Breast Mass Detection in Mammography Based on Image Template Matching and CNN
Abstract
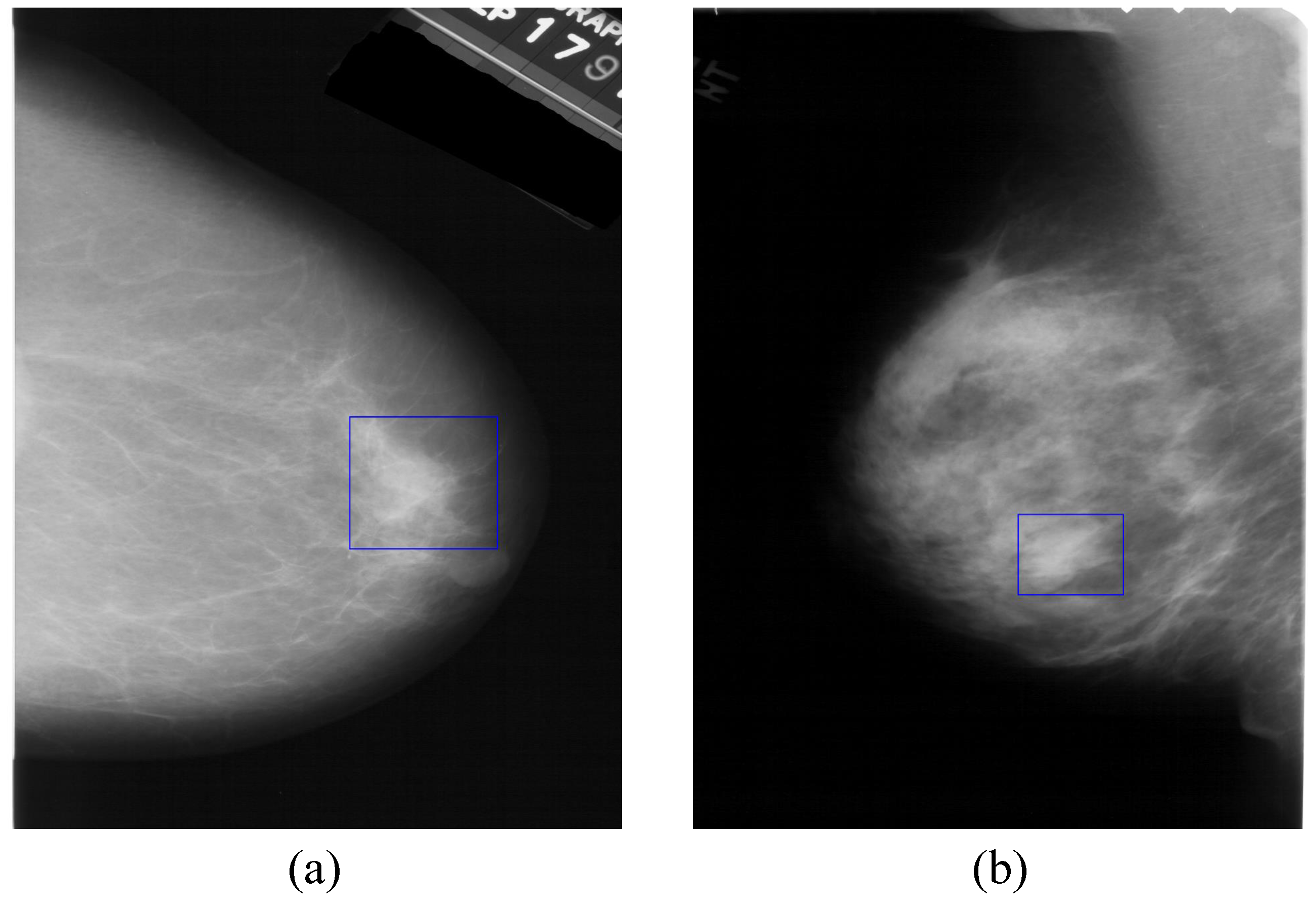
1. Introduction
2. Related Work
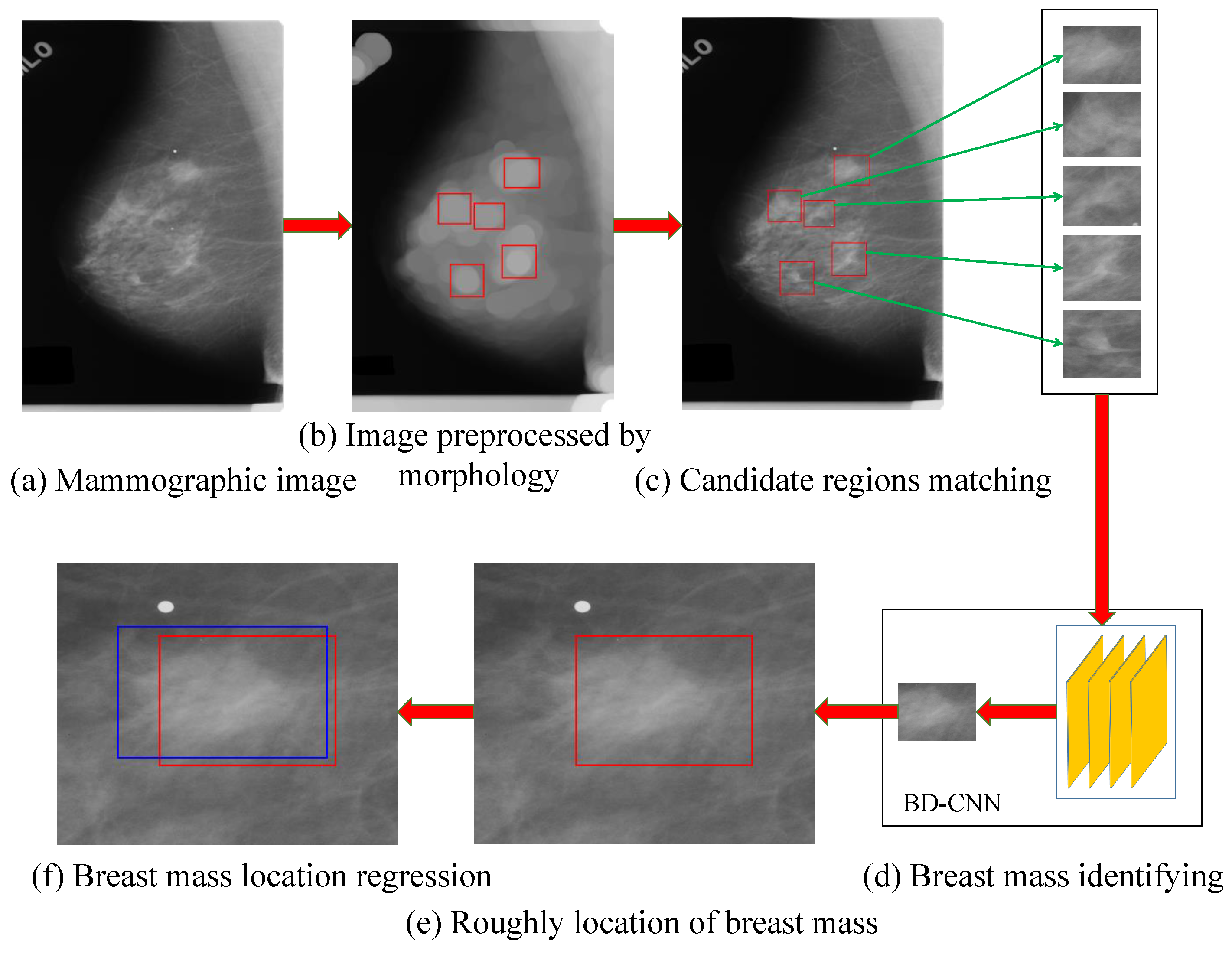
3. The Proposed Method
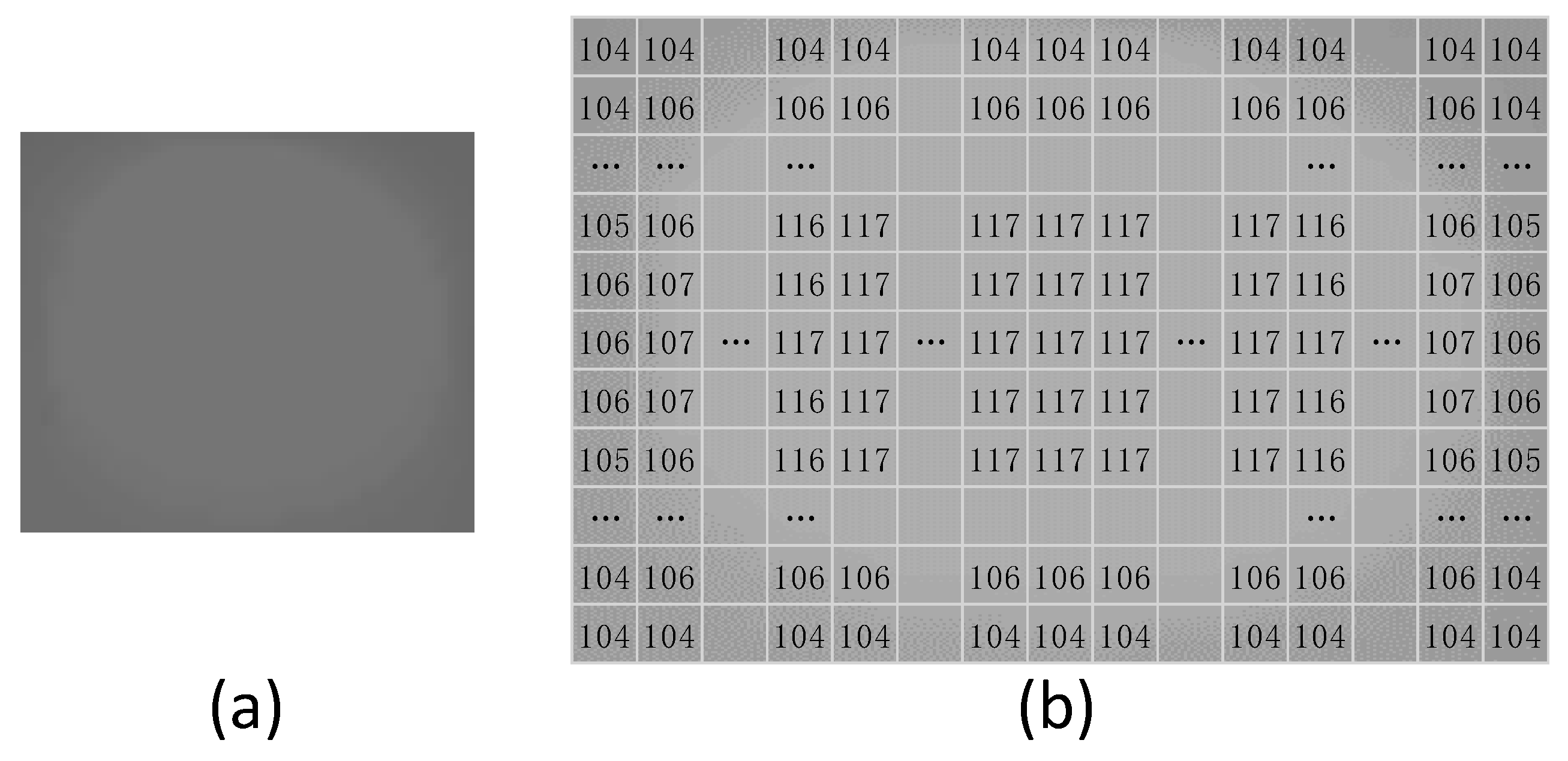
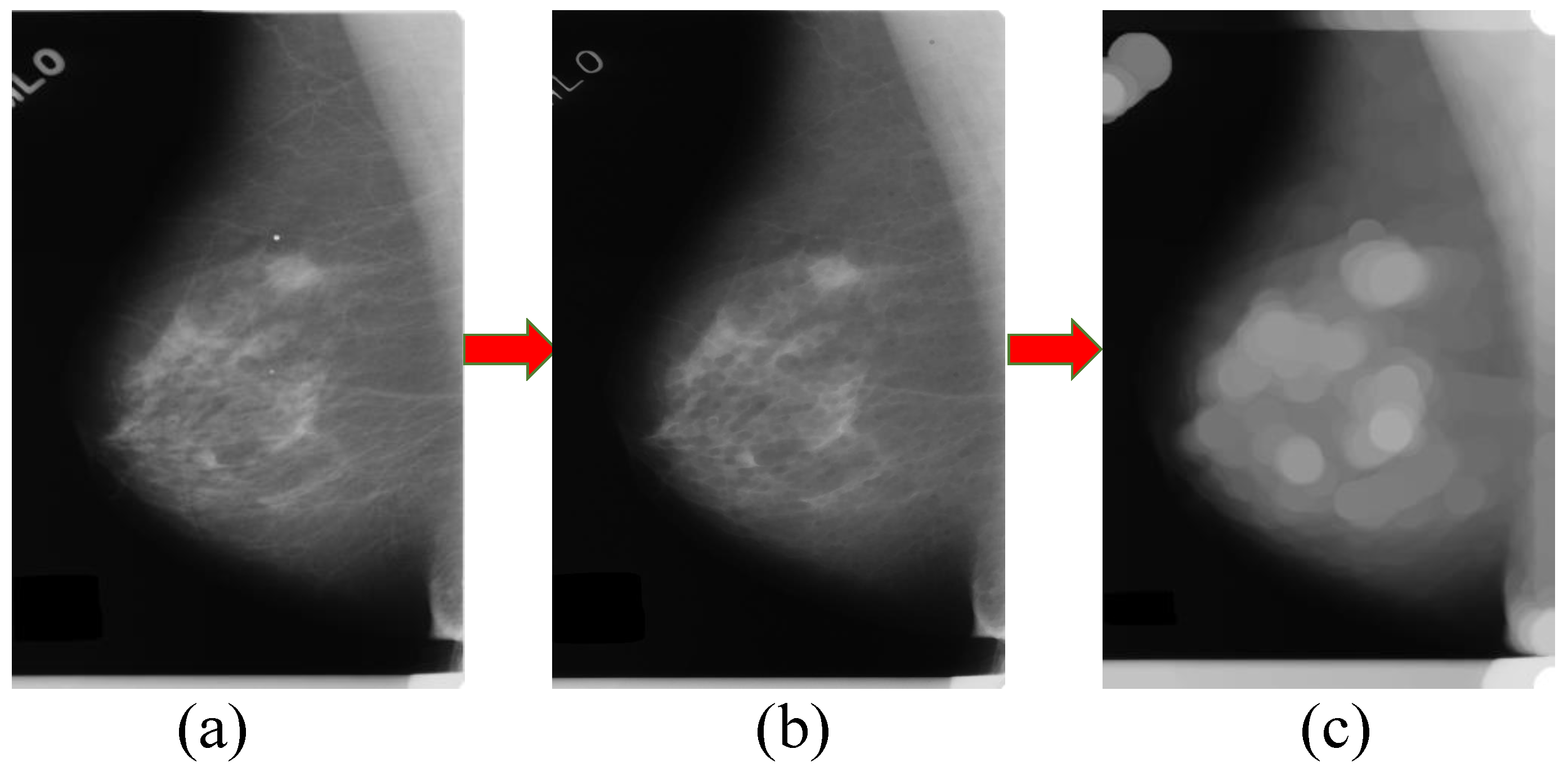
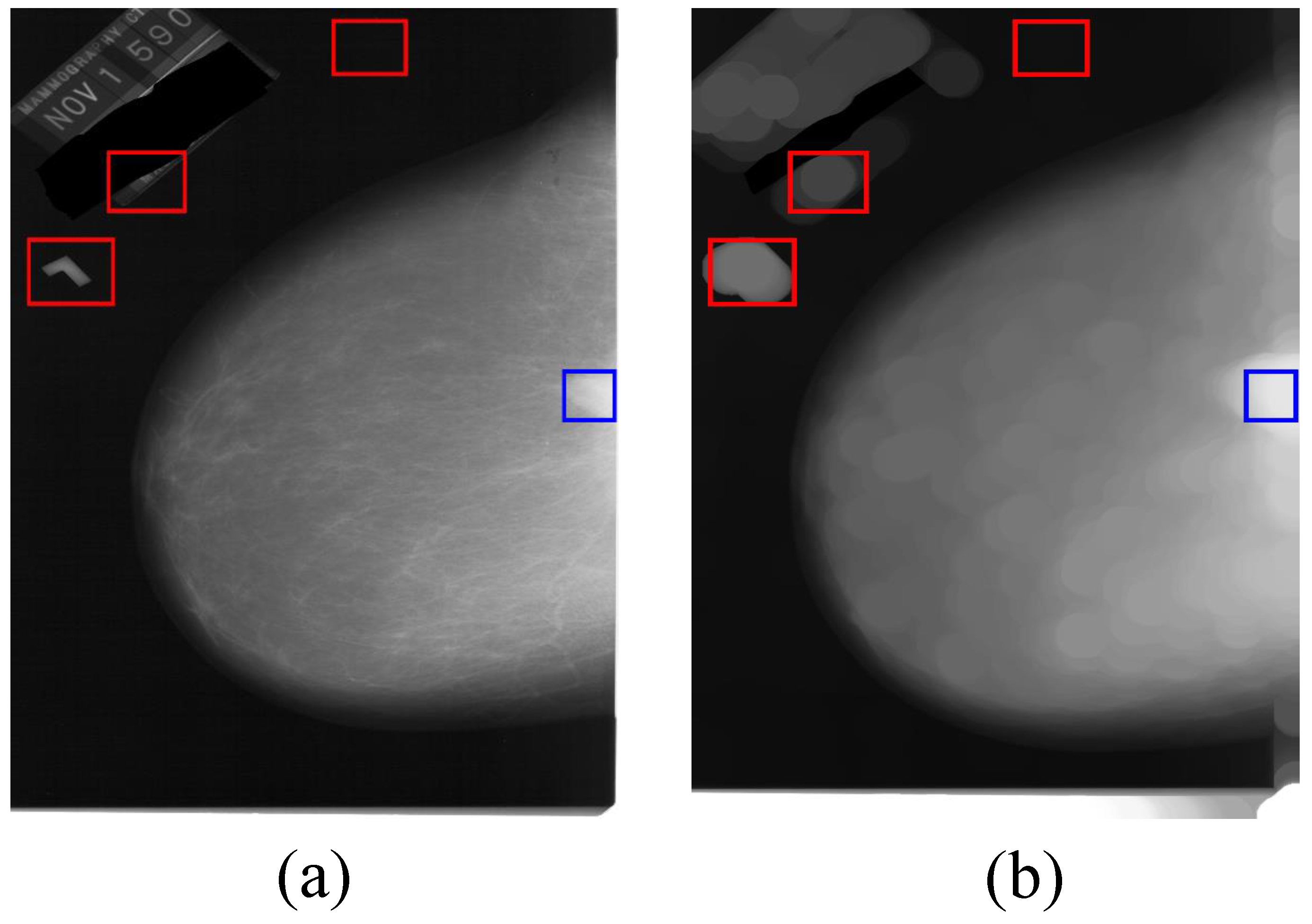
3.1. Processing of Mammographic Images by the Mathematical Morphology Method
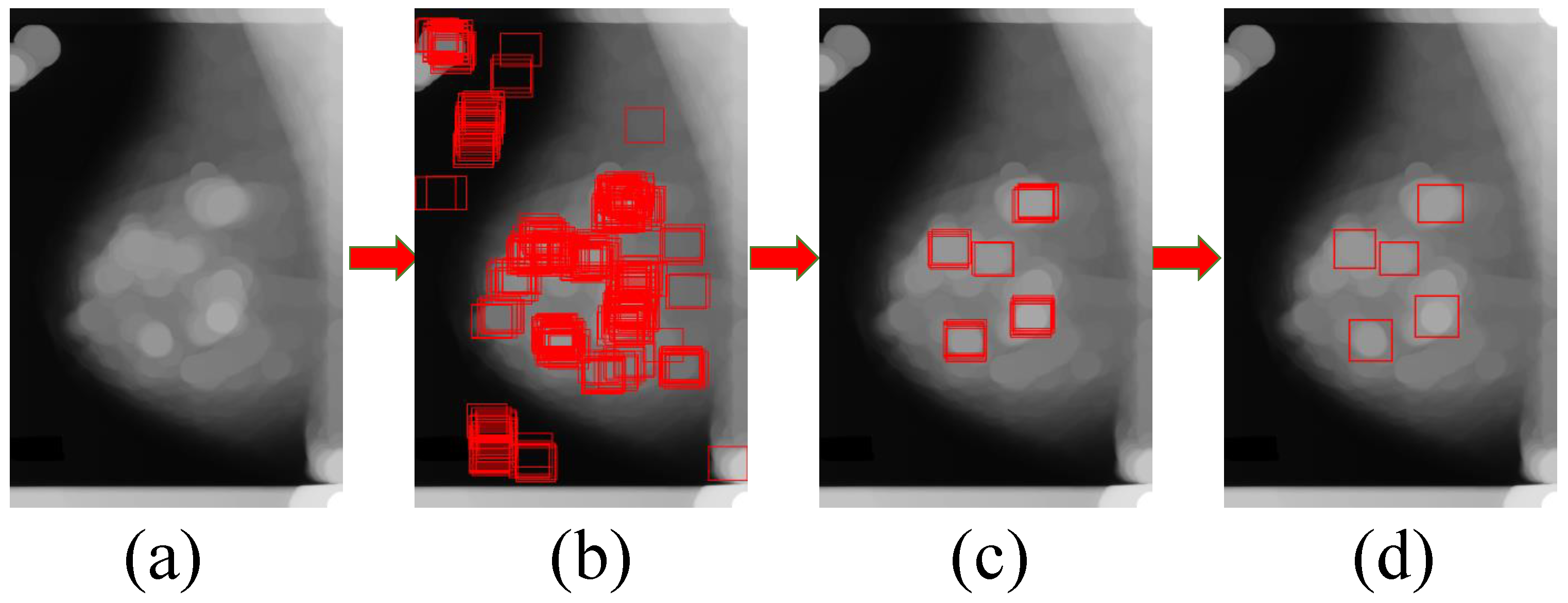
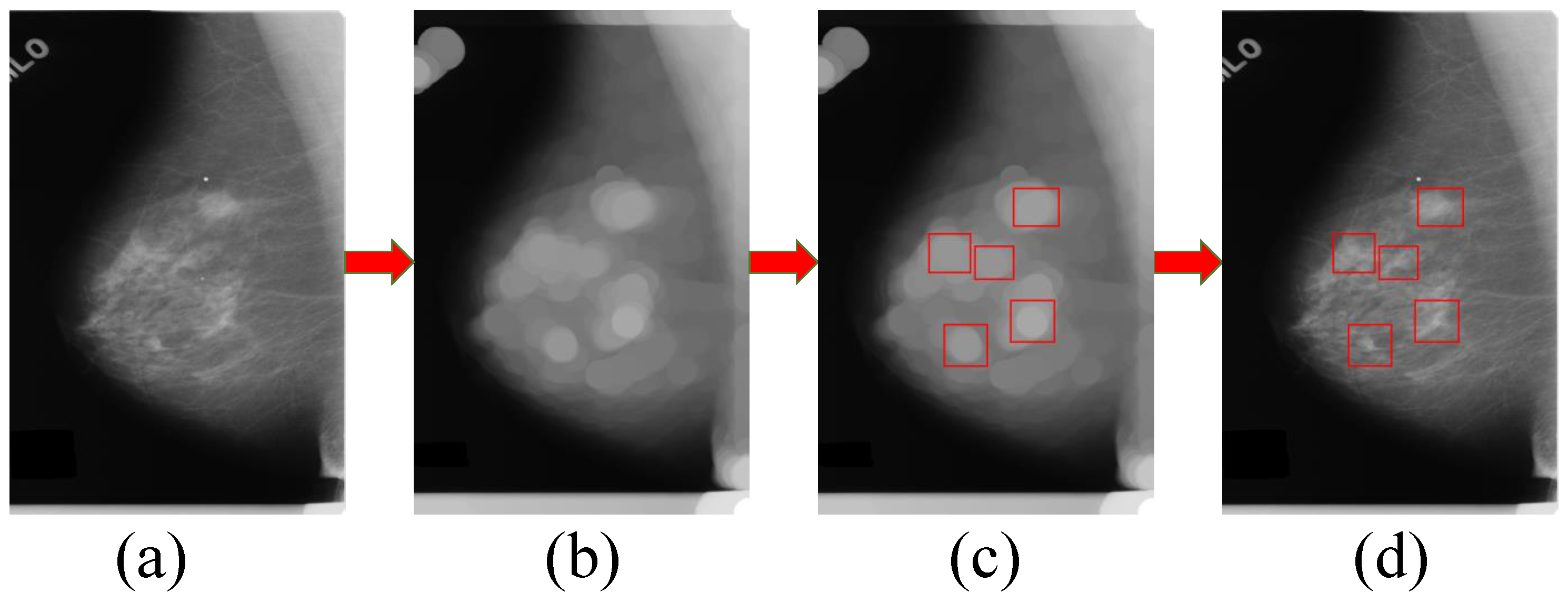
3.2. The Generation Model for Candidate Region of Breast Mass
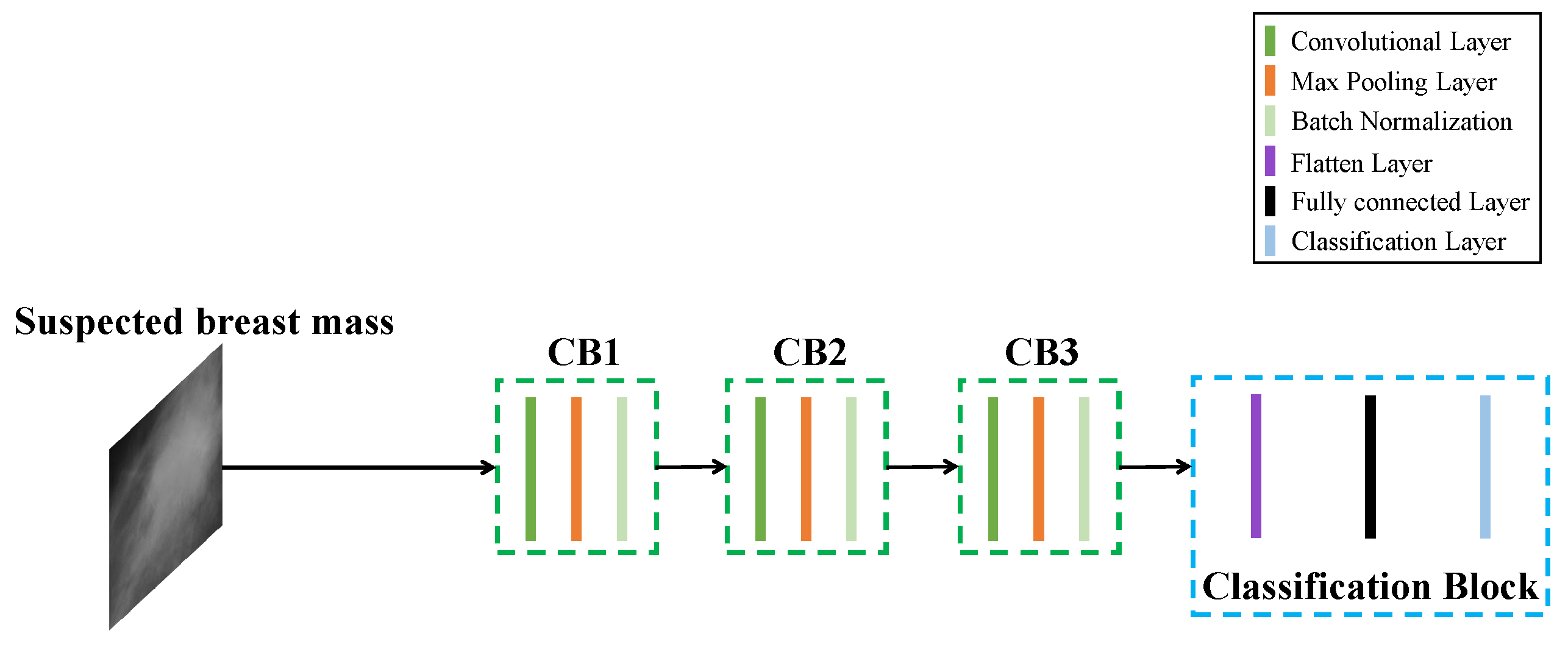
3.3. Candidate Regions Identification Model
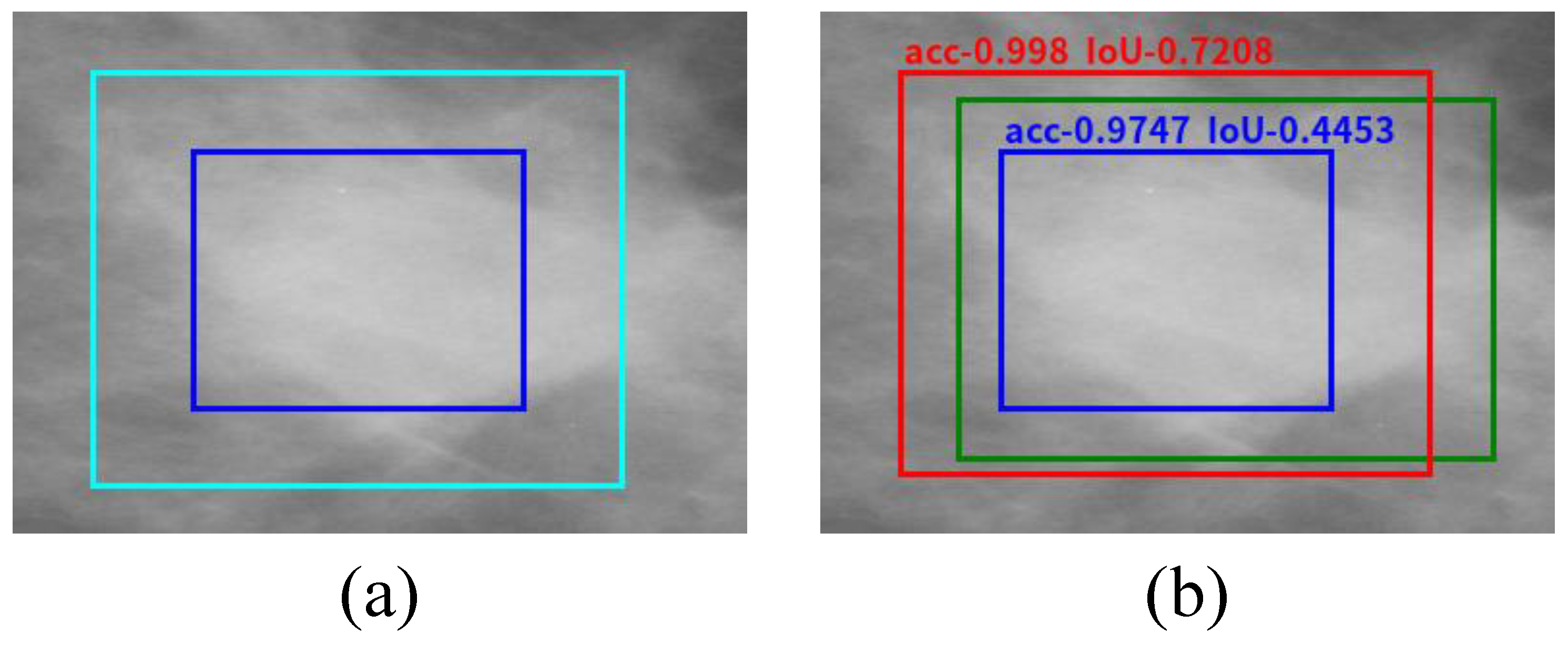
3.4. Regression Model for the Location and Bounding Box of Breast Mass
| Algorithm 1. PSO for breast mass bounding box refinement. |
| Input: Number of particles, maximum iteration number, classification model of breast mass based on CNN: Prediction_Model, rough bounding box of breast mass Output: for to number of particles do if then for to Maximum iteration number do for to Number of particles do 1. Update 2. Update 3. Get fitness from by 4. Update if then 5. Update and for to number of particles do if then |
4. Experiments
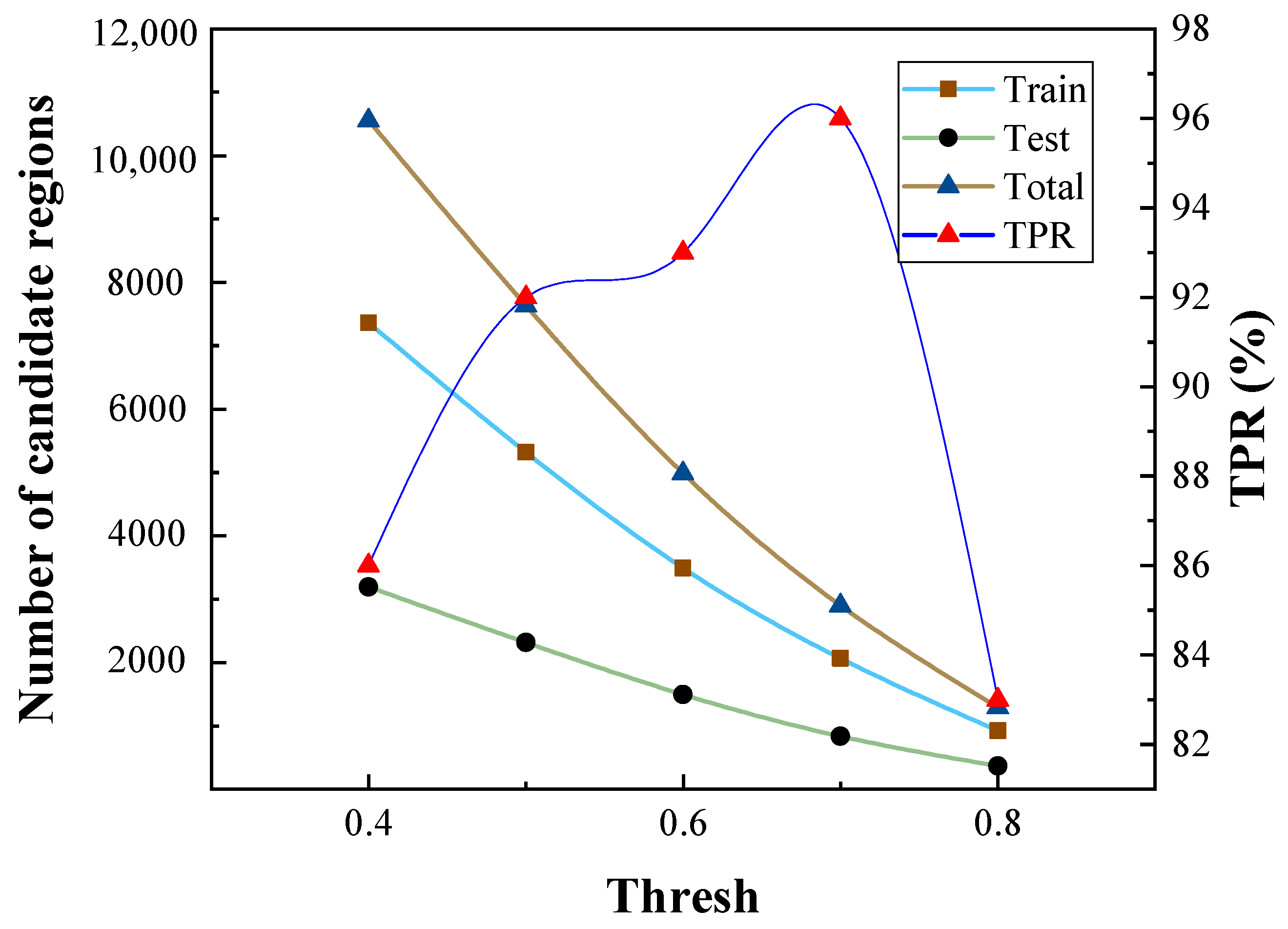
4.1. Dataset and Experimental Setting
4.2. Accuracy Comparison and Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Katanoda, K.; Matsuda, T. Five-year relative survival rate of breast cancer in the USA, Europe and Japan. Jpn. J. Clin. Oncol. 2014, 44, 611. [Google Scholar] [CrossRef] [PubMed]
