Multilayer Network Approach in EEG Motor Imagery with an Adaptive Threshold
Abstract
1. Introduction
2. Materials and Methods
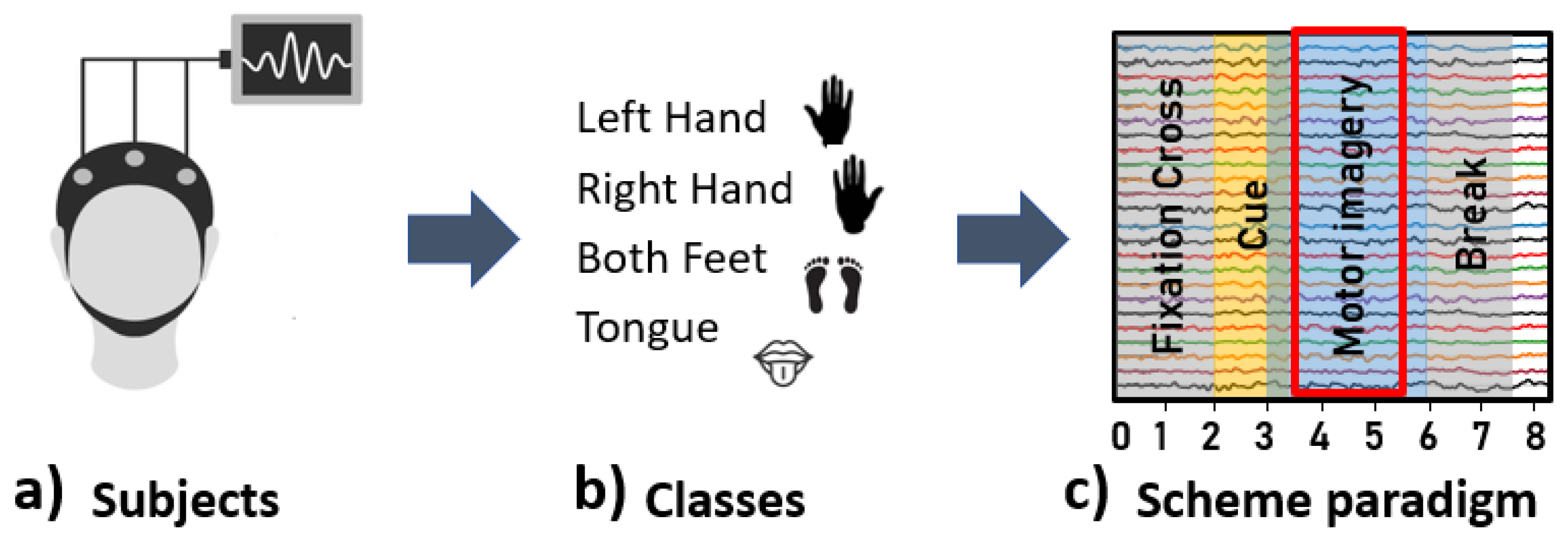
2.1. Database
2.2. Preprocessing
2.3. Connectivity Estimation
3. EEG Processing
3.1. Layers Construction
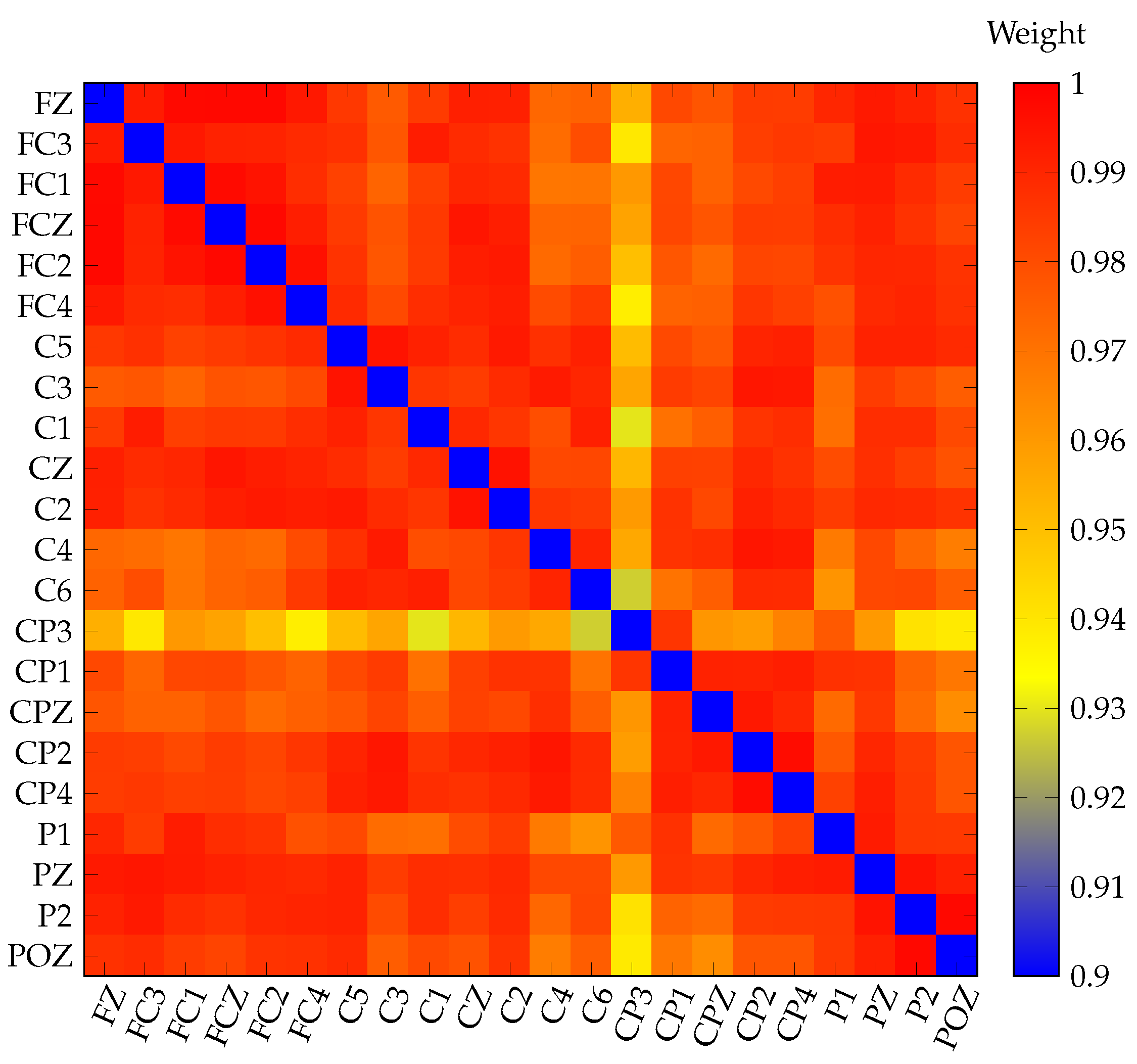
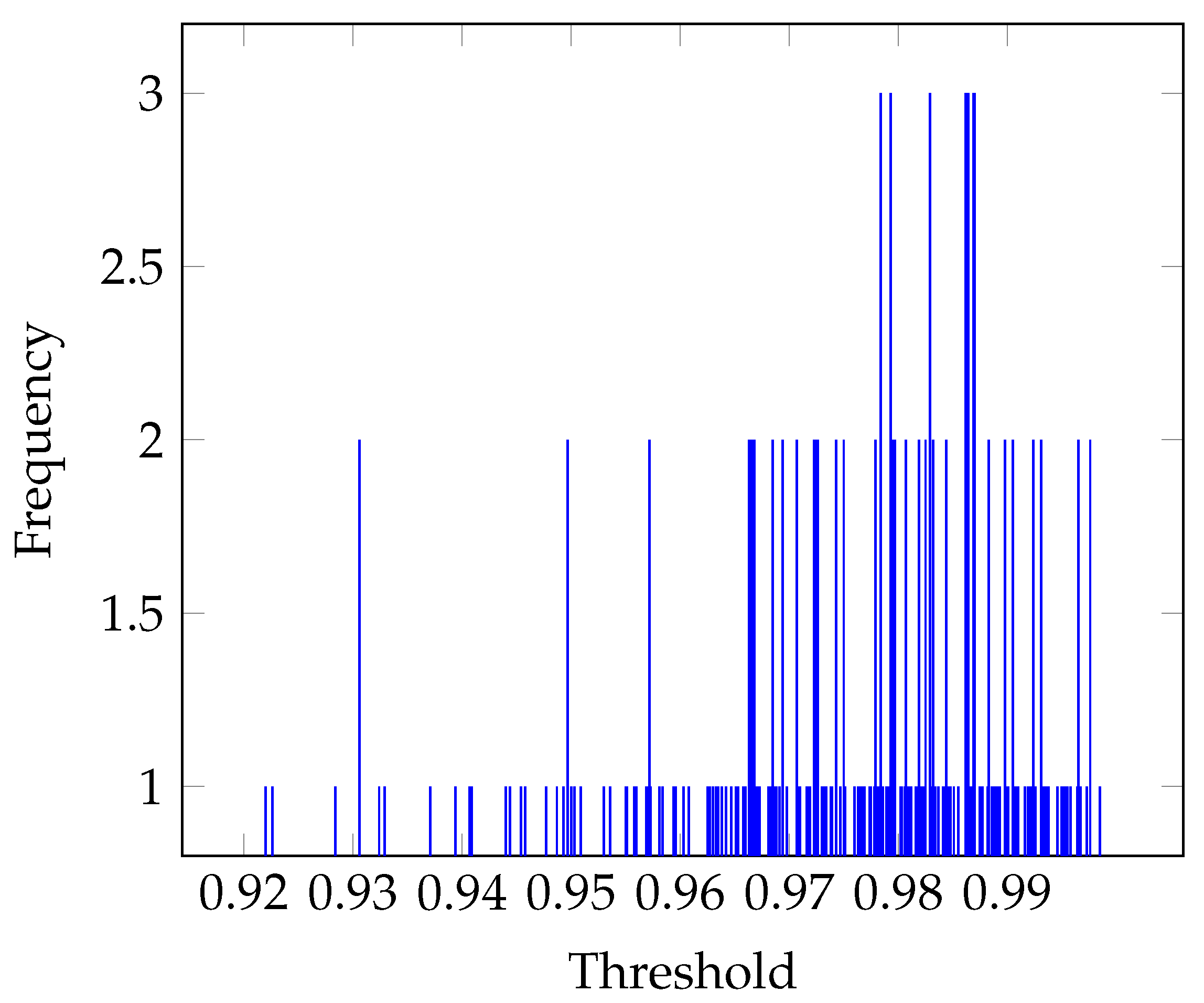
3.2. Threshold Estimation
Otsu’s Threshold
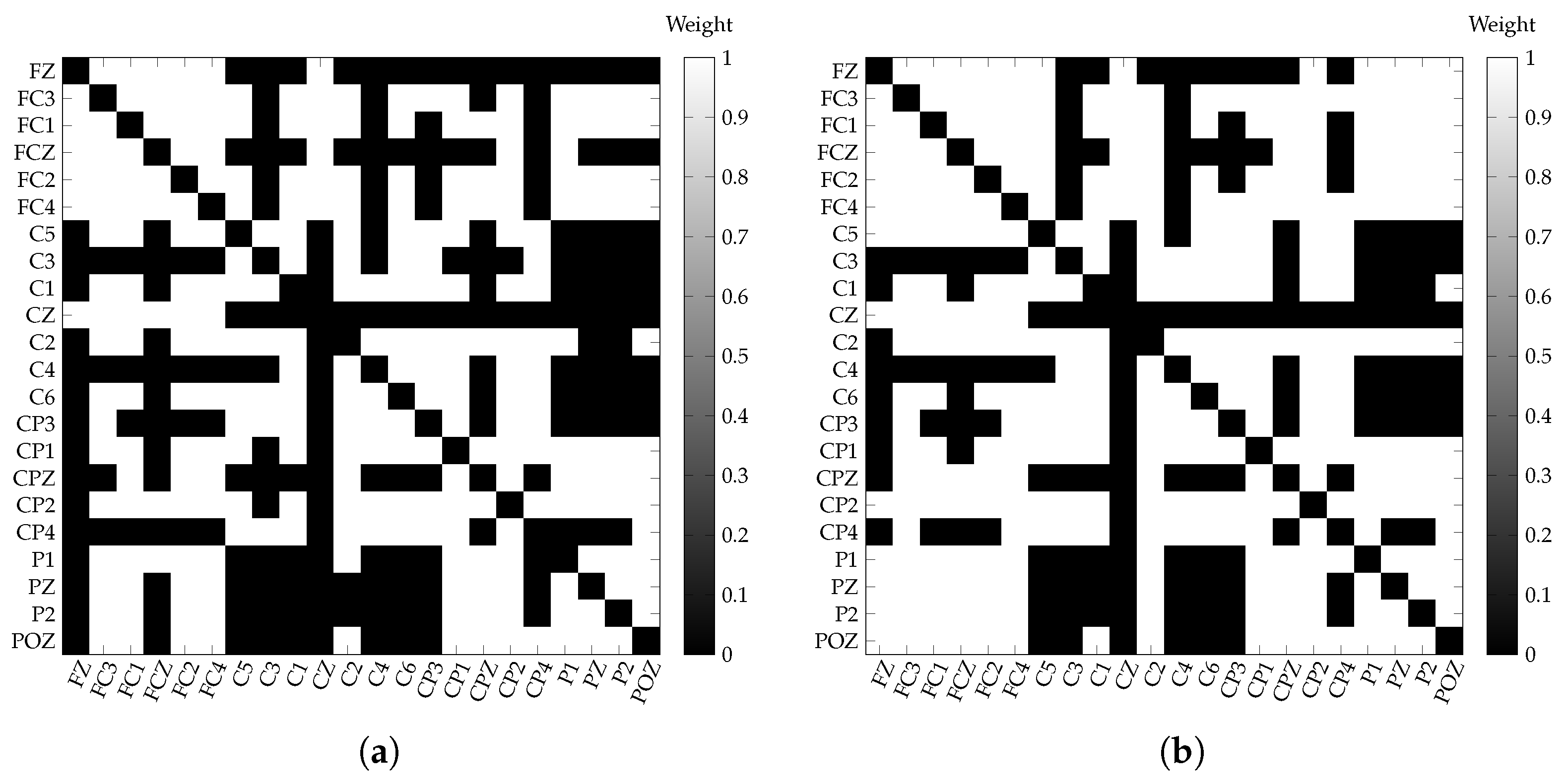
3.3. Single-Layer Network Estimation
3.4. Multilayer Network Estimation
4. Results and Discussion
4.1. Single-Layer Network
4.2. Multilayer Network
4.2.1. Clustering
4.2.2. Key Electrodes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, H.J.; Friston, K. Structural and functional brain networks: From connections to cognition. Science 2013, 342, 6158. [Google Scholar] [CrossRef] [PubMed]
- Subasi, A.; Ercelebi, E. Classification of EEG signals using neural network and logistic regression. Comput. Methods Programs Biomed. 2005, 78, 87–99. [Google Scholar] [CrossRef]
- Sanei, S.; Chambers, J. Brain–Computer Interfacing. EEG Signal Process. 2007, 239–265. [Google Scholar] [CrossRef]
- Mandke, K.; Meier, J.; Brookes, M.J.; O’Dea, R.D.; Van Mieghem, P.; Stam, C.J.; Hillebrand, A.; Tewarie, P. Comparing multilayer brain networks between groups: Introducing graph metrics and recommendations. NeuroImage 2018, 166, 371–384. [Google Scholar] [CrossRef]
- Klados, M.A.; Pandria, N.; Micheloyannis, S.; Margulies, D.; Bamidis, P.D. Math anxiety: Brain cortical network changes in anticipation of doing mathematics. Int. J. Psychophysiol. 2017, 122, 24–31. [Google Scholar] [CrossRef]
