Remote Arrhythmia Detection for Eldercare in Malaysia
Abstract
:1. Introduction
2. Existing Works
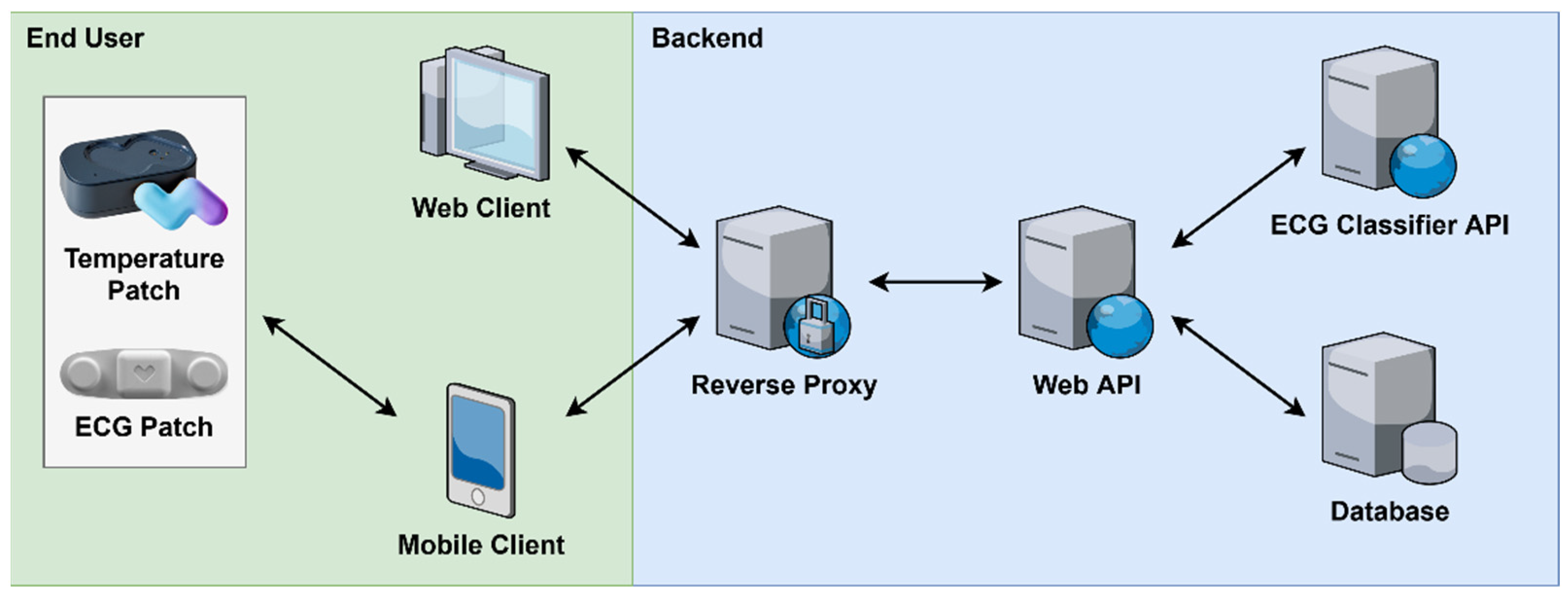
3. System Architecture
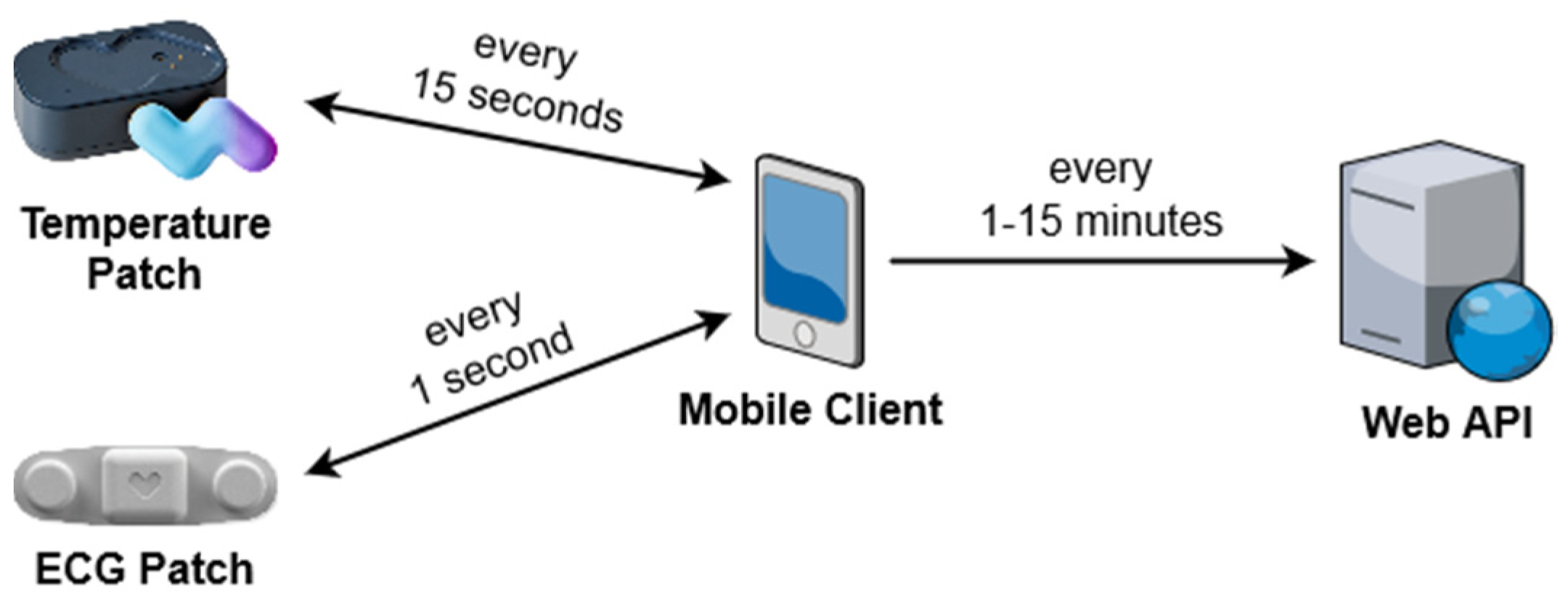
3.1. Module System
3.2. Alert Generation
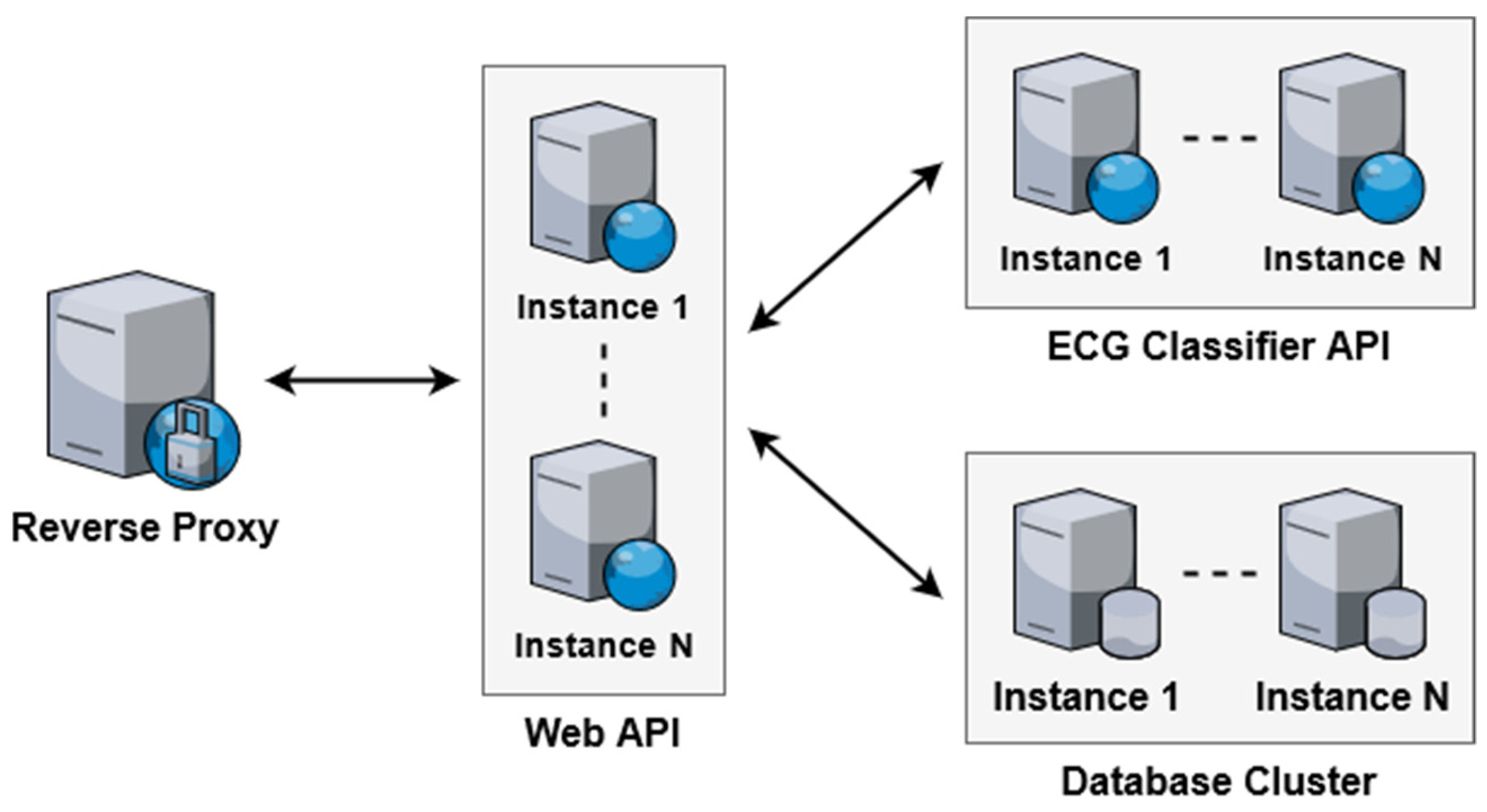
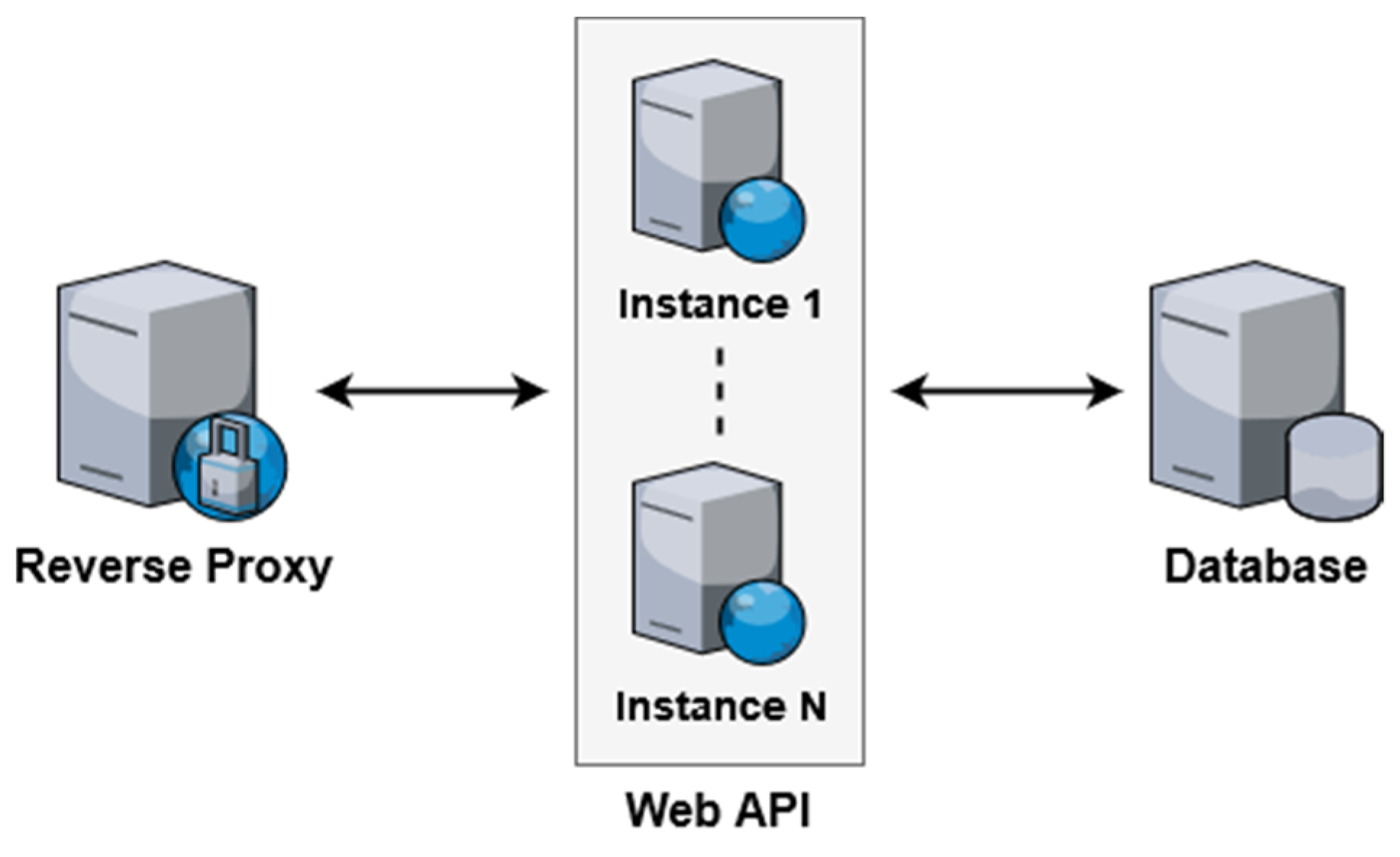
3.3. Scalability
3.4. Prototype Setup
4. Methodology
4.1. ECG Classification Algorithm
4.2. Dataset Preparation
4.2.1. Data Selection
4.2.2. Data Preparation