- Aličković, E.; Subasi, A. Breast cancer diagnosis using GA feature selection and Rotation Forest. Neural Comput. Appl. 2017, 28, 753–763. [Google Scholar] [CrossRef]
- Omondiagbe, D.A.; Veeramani, S.; Sidhu, A.S. Machine Learning Classification Techniques for Breast Cancer Diagnosis. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2019; Volume 495, p. 012033. [Google Scholar]
- Yang, Z.; Cao, Z.; Zhang, Y.; Han, M.; Xiao, J.; Huang, L.; Wu, S.; Ma, J.; Chang, P. MommiNet: Mammographic Multi-View Mass Identification Networks. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 200–210. [Google Scholar]
- Kerhet, A.; Raffetto, M.; Boni, A.; Massa, A. A SVM-based approach to microwave breast cancer detection. Eng. Appl. Artif. Intell. 2006, 19, 807–818. [Google Scholar] [CrossRef]
- Kom, G.; Tiedeu, A.; Kom, M. Automated detection of masses in mammograms by local adaptive thresholding. Comput. Biol. Med. 2007, 37, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xu, S.; Jin, L.; Zhang, S. Using PSO to improve dynamic programming based algorithm for breast mass segmentation. In Proceedings of the 2010 IEEE Fifth International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA), Changsha, China, 23–26 September 2010; pp. 485–488. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar]