- González-Garrido, A.A.; Gómez-Velázquez, F.R.; Salido-Ruiz, R.A.; Espinoza-Valdez, A.; Vélez-Pérez, H.; Romo-Vazquez, R.; Gallardo-Moreno, G.B.; Ruiz-Stovel, V.D.; Martínez-Ramos, A.; Berumen, G. The analysis of EEG coherence reflects middle childhood differences in mathematical achievement. Brain Cogn. 2018, 124, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Sakkalis, V. Review of advanced techniques for the estimation of brain connectivity measured with EEG/MEG. Comput. Biol. Med. 2011, 41, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Bowyer, S.M. Coherence a measure of the brain networks: Past and present. Neuropsychiatr. Electrophysiol. 2016, 2, 1. [Google Scholar] [CrossRef]
- Jiang, Z. Study on EEG power and coherence in patients with mild cognitive impairment during working memory task. J. Zhejiang Univ. Sci. B 2005, 6, 1213–1219. [Google Scholar] [CrossRef]
- Vecchiato, G.; Susac, A.; Margeti, S.; Fallani, F.D.V.; Maglione, A.G.; Supek, S.; Planinic, M.; Babiloni, F. High-resolution EEG analysis of power spectral density maps and coherence networks in a proportional reasoning task. Brain Topogr. 2013, 26, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Özerdem, A.; Güntekin, B.; İlhan, A.; Turp, B.; Başar, E. Reduced long distance gamma (28–48 Hz) coherence in euthymic patients with bipolar disorder. J. Affect. Disord. 2011, 132, 325–332. [Google Scholar] [CrossRef]
- Moazami-Goudarzi, M.; Sarnthein, J.; Michels, L.; Moukhtieva, R.; Jeanmonod, D. Enhanced frontal low and high frequency power and synchronization in the resting EEG of parkinsonian patients. NeuroImage 2008, 41, 985–997. [Google Scholar] [CrossRef]
- Hidasi, Z.; Czigler, B.; Salacz, P.; Csibri, É.; Molnár, M. Changes of EEG spectra and coherence following performance in a cognitive task in Alzheimer’s disease. Int. J. Psychophysiol. 2007, 65, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Tucker, D.; Gilbert, T.; Hou, J.; Mattson, C.; Luu, P.; Holmes, M. Methods for Examining Electrophysiological Coherence in Epileptic Networks. Front. Neurol. 2013, 4, 55. [Google Scholar] [CrossRef]
