological signals will be affected by the resolutions of the encodings. Suppose that a single latent biological factor impacts the behavioral responses collected by both PLF data streams, creating informational redundancy between the two encodings. If we cut the trees above the intrinsic magnitude of its impact on a given behavior, its influence may be overlooked and mutual information underestimated. On the other hand, if we prune the tree far below the magnitude of its impact, our inferences can lose power, as bin sizes in the joint encoding become progressively smaller, weakening the empirical estimation of the joint probability distribution and thereby increasing estimation error in the MI estimator. The resolution of our encodings must, therefore, be optimized to match the dynamics of the system, or a false negative result may be returned. To further complicate matters, however, we cannot necessarily assume that the magnitude of impact of a given latent factor will be uniform across behaviors, nor should we expect in a complex farm environment that behaviors will be influenced by a single latent factor.
To overcome this logistical challenge without falling back on dubious
a priori assumptions, the LIT package implements mutual information-based permutation tests on a grid, varying the cluster resolutions across both behavioral axes [
7]. Under the null hypothesis that no significant bivariate relationship exists between data streams, cow ID labels are randomly permuted within each tree, preserving the marginal distribution of the data along each axis, but destroying any latent bivariate relationships. These permuted trees are then cut, and the mutual information of the joint encoding estimated for each combination of cluster counts on the grid. A p-value is then generated by comparing the observed MI value of the joint encoding at each grid point against the corresponding distribution of MI values simulated under the null. Just as a scientist varies the focus of a microscope to bring microbes of different size into resolution, we can expect that geometric features of the joint probability distribution imposed by latent deterministic variables, that vary in scale of impact, will come into and fall out resolution as these meta-parameters are varied across the grid of cluster counts. To help the user visually identify where such features have come into resolution, the LIT package also returns a heatmap visualization of the observed MI value for each grid point that is centered and scaled, relative to the distribution of MI values under the null. For behavioral measurements subject to the influence of multiple biological and environmental factors operating simultaneously, this exhaustive approach to parameterization enables users not only to build a more complete picture of a complex behavioral system, but may also provide insight into the hierarchy of these behavioral responses.
Unfortunately, as the resolution of the encodings is increased, MI estimates not only become less precise, but they may also become less accurate. Bias is introduced when empirical estimates of the joint probability distribution become so granular (i.e.,a high number of bins relative to the total sample size) that regions with low but nonzero probabilities go unsampled. These zero-count bins cause the total entropy calculated from the empirical joint probability distribution to be underestimated which, in turn, causes the relative amount of redundant information to be overestimated. Although the magnitude of this bias is partially dependent on the total sample size, it is also contingent on the structure of the joint probability distribution itself, namely the number of low-probability cells. Given that the joint probability distribution under the null, which is randomly permuted to intentionally remove any nonrandom features in the sample, can be expected to have a more uniform distribution of probability than the observed dataset, we can anticipate that the magnitude of the bias may differ between these two distributions as the sample becomes more granular, preventing MI estimates from being directly comparable. To overcome this issue, the LIT package by default provides entropy estimators based on the Maximum Likelihood frequency estimates, but allows users to select from a range of bias-corrected frequency estimates available in the
entropy package [
35]. Based on the simulation work by Hausser and Strimmer (2009), the JS “shrink” estimator was used in our study to conduct bias-corrected mutual information permutation tests [
36].