- Kuo, Y.C.; Lin, W.C.; Hsu, S.C.; Cheng, A.C. Mass detection in digital mammograms system based on PSO algorithm. In Proceedings of the 2014 International Symposium on Computer, Consumer and Control, Taichung, Taiwan, 10–12 June 2014; pp. 662–668. [Google Scholar]
- StojiC, T.; Reljin, I.; Reljin, B. Local contrast enhancement in digital mammography by using mathematical morphology. In Proceedings of the International Symposium on Signals, Circuits and Systems, 2005 ISSCS 2005, Iasi, Romania, 14–15 July 2005; Volume 2, pp. 609–612. [Google Scholar]
- Amutha, S.; Babu, D.R.; Shankar, M.R.; Kumar, N.H. Mammographic image enhancement using modified mathematical morphology and Bi-orthogonal wavelet. In Proceedings of the 2011 IEEE International Symposium on IT in Medicine and Education, Guangzhou, China, 9–11 December 2011; Volume 1, pp. 548–553. [Google Scholar]
- Liu, F.; Zhang, F.; Gong, Z.; Chen, Y.; Chai, W. A fully automated scheme for mass detection and segmentation in mammograms. In Proceedings of the 2012 5th International Conference on BioMedical Engineering and Informatics, Chongqing, China, 16–18 October 2012; pp. 140–144. [Google Scholar]
- Wen, J.; Zhang, Z.; Zhang, Z.; Fei, L.; Wang, M. Generalized incomplete multiview clustering with flexible locality structure diffusion. IEEE Trans. Cybern. 2020, 51, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Martel, A.L.; Abolmaesumi, P.; Stoyanov, D.; Mateus, D.; Zuluaga, M.A.; Zhou, S.K.; Racoceanu, D.; Joskowicz, L. Medical Image Computing and Computer Assisted Intervention-MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part I; Springer Nature: Berlin/Heidelberg, Germany, 2020; Volume 12261. [Google Scholar]