- Gaxiola-Tirado, J.A.; Salazar-Varas, R.; Gutiérrez, D. Using the partial directed coherence to assess functional connectivity in electroencephalography data for brain–computer interfaces. IEEE Trans. Cogn. Dev. Syst. 2017, 10, 776–783. [Google Scholar] [CrossRef]
- Dennis, E.L.; Jahanshad, N.; Toga, A.W.; McMahon, K.L.; De Zubicaray, G.I.; Martin, N.G.; Wright, M.J.; Thompson, P.M. Test-retest reliability of graph theory measures of structural brain connectivity. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2012; pp. 305–312. [Google Scholar]
- Kinsley, A.C.; Rossi, G.; Silk, M.J.; VanderWaal, K. Multilayer and Multiplex Networks: An Introduction to Their Use in Veterinary Epidemiology. Front. Vet. Sci. 2020, 7, 596. [Google Scholar] [CrossRef]
- Ahmadlou, M.; Adeli, H. Functional community analysis of brain: A new approach for EEG-based investigation of the brain pathology. Neuroimage 2011, 58, 401–408. [Google Scholar] [CrossRef]
- Zippo, A.G.; Della Rosa, P.A.; Castiglioni, I.; Biella, G.E. Alternating dynamics of segregation and integration in human EEG functional networks during working-memory task. Neuroscience 2018, 371, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Puxeddu, M.; Petti, M.; Mattia, D.; Astolfi, L. The optimal setting for multilayer modularity optimization in multilayer brain networks. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 624–627. [Google Scholar]
- Puxeddu, M.G.; Petti, M.; Astolfi, L. A Comprehensive Analysis of Multilayer Community Detection Algorithms for Application to EEG-Based Brain Networks. Front. Syst. Neurosci. 2021, 15, 624183. [Google Scholar] [CrossRef] [PubMed]
- Muldoon, S.F.; Bassett, D.S. Network and multilayer network approaches to understanding human brain dynamics. Philos. Sci. 2016, 83, 710–720. [Google Scholar] [CrossRef]