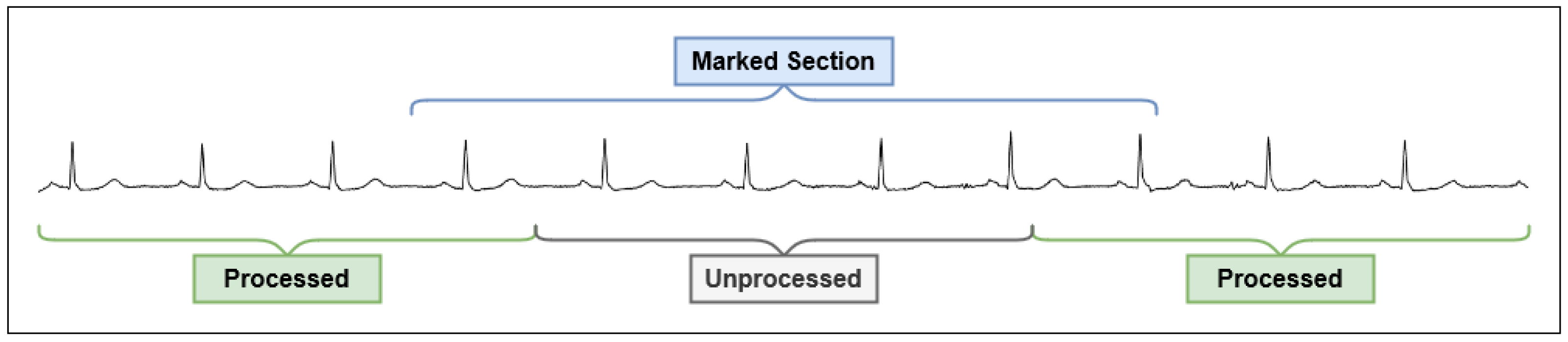
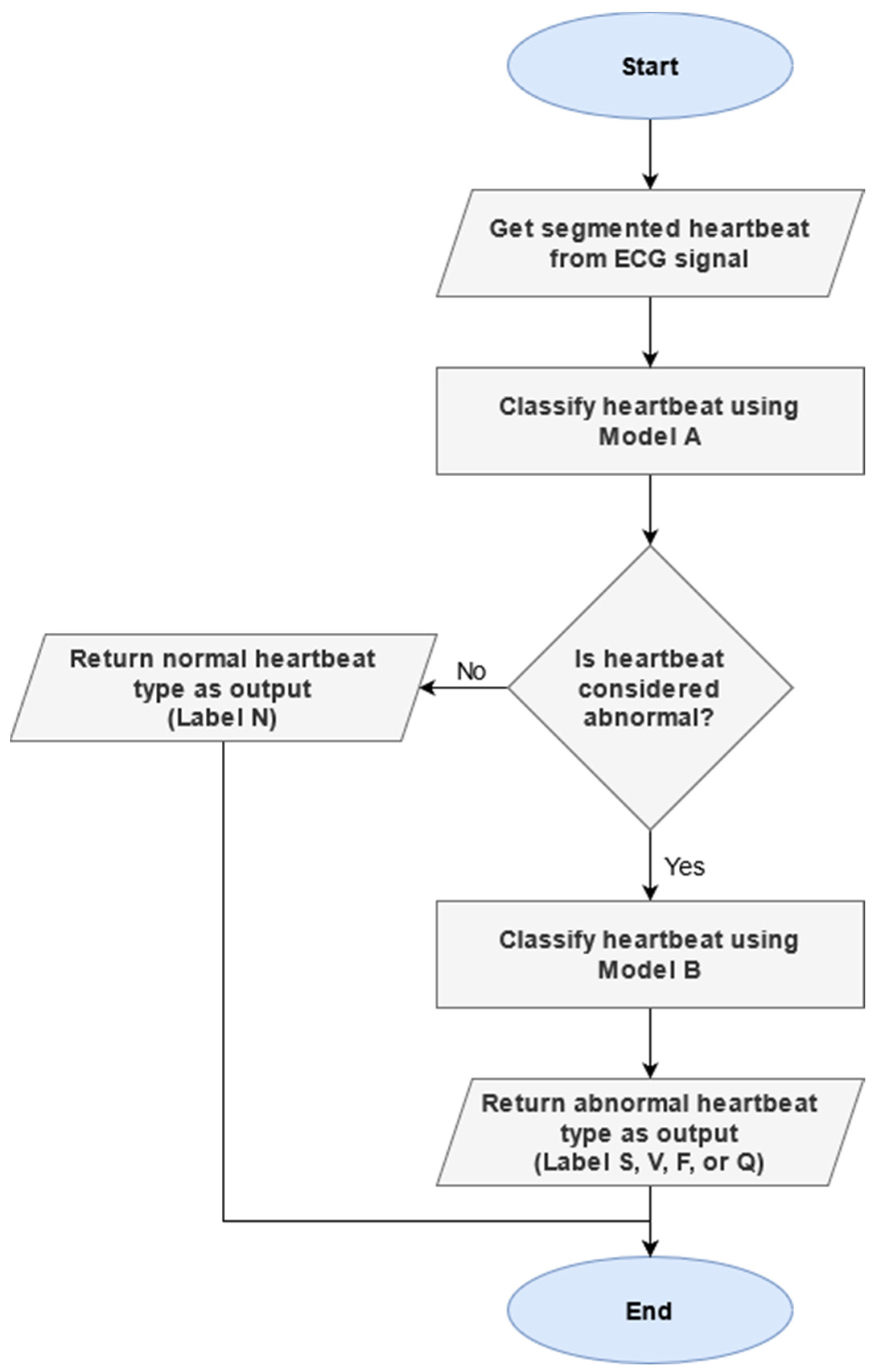
4.3. Two-Phase Classification Scheme
4.3.1. Motivation
4.3.2. ECG Classification Process
4.4. Training and Testing Procedure
5. Evaluation & Results
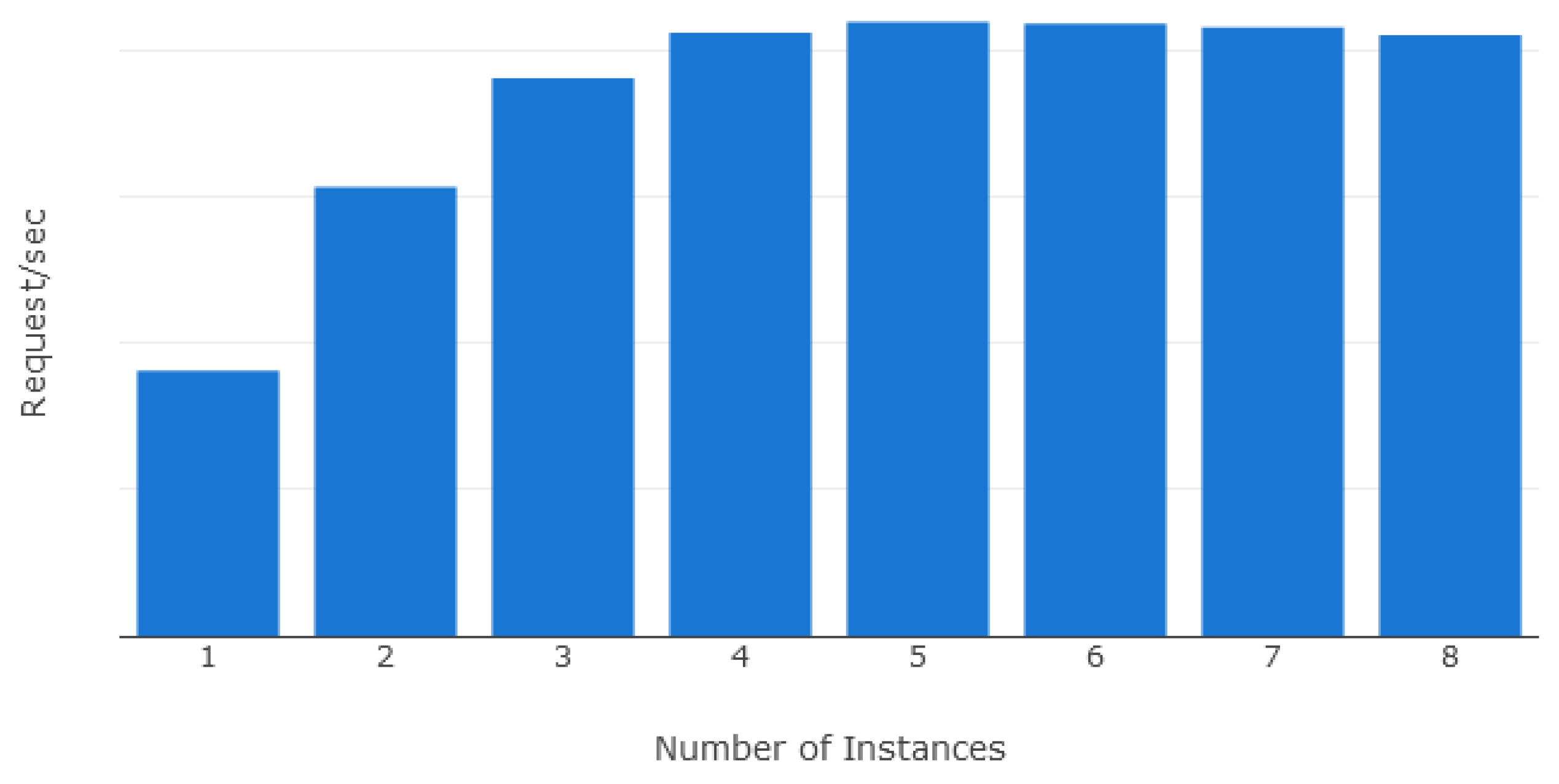
5.1. Scalability Evaluation
5.2. Prototype Deployment
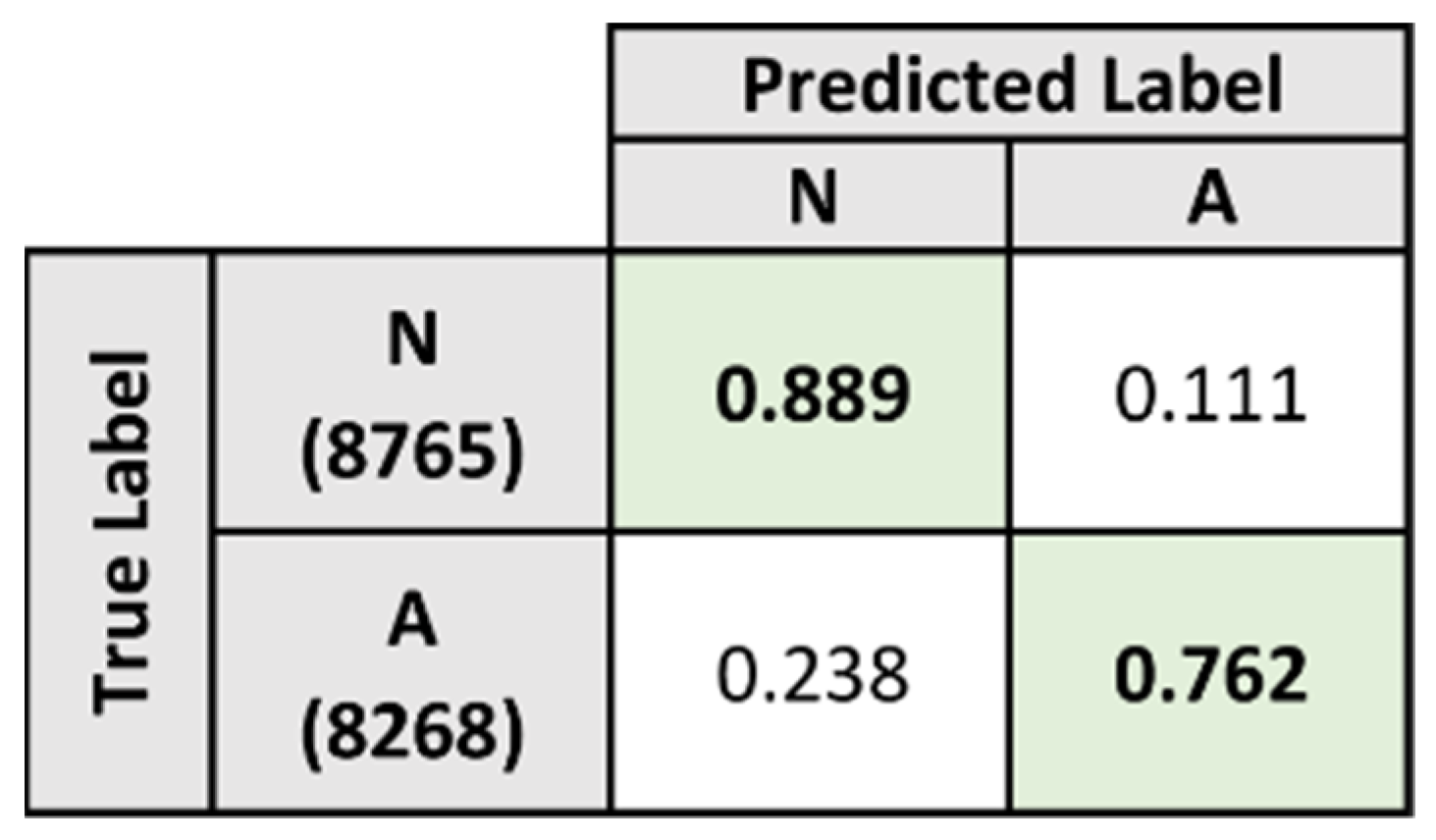
5.3. Baseline Evaluation
5.4. Two-Phase Classification Scheme
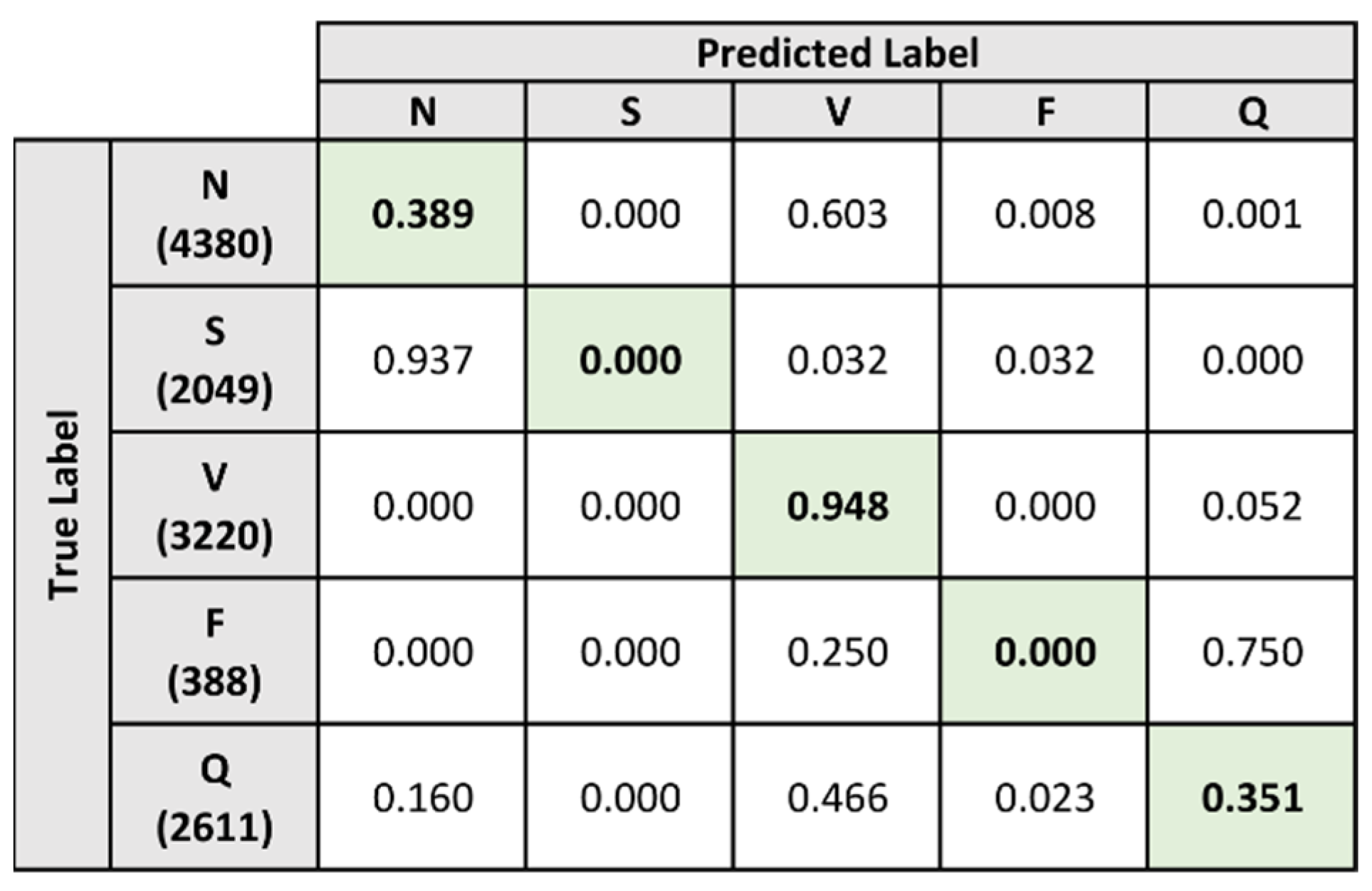