- Woźniak, M.; Siłka, J.; Wieczorek, M. Deep neural network correlation learning mechanism for CT brain tumor detection. Neural Comput. Appl. 2021, 1–16. [Google Scholar]
- Ke, Q.; Zhang, J.; Wei, W.; Połap, D.; Woźniak, M.; Kośmider, L.; Damaševĭ, R. A neuro-heuristic approach for recognition of lung diseases from X-ray images. Expert. Syst. Appl. 2019, 126, 218–232. [Google Scholar] [CrossRef]
- Wen, J.; Zhang, Z.; Xu, Y.; Zhang, B.; Fei, L.; Xie, G.S. CDIMC-Net: Cognitive Deep Incomplete Multiview Clustering Network. In International Joint Conference on Artificial Intelligence; AAAI Press: Palo Alto, CA, USA, 2020; pp. 3230–3236. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Uijlings, J.; Sande, K.E.; Gevers, T.; Smeulders, A.W.M. Selective Search for Object Recognition. Int. J. Comput. Vis. 2013, 104, 154–171. [Google Scholar] [CrossRef]
- Almasni, M.A.; Alantari, M.A.; Park, J.; Gi, G.; Kim, T.; Rivera, P.; Valarezo, E.; Choi, M.; Han, S.; Kim, T. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput. Methods Programs Biomed. 2018, 157, 85–94. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference On Computer Vision And Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Heath, M.; Bowyer, K.; Kopans, D.; Moore, R.; Kegelmeyer, W.P. The Digital Database for Screening Mammography. In Proceedings of the 5th International Workshop on Digital Mammography; Medical Physics Publishing: Madison, WI, USA, 2000; pp. 212–218. [Google Scholar]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Phang, J.; Park, J.; Shen, Y.; Huang, Z.; Zorin, M.; Jastrzebski, S.; Fevry, T.; Katsnelson, J.; Kim, E.; et al. Deep Neural Networks Improve Radiologists’ Performance in Breast Cancer Screening. IEEE Trans. Med. Imaging 2019, 39, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Eltonsy, N.H.; Tourassi, G.D.; Elmaghraby, A. A Concentric Morphology Model for the Detection of Masses in Mammography. IEEE Trans. Med Imaging 2007, 26, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Nagi, J.; Kareem, S.A.; Nagi, F.; Ahmed, S.K. Automated breast profile segmentation for ROI detection using digital mammograms. In Proceedings of the 2010 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 30 November–2 December 2010; pp. 87–92. [Google Scholar]
- Ciecholewski, M. Microcalcification segmentation from mammograms: A morphological approach. J. Digit. Imaging 2017, 30, 172–184. [Google Scholar] [CrossRef]
- Tourassi, G.D.; Vargasvoracek, R.; Catarious, D.M.; Floyd, C.E. Computer-assisted detection of mammographic masses: A template matching scheme based on mutual information. Med. Phys. 2003, 30, 2123–2130. [Google Scholar] [CrossRef] [PubMed]