- Vaiana, M.; Muldoon, S.F. Multilayer brain networks. J. Nonlinear Sci. 2018, 30, 2147–2169. [Google Scholar] [CrossRef]
- Braun, U.; Schäfer, A.; Walter, H.; Erk, S.; Romanczuk-Seiferth, N.; Haddad, L.; Schweiger, J.I.; Grimm, O.; Heinz, A.; Tost, H.; et al. Dynamic reconfiguration of frontal brain networks during executive cognition in humans. Proc. Natl. Acad. Sci. USA 2015, 112, 11678–11683. [Google Scholar] [CrossRef] [PubMed]
- De Domenico, M.; Sasai, S.; Arenas, A. Mapping multiplex hubs in human functional brain networks. Front. Neurosci. 2016, 10, 326. [Google Scholar] [CrossRef]
- Brookes, M.J.; Tewarie, P.K.; Hunt, B.A.; Robson, S.E.; Gascoyne, L.E.; Liddle, E.B.; Liddle, P.F.; Morris, P.G. A multi-layer network approach to MEG connectivity analysis. Neuroimage 2016, 132, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Paredes, O.; López, J.B.; Covantes-Osuna, C.; Ocegueda-Hernández, V.; Romo-Vázquez, R.; Morales, J.A. A Transcriptome Community-and-Module Approach of the Human Mesoconnectome. Entropy 2021, 23, 1031. [Google Scholar] [CrossRef]
- Frolov, N.; Maksimenko, V.; Hramov, A. Revealing a multiplex brain network through the analysis of recurrences. Chaos Interdiscip. J. Nonlinear Sci. 2020, 30, 121108. [Google Scholar] [CrossRef]
- Naro, A.; Maggio, M.G.; Leo, A.; Calabrò, R.S. Multiplex and Multilayer Network EEG Analyses: A Novel Strategy in the Differential Diagnosis of Patients with Chronic Disorders of Consciousness. Int. J. Neural Syst. 2020, 31, 2050052. [Google Scholar] [CrossRef] [PubMed]
- Havlin, S.; Stanley, H.E.; Bashan, A.; Gao, J.; Kenett, D.Y. Percolation of interdependent network of networks. Chaos Solitons Fractals 2015, 72, 4–19. [Google Scholar] [CrossRef]