5.4.1. Models A and B
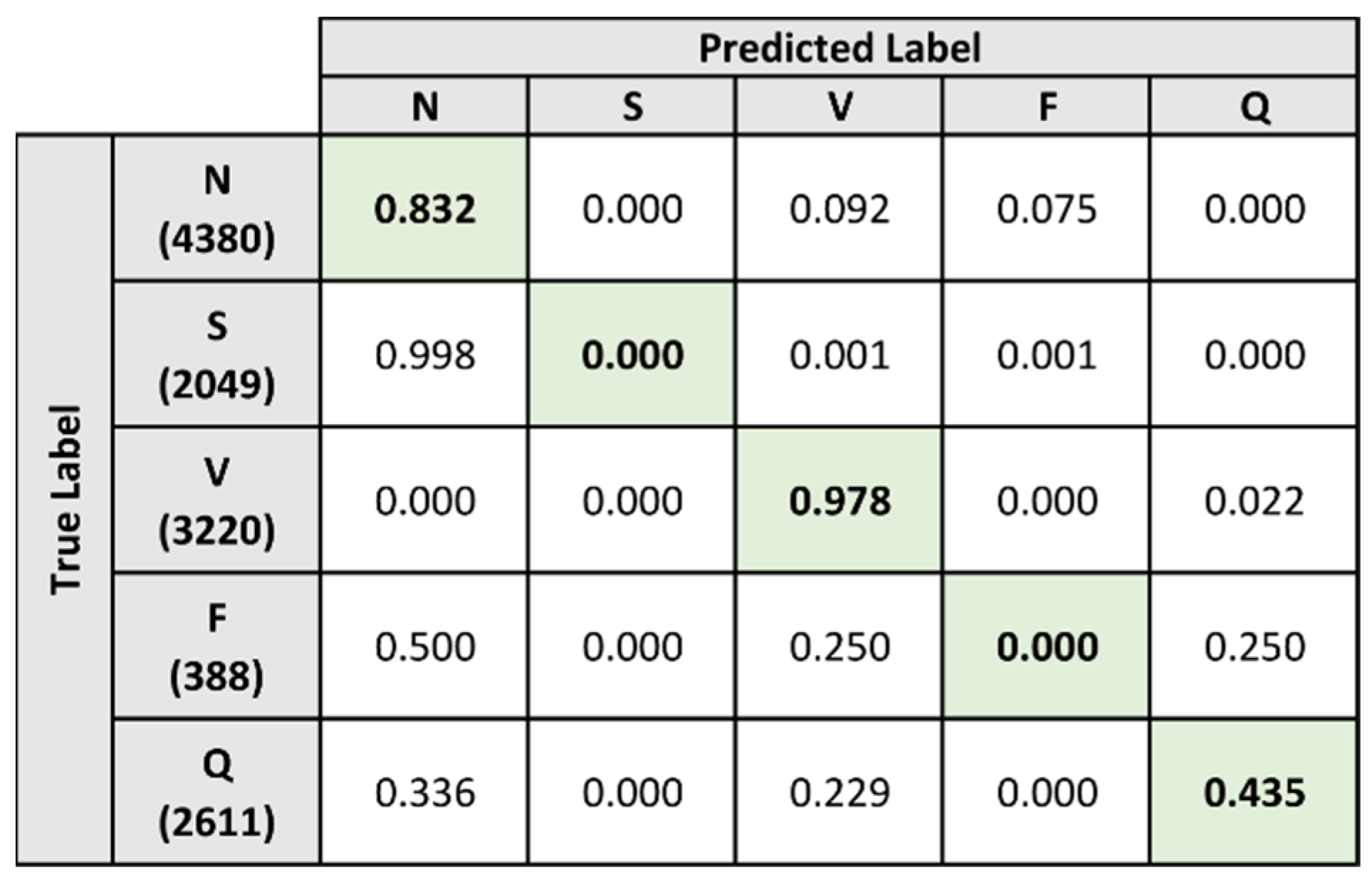
5.4.2. Composite Model
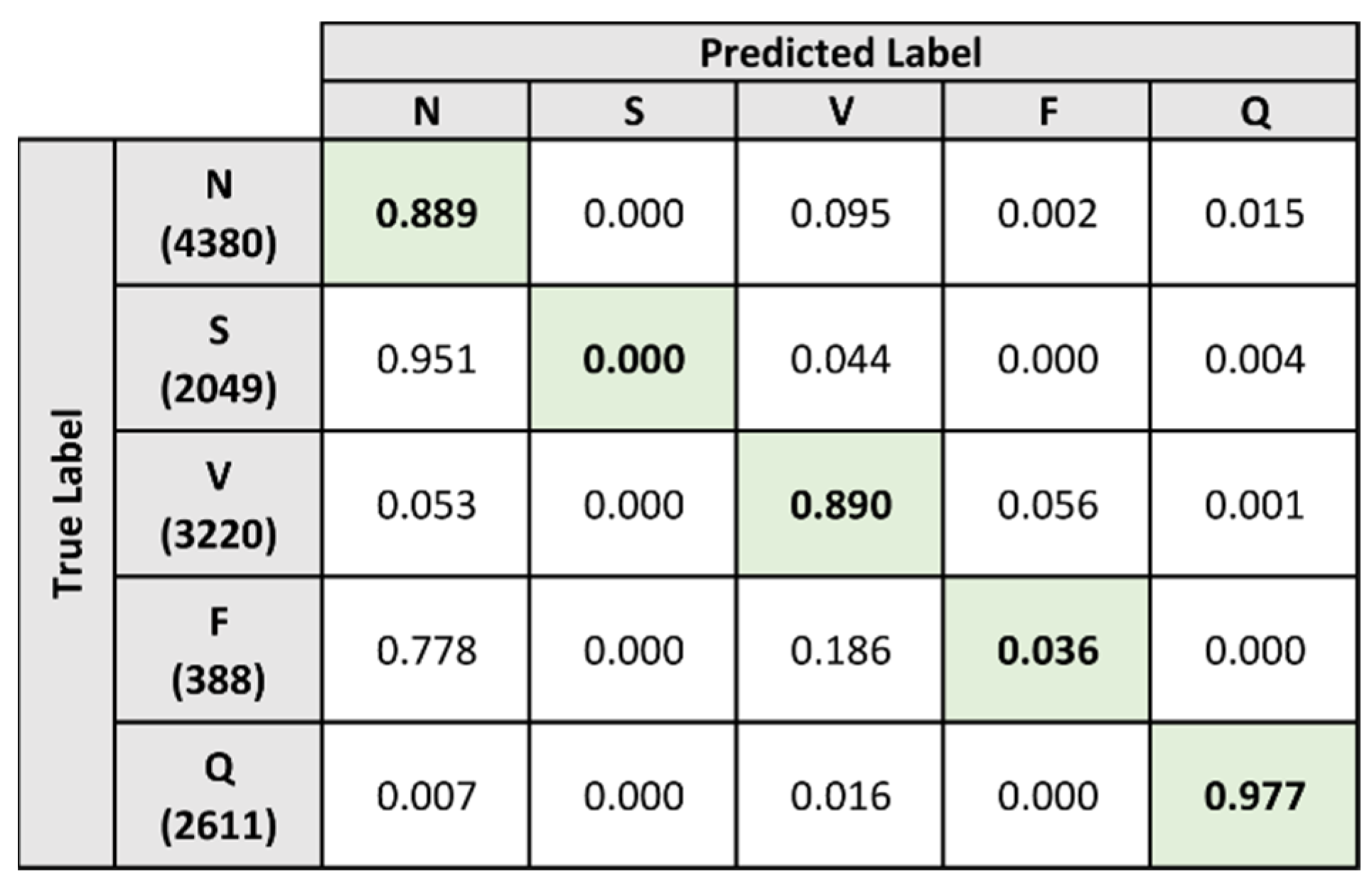
5.5. Evaluation Results
5.6. Evaluation on Single-Lead ECG Signals
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardiovascular Diseases (CVDs). May 2017. Available online: https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 6 November 2021).
- Statistics on Causes of Death, Malaysia. 