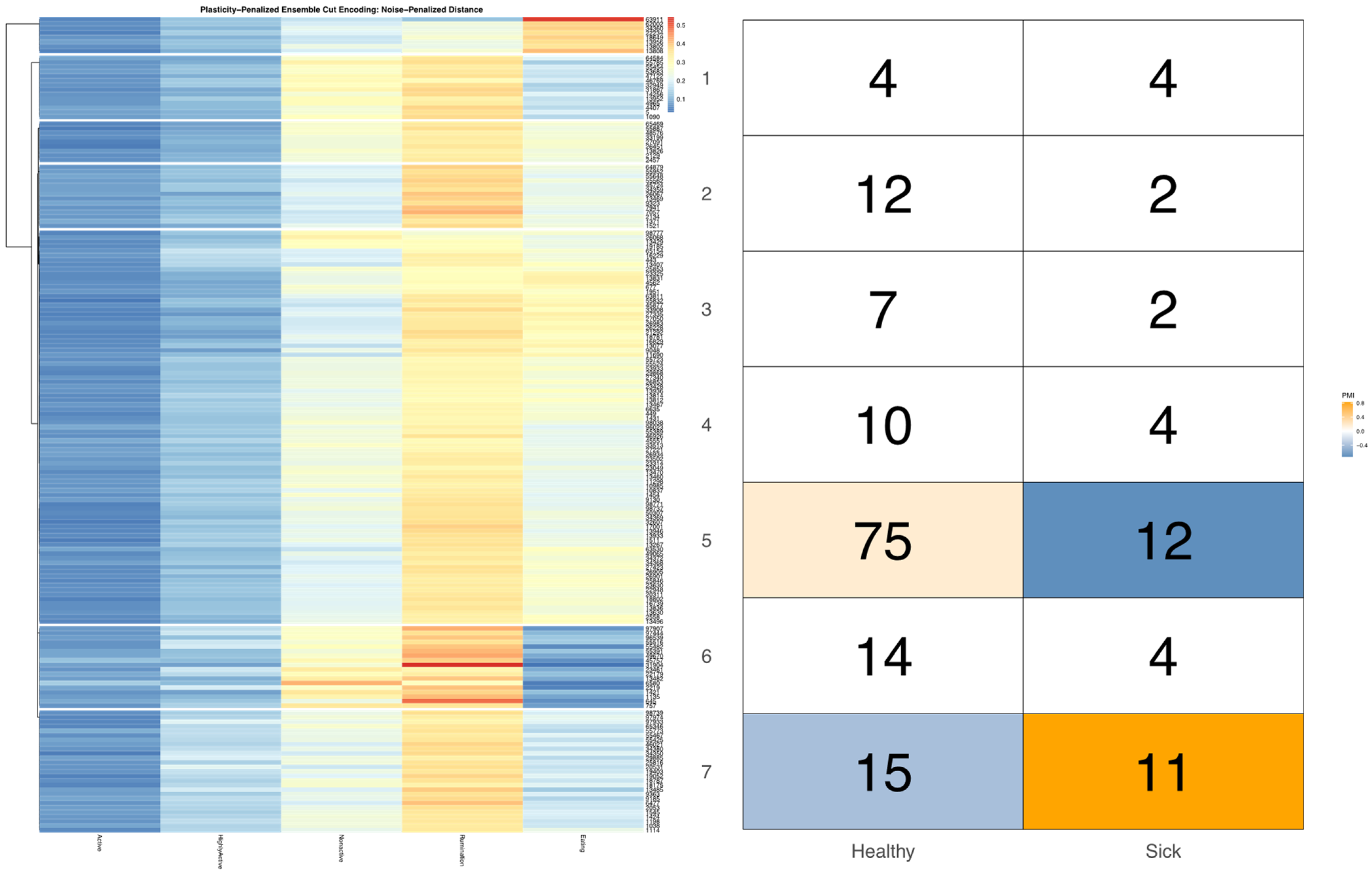
Not only can the impact of latent factors on behavioral measures differ in magnitude, we can also anticipate that responses may differ in both strength and direction for different subgroups within the herd. Such nonlinear dynamics are easily captured in a model-free MI test, but further inspection of the contingency table is needed to fully characterize such complex bivariate relationships between sensor outputs. If either marginal encoding has roughly the same number of observations in each bin, then the cell counts in the joint contingency table can be directly compared, as under the null we would expect each cells to be equiprobable. For empirically defined encodings, however, bin sizes can vary significantly to better capture the underlying geometry of the univariate data distribution. Such differences in marginal probabilities prevent the raw cell counts from being directly compared. To better identify which cells in an empirically defined joint encoding are driving a significant overall relationship between two data streams, mutual information can be decomposed into pointwise mutual information (PMI) values [
37]. The LIT package provides users the option in the
compareEncodings plotting utility to color cells in the joint contingency table by PMI estimate, to better facilitate direct visual comparisons of the encodings. To further enhance visualizations of the joint probability distribution that significantly differs from expected cell counts under the null, users may also specify a probability threshold above which PMI values should not be displayed, which was determined here by simulating PMI estimates under the null by redrawing from a multinomial distribution using the outer product of the marginal distributions.
Bivariate tree tests were applied to the time budget encodings, using both the noise- and plasticity-penalized dissimilarity metrics, and pruned using the more conservative plasticity-penalized mimicry, against the encoding of parlor entry order data produced using data mechanics clustering from our previous work [
7]. A 2:10 × 2:10 grid was used to determine the optimal resolution for the bivariate relationship, with the optimal meta-parameters used to create visualizations of the joint encoding, wherein pointwise mutual information values were used to color cell counts that were significant at the alpha = 0.05 significance level. To further explore latent factors that might explain significant associations between entry position and time budgets, bivariate tree tests and pointwise mutual information tests were also applied separately to the encodings of both PLF data streams and health records.