- Yu, M.; Engels, M.M.; Hillebrand, A.; Van Straaten, E.C.; Gouw, A.A.; Teunissen, C.; Van Der Flier, W.M.; Scheltens, P.; Stam, C.J. Selective impairment of hippocampus and posterior hub areas in Alzheimer’s disease: An MEG-based multiplex network study. Brain 2017, 140, 1466–1485. [Google Scholar] [CrossRef] [PubMed]
- Van Den Heuvel, M.P.; Fornito, A. Brain networks in schizophrenia. Neuropsychol. Rev. 2014, 24, 32–48. [Google Scholar] [CrossRef]
- Van Wijk, B.C.; Stam, C.J.; Daffertshofer, A. Comparing brain networks of different size and connectivity density using graph theory. PLoS ONE 2010, 5, e13701. [Google Scholar] [CrossRef]
- Sezgin, M.; Sankur, B. Survey over image thresholding techniques and quantitative performance evaluation. J. Electron. Imaging 2004, 13, 146–166. [Google Scholar]
- Roy, S.; Nag, S.; Maitra, I.K.; Bandyopadhyay, S.K. A review on automated brain tumor detection and segmentation from MRI of brain. arXiv 2013, arXiv:1312.6150. [Google Scholar]
- Sandhya, G.; Babu Kande, G.; Savithri, T.S. Multilevel thresholding method based on electromagnetism for accurate brain MRI segmentation to detect white matter, gray matter, and CSF. BioMed Res. Int. 2017, 2017, 6783209. [Google Scholar] [CrossRef]
- Fortin, M.; Omidyeganeh, M.; Battié, M.C.; Ahmad, O.; Rivaz, H. Evaluation of an automated thresholding algorithm for the quantification of paraspinal muscle composition from MRI images. Biomed. Eng. Online 2017, 16, 61. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, S.; Singh, S.N. Brain tumor segmentation using DE embedded OTSU method and neural network. Multidimens. Syst. Signal Process. 2019, 30, 1263–1291. [Google Scholar] [CrossRef]
- Geetha, G.; Geethalakshmi, S. Artifact removal from eeg using spatially constrained independent component analysis and wavelet denoising with otsu’s thresholding technique. Procedia Eng. 2012, 30, 1064–1071. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Astolfi, L.; Cincotti, F.; Mattia, D.; Marciani, M.G.; Baccala, L.A.; de Vico Fallani, F.; Salinari, S.; Ursino, M.; Zavaglia, M.; Ding, L.; et al. Comparison of different cortical connectivity estimators for high-resolution EEG recordings. Hum. Brain Mapp. 2007, 28, 143–157. [Google Scholar] [CrossRef]