- Cover, T.M.; Allen, T. Elements of Information Theory, Wiley Series in Telecommunications; Tsinghua University Press: Beijing, China, 1991. [Google Scholar]
- Divyashree, B.; Kumar, G.H. Breast Cancer Mass Detection in Mammograms Using Gray Difference Weight and MSER Detector. SN Comput. Sci. 2021, 2, 1–13. [Google Scholar] [CrossRef]
- Pizer, S.M.; Johnston, R.E.; Ericksen, J.P.; Yankaskas, B.C.; Muller, K.E. Contrast-limited adaptive histogram equalization: Speed and effectiveness. In Proceedings of the First Conference on Visualization in Biomedical Computing, IEEE Computer Society, Atlanta, GA, USA, 22–25 May 1990; pp. 337–338. [Google Scholar]
- Matas, J.; Chum, O.; Urban, M.; Pajdla, T. Robust wide-baseline stereo from maximally stable extremal regions. Image Vis. Comput. 2004, 22, 761–767. [Google Scholar] [CrossRef]
- Lbachir, I.A.; Daoudi, I.; Tallal, S. Automatic computer-aided diagnosis system for mass detection and classification in mammography. Multimed. Tools Appl. 2021, 80, 9493–9525. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man. Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 448–456. [Google Scholar]
- Sampat, M.P.; Bovik, A.C.; Whitman, G.J.; Markey, M.K. A model-based framework for the detection of spiculated masses on mammography. Med. Phys. 2008, 35, 2110–2123. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, W.; Cui, L.; Wang, H.; Bu, Q.; Feng, J. Automatic Mass Detection from Mammograms with Region-Based Convolutional Neural Network. In Chinese Conference on Image and Graphics Technologies; Springer: Berlin/Heidelberg, Germany, 2018; pp. 442–450. [Google Scholar]