2019. Available online: https://www.dosm.gov.my/v1/index.php?r=column/pdfPrev&id=RUxlSDNkcnRVazJnakNCNVN2VGgrdz09 (accessed on 6 November 2021).
- He, W.; Goodkind, D.; Kowal, P. An Aging World: 2015. March 2016. Available online: https://www.census.gov/content/dam/Census/library/publications/2016/demo/p95-16-1.pdf (accessed on 6 November 2021).
- Plöthner, M.; Schmidt, K.; De Jong, L.; Zeidler, J.; Damm, K. Needs and preferences of informal caregivers regarding outpatient care for the elderly: A systematic literature review. BMC Geriatr. 2019, 19, 82. [Google Scholar] [CrossRef] [PubMed]
- Karim, H.A. The elderly in Malaysia: Demographic trends. Med. J. Malays. 1997, 52, 206–212. [Google Scholar]
- Ageing and Health. 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (accessed on 6 November 2021).
- Chen, P.C.Y.; Arokiasamy, J.T.; Gan, C.Y. The prevalence, nature and severity of disabilities in a Malaysian community. Med. J. Malays. 1983, 38, 206–211. [Google Scholar]
- Guinan, P. Frailty and Old Age. Linacre Q. 2016, 83, 131–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, W.J.; Harrigan, R.A.; Chan, T.C. Basic electrocardiographic techniques. In Roberts and Hedges’ Clinical Procedures in Emergency Medicine and Acute Care, 7th ed.; Roberts, J.R., Custalow, C.B., Thomsen, T.W., Eds.; Elsevier: Philadelphia, PA, USA, 2019; Chapter 14. [Google Scholar]
- Ganz, L.; Link, M.S. Electrocardiography. In Goldman-Cecil Medicine, 26th ed.; Goldman, L., Schafer, A.I., Eds.; Elsevier: Philadelphia, PA, USA, 2020; Chapter 48. [Google Scholar]
- Jambukia, S.H.; Dabhi, V.K.; Prajapati, H.B. Classification of ECG signals using machine learning techniques: A survey. In Proceedings of the 2015 International Conference on Advances in Computer Engineering and Applications, Ghaziabad, India, 19–20 March 2015; pp. 714–721. [Google Scholar]
- Arrhythmia. 2020. Available online: https://www.nhlbi.nih.gov/health-topics/arrhythmia (accessed on 6 November 2021).