- Rathee, D.; Cecotti, H.; Prasad, G. Estimation of effective fronto-parietal connectivity during motor imagery using partial granger causality analysis. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2055–2062. [Google Scholar]
- Cardoso, V.F.; Delisle-Rodriguez, D.; Romero-Laiseca, M.A.; Loterio, F.A.; Gurve, D.; Floriano, A.; Valadão, C.; Silva, L.; Krishnan, S.; Frizera-Neto, A.; et al. Effect of a Brain–Computer Interface Based on Pedaling Motor Imagery on Cortical Excitability and Connectivity. Sensors 2021, 21, 2020. [Google Scholar] [CrossRef]
- Alanis-Espinosa, M.; Gutiérrez, D. On the assessment of functional connectivity in an immersive brain-computer interface during motor imagery. Front. Psychol. 2020, 11, 1301. [Google Scholar] [CrossRef]
- Tangermann, M.; Müller, K.R.; Aertsen, A.; Birbaumer, N.; Braun, C.; Brunner, C.; Leeb, R.; Mehring, C.; Miller, K.J.; Mueller-Putz, G.; et al. Review of the BCI competition IV. Front. Neurosci. 2012, 6, 55. [Google Scholar]
- Brunner, C.; Leeb, R.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set A; Institute for Knowledge Discovery, Graz University of Technology: Graz, Austria, 2008; Volume 16, pp. 1–6. [Google Scholar]
- De Vico Fallani, F.; Astolfi, L.; Cincotti, F.; Mattia, D.; la Rocca, D.; Maksuti, E.; Salinari, S.; Babiloni, F.; Vegso, B.; Kozmann, G.; et al. Evaluation of the Brain Network Organization From EEG Signals: A Preliminary Evidence in Stroke Patient. Anat. Rec. 2009, 292, 2023–2031. [Google Scholar] [CrossRef]
- Tsai, S.Y. Reproducibility of structural brain connectivity and network metrics using probabilistic diffusion tractography. Sci. Rep. 2018, 8, 11562. [Google Scholar] [CrossRef]