- Junior, G.B.; Rocha, S.V.D.; De Almeida, J.D.S.; De Paiva, A.C.; Silva, A.C.; Gattass, M. Breast cancer detection in mammography using spatial diversity, geostatistics, and concave geometry. Multimed. Tools Appl. 2019, 78, 13005–13031. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, F.; Zhang, Q.; Wang, S.; Wang, Y.; Yu, Y. Cross-View Correspondence Reasoning Based on Bipartite Graph Convolutional Network for Mammogram Mass Detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 3812–3822. [Google Scholar]
- Cao, H. Breast mass detection in digital mammography based on anchor-free architecture. arXiv 2020, arXiv:2009.00857. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 42, 2999–3007. [Google Scholar]
- Zhu, C.; He, Y.; Savvides, M. Feature selective anchor-free module for single-shot object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 840–849. [Google Scholar]
- Kong, T.; Sun, F.; Liu, H.; Jiang, Y.; Li, L.; Shi, J. FoveaBox: Beyound Anchor-Based Object Detection. IEEE Trans. Image Process. 2020, 29, 7389–7398. [Google Scholar] [CrossRef]



| Method | Tradition /Deep | TPR (%) | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) | FPI | Year |
|---|---|---|---|---|---|---|---|---|
| Eltonsy [26] | Tradition | 92.1 | - | - | - | - | 5.4 | 2007 |