- Stefan, S.; Schorr, B.; Lopez-Rolon, A.; Kolassa, I.T.; Shock, J.P.; Rosenfelder, M.; Heck, S.; Bender, A. Consciousness indexing and outcome prediction with resting-state EEG in severe disorders of consciousness. Brain Topogr. 2018, 31, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Domenico, M.D.; Porter, M.A.; Arenas, A. MuxViz: A tool for multilayer analysis and visualization of networks. J. Complex Networks 2014, 3, 159–176. [Google Scholar] [CrossRef]
- Brin, S.; Page, L. The anatomy of a large-scale hypertextual Web search engine. Comput. Netw. ISDN Syst. 1998, 30, 107–117. [Google Scholar] [CrossRef]
- Domenico, M.D.; Solé-Ribalta, A.; Omodei, E.; Gómez, S.; Arenas, A. Ranking in interconnected multilayer networks reveals versatile nodes. Nat. Commun. 2015, 6, 6868. [Google Scholar] [CrossRef]
- Azimi-Tafreshi, N.; Gómez-Gardeñes, J.; Dorogovtsev, S.N. k-core percolation on multiplex networks. Phys. Rev. E 2014, 90, 032816. [Google Scholar] [CrossRef]
- Shenoy, H.V.; Vinod, A.P. An iterative optimization technique for robust channel selection in motor imagery based brain computer interface. In Proceedings of the 2014 IEEE International Conference on Systems, Man, and Cybernetics (SMC), San Diego, CA, USA, 5–8 October 2014; pp. 1858–1863. [Google Scholar]
- Babiloni, C.; Brancucci, A.; Vecchio, F.; Arendt-Nielsen, L.; Chen, A.C.; Rossini, P.M. Anticipation of somatosensory and motor events increases centro-parietal functional coupling: An EEG coherence study. Clin. Neurophysiol. 2006, 117, 1000–1008. [Google Scholar] [CrossRef]


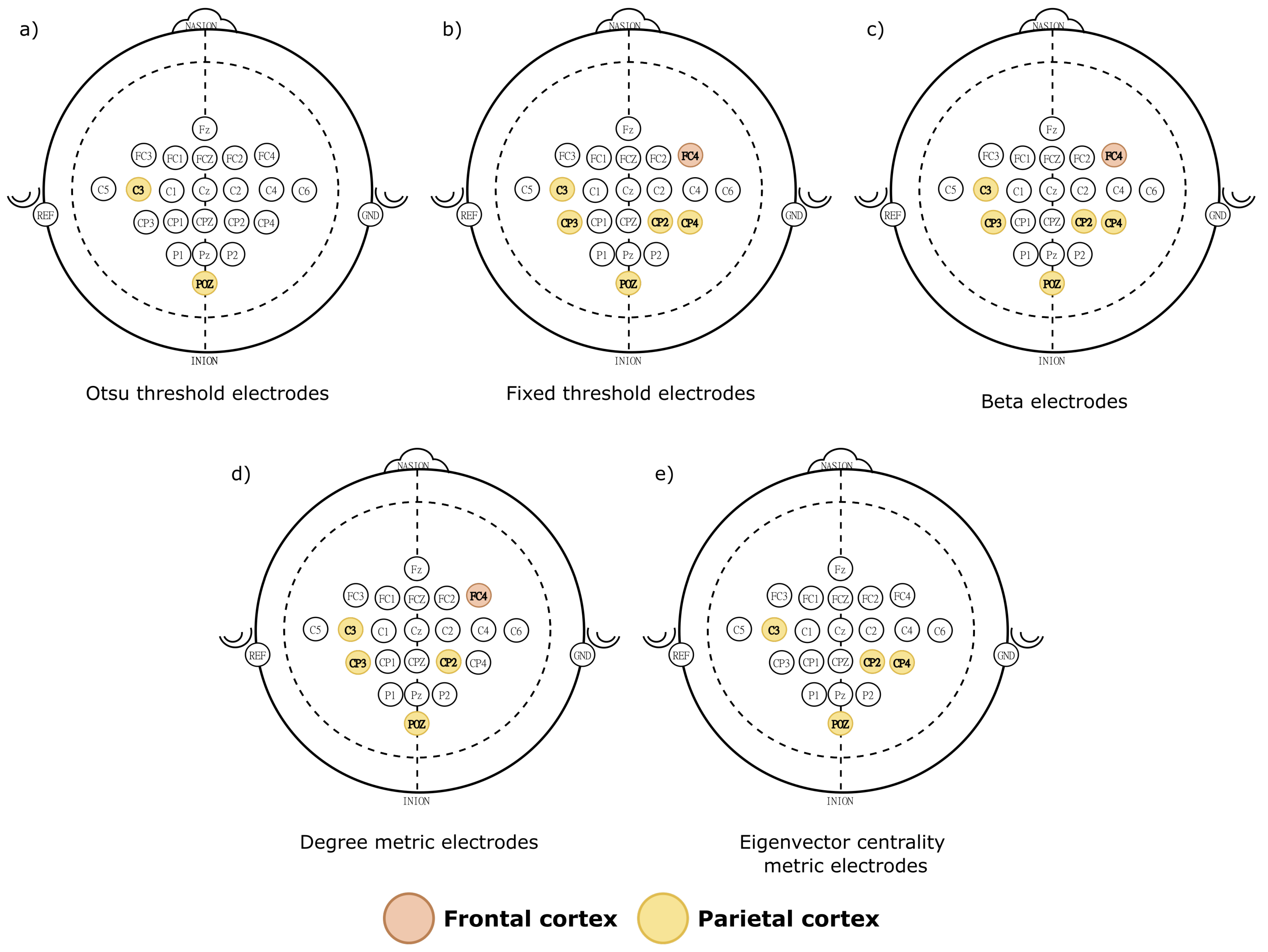
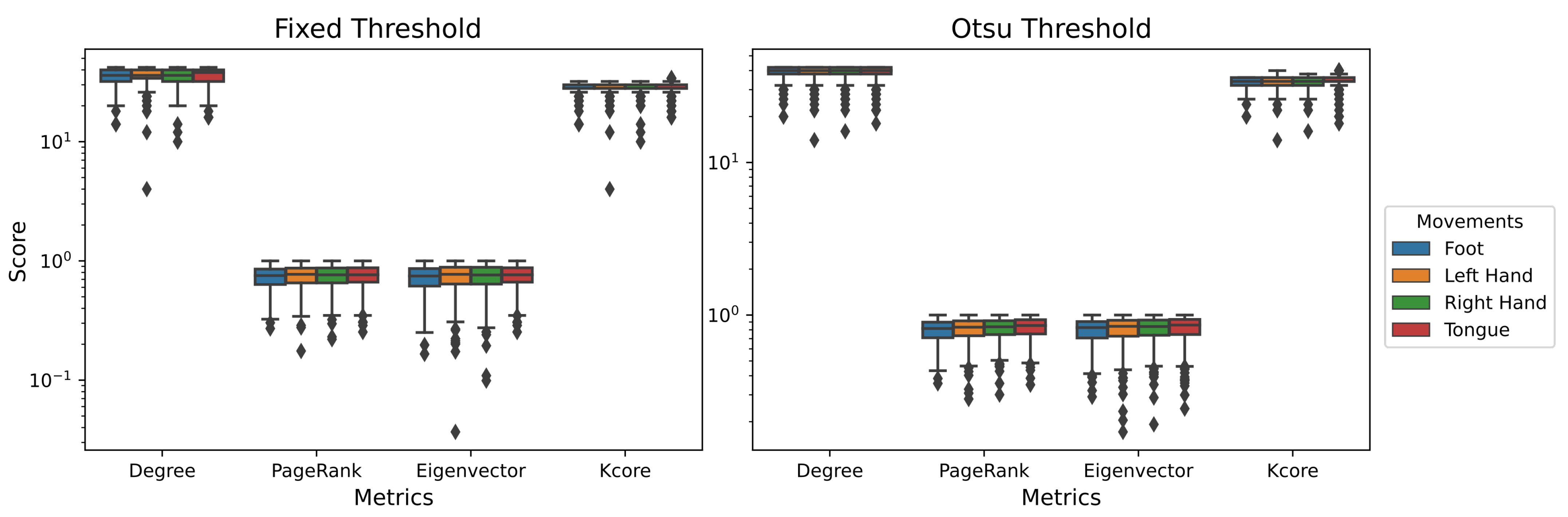



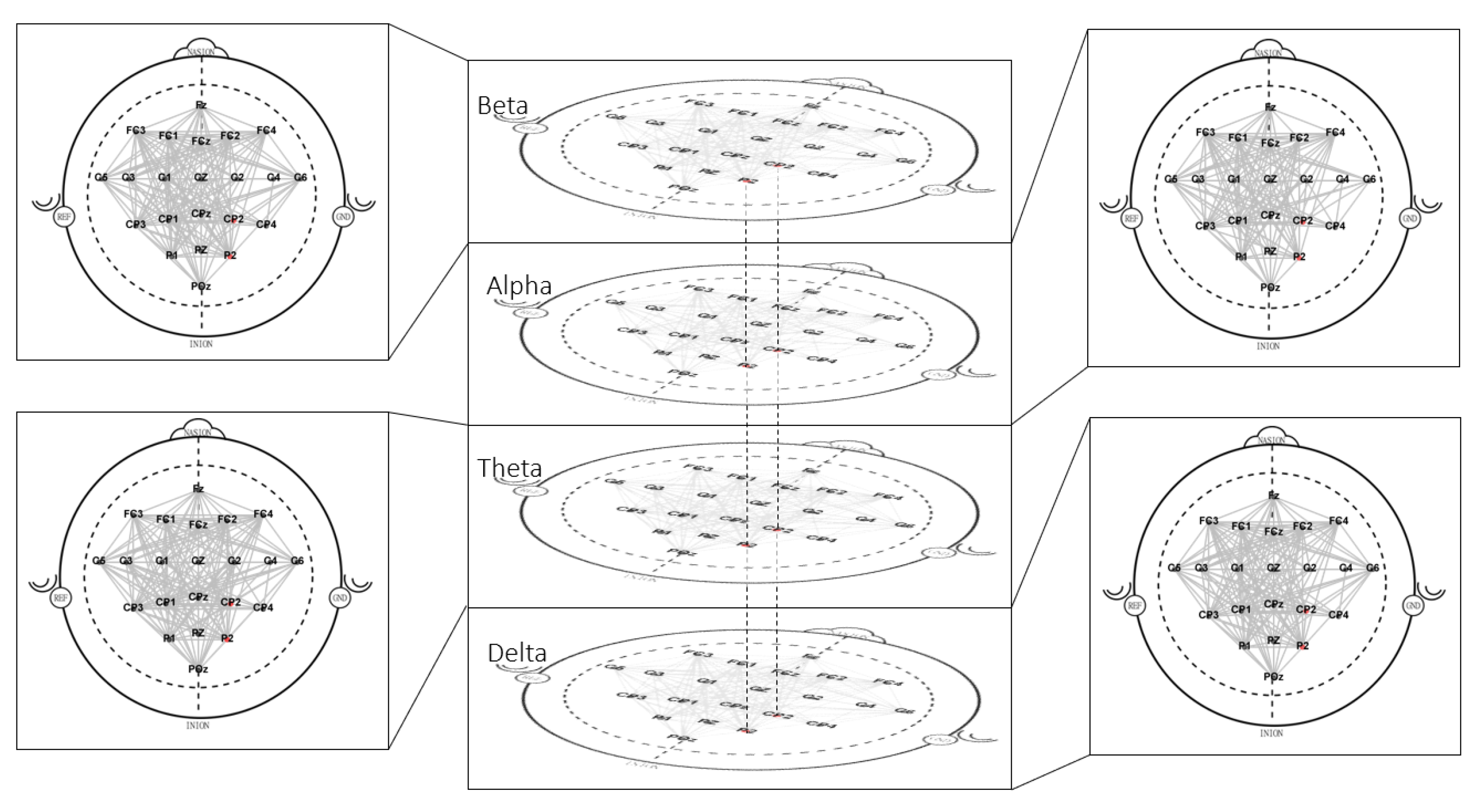
| Metric | Band | Electrode | Movement | Movement | Threshold | |
|---|---|---|---|---|---|---|
| 1 | 2 | Fixed | Otsu | |||
| Degree | Beta | C3 | Foot | Right Hand | * | * |
| POz | Right Hand | Tongue | * | * | ||
| CP2 | Left Hand | Right Hand | * | − | ||
| CP3 | Right Hand | Tongue | * | − | ||
| FC4 | Right Hand | Tongue | * | − | ||
| Eigenvector | Beta | C3 | Foot | Right Hand | * | * |
| POz | Right Hand | Tongue | * | * | ||
| CP2 | Left Hand | Right Hand | * | − | ||
| CP4 | Foot | Left Hand | * | − | ||
| Fixed | Otsu | |
|---|---|---|
| Degree | ||
| PageRank | ||
| Eigenvector | ||
| Kcore |
| Fixed | Otsu | |
|---|---|---|
| Degree | ||
| PageRank | ||
| Eigenvector | ||
| Kcore | Error | Error |
| Electrode | Movement | Movement | Metric | Threshold | |||
|---|---|---|---|---|---|---|---|
| 1 | 2 | Degree | Eigenvector | PageRank | Fixed | Otsu | |
| CP2 | Left Hand | Tongue | − | * | − | * | − |
| P2 | Foot | Left Hand | − | * | − | − | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Covantes-Osuna, C.; López, J.B.; Paredes, O.; Vélez-Pérez, H.; Romo-Vázquez, R. Multilayer Network Approach in EEG Motor Imagery with an Adaptive Threshold. Sensors 2021, 21, 8305. https://doi.org/10.3390/s21248305
Covantes-Osuna C, López JB, Paredes O, Vélez-Pérez H, Romo-Vázquez R. Multilayer Network Approach in EEG Motor Imagery with an Adaptive Threshold. Sensors. 2021; 21(24):8305. https://doi.org/10.3390/s21248305
Chicago/Turabian StyleCovantes-Osuna, César, Jhonatan B. López, Omar Paredes, Hugo Vélez-Pérez, and Rebeca Romo-Vázquez. 2021. "Multilayer Network Approach in EEG Motor Imagery with an Adaptive Threshold" Sensors 21, no. 24: 8305. https://doi.org/10.3390/s21248305
APA StyleCovantes-Osuna, C., López, J. B., Paredes, O., Vélez-Pérez, H., & Romo-Vázquez, R. (2021). Multilayer Network Approach in EEG Motor Imagery with an Adaptive Threshold. Sensors, 21(24), 8305. https://doi.org/10.3390/s21248305