| Sampat [37] | Tradition | 88 | - | - | - | - | 2.7 | 2008 |
| Wu [38] | Deep | 81 | - | - | - | - | 1.1 | 2018 |
| Junior [39] | Tradition | 91.63 | - | - | - | - | 0.86 | 2019 |
| Liu [40] | Deep | 95 | - | - | - | - | 4.4 | 2020 |
| Cao [41] | Deep | 94.3 | - | - | - | - | 0.599 | 2020 |
| RetinaNet [42] | Deep | 91.95 | 88.41 | 56.39 | 91.53 | 69.79 | 1.18 | 2018 |
| FSAF [43] | Deep | 85.05 | 63.66 | 26.74 | 85.38 | 40.73 | 1.04 | 2019 |
| Foveabox [44] | Deep | 89.65 | 76.82 | 37.66 | 89.23 | 52.96 | 1.18 | 2020 |
| Our method | Deep | 96 | 85.82 | 50.81 | 95.38 | 66.31 | 0.53 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Sun, H.; Wang, J.; Wu, S.; Zhao, Y.; Xu, Y. Breast Mass Detection in Mammography Based on Image Template Matching and CNN. Sensors 2021, 21, 2855. https://doi.org/10.3390/s21082855
Sun L, Sun H, Wang J, Wu S, Zhao Y, Xu Y. Breast Mass Detection in Mammography Based on Image Template Matching and CNN. Sensors. 2021; 21(8):2855. https://doi.org/10.3390/s21082855
Chicago/Turabian StyleSun, Lilei, Huijie Sun, Junqian Wang, Shuai Wu, Yong Zhao, and Yong Xu. 2021. "Breast Mass Detection in Mammography Based on Image Template Matching and CNN" Sensors 21, no. 8: 2855. https://doi.org/10.3390/s21082855
APA StyleSun, L., Sun, H., Wang, J., Wu, S., Zhao, Y., & Xu, Y. (2021). Breast Mass Detection in Mammography Based on Image Template Matching and CNN. Sensors, 21(8), 2855. https://doi.org/10.3390/s21082855