- Samol, A.; Bischof, K.; Luani, B.; Pascut, D.; Wiemer, M.; Kaese, S. Single-Lead ECG Recordings Including Einthoven and Wilson Leads by a Smartwatch: A New Era of Patient Directed Early ECG Differential Diagnosis of Cardiac Diseases? Sensors 2019, 19, 4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Chen, J.; Bao, N.; Gupta, B.B.; Lv, Z. Survey on atrial fibrillation detection from a single-lead ECG wave for Internet of Medical Things. Comput. Commun. 2021, 178, 245–258. [Google Scholar] [CrossRef]
- Khan, J.Y.; Yuce, M.R.; Karami, F. Performance evaluation of a Wireless Body Area sensor network for remote patient monitoring. In Proceedings of the 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS’08—Personalized Healthcare through Technology, Vancouver, BC, Canada, 20–25 August 2008. [Google Scholar]
- González, F.C.J.; Villegas, O.O.V.; Ramírez, D.E.T.; Sánchez, V.G.C.; Domínguez, H.O. Smart Multi-Level Tool for Remote Patient Monitoring Based on a Wireless Sensor Network and Mobile Augmented Reality. Sensors 2014, 14, 17212–17234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollonini, L.; Rajan, N.O.; Xu, S.; Madala, S.; Dacso, C.C. A Novel Handheld Device for Use in Remote Patient Monitoring of Heart Failure Patients—Design and Preliminary Validation on Healthy Subjects. J. Med. Syst. 2010, 36, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Szydło, T.; Konieczny, M. Mobile and wearable devices in an open and universal system for remote patient monitoring. Microprocess. Microsyst. 2016, 46, 44–54. [Google Scholar] [CrossRef]
- de Chazal, P.; O’Dwyer, M.; Reilly, R. Automatic Classification of Heartbeats Using ECG Morphology and Heartbeat Interval Features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.; Coimbra, M.T.; Kumar, B.V. Arrhythmia detection and classification using morphological and dynamic features of ECG signals. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; Volume 2010, pp. 1918–1921. [Google Scholar]
- Chauhan, S.; Vig, L. Anomaly detection in ECG time signals via deep long short-term memory networks. In Proceedings of the 2015 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Paris, France, 19–21 October 2015; pp. 1–7. [Google Scholar]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Liang, S.; Zhang, Y. Simultaneous ECG Heartbeat Segmentation and Classification with Feature Fusion and Long Term Context Dependencies. In Advances in Knowledge Discovery and Data Mining; Lecture Notes in Computer Science; Springer: Singapore, 2020; Volume 12085, pp. 371–383. [Google Scholar]
- Luz, E.; Menotti, D. How the choice of samples for building arrhythmia classifiers impact their performances. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 4988–4991. [Google Scholar]
- Mahajan, R.; Kamaleswaran, R.; Howe, J.A.; Akbilgic, O. Cardiac Rhythm Classification from a Short Single Lead ECG Recording via Random Forest. In Proceedings of the 2017 Computing in Cardiology Conference (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Moody, G.; Mark, R. The impact of the MIT-BIH Arrhythmia Database. IEEE Eng. Med. Boil. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.D.; Liu, C.; Moody, B.; Lehman, L.H.; Silva, I.; Li, Q.; Johnson, A.E.; Mark, R.G. AF classification from a short single lead ECG recording: The PhysioNet/Computing in Cardiology Challenge 2017. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar] [CrossRef]






| Transmission Interval | Description |
|---|---|
| Every 15 min | Default data transmission interval when idle |
| Every 5 min | Interval when actively monitoring vital signs |
| Every 1 min | Interval when vital signs exceed regular levels |
| Original Label | Final Label | Heartbeat Type |
|---|---|---|
| N | N | Normal beat |
| L | Left bundle branch block beat | |
| R | Right bundle branch block beat | |
| A | S | Atrial premature beat |
| a | Aberrated atrial premature beat | |
| J | Nodal (junctional) premature beat | |
| S | Supraventricular premature or ectopic beat (atrial or nodal) | |
| e | Atrial escape beat | |
| j | Nodal (junctional) escape beat | |
| V | V | Premature ventricular contraction |
| E | Ventricular escape beat | |
| F | F | Fusion of ventricular and normal beat |
| / | Q | Paced beat |
| f | Fusion of paced and normal beat | |
| Q | Unclassifiable beat |
| Record ID | Dataset Assignment |
|---|---|
| A00022 | Training |
| A00034 | Test |
| A00056 | Training |
| A00106 | Test |
| … | … |
| Metric (%) | Overall | Labels | ||||
|---|---|---|---|---|---|---|
| N | S | V | F | Q | ||
| Accuracy | 89.481 | 76.866 | 83.799 | 92.307 | 95.533 | 98.901 |
| Sensitivity | 55.827 | 88.858 | 0 | 89.006 | 3.608 | 97.664 |
| Specificity | 92.322 | 70.513 | 100 | 93.435 | 98.442 | 99.223 |
| Metric (%) | Overall |
|---|---|
| Accuracy | 82.745 |
| Sensitivity | 76.173 |
| Specificity | 88.945 |
| Metric (%) | Overall | Labels | |||
|---|---|---|---|---|---|
| S | V | F | Q | ||
| Accuracy | 95.391 | 94.981 | 94.219 | 93.977 | 98.391 |
| Sensitivity | 83.775 | 87.506 | 92.019 | 59.021 | 96.553 |
| Specificity | 97.001 | 97.443 | 95.622 | 95.698 | 99.240 |
| Metric (%) | Overall | Labels | ||||
|---|---|---|---|---|---|---|
| N | S | V | F | Q | ||
| Accuracy | 91.385 | 80.898 | 89.500 | 92.876 | 96.149 | 97.502 |
| Sensitivity | 61.959 | 89.817 | 45.583 | 82.516 | 0 | 91.881 |
| Specificity | 93.747 | 76.173 | 97.990 | 96.415 | 99.192 | 98.964 |
| Method | Overall Metrics (%) | ||
|---|---|---|---|
| Accuracy | Sensitivity | Specificity | |
| Baseline Model | 89.841 | 55.827 | 92.322 |
| Composite Model | 91.385 | 61.959 | 93.747 |
| Label | Heartbeat Count |
|---|---|
| N | 18,801 |
| S | 1392 |
| V | 135 |
| F | 4 |
| Q | 131 |
| Metric (%) | Overall | Labels | ||||
|---|---|---|---|---|---|---|
| N | S | V | F | Q | ||
| Accuracy | 74.622 | 37.350 | 93.198 | 44.036 | 99.047 | 99.477 |
| Sensitivity | 33.758 | 38.860 | 0 | 94.815 | 0 | 35.115 |
| Specificity | 72.587 | 20.277 | 100 | 43.698 | 99.066 | 99.892 |
| Metric (%) | Overall | Labels | ||||
|---|---|---|---|---|---|---|
| N | S | V | F | Q | ||
| Accuracy | 90.948 | 77.555 | 93.198 | 91.360 | 93.051 | 99.575 |
| Sensitivity | 44.898 | 83.203 | 0 | 97.778 | 0 | 43.512 |
| Specificity | 79.596 | 13.658 | 100 | 91.317 | 93.069 | 99.936 |
| Method | Overall Metrics (%) | ||
|---|---|---|---|
| Accuracy | Sensitivity | Specificity | |
| Baseline Model | 74.622 | 33.758 | 72.587 |
| Composite Model | 90.948 | 44.898 | 79.596 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chew, K.T.; Raman, V.; Then, P.H.H. Remote Arrhythmia Detection for Eldercare in Malaysia. Sensors 2021, 21, 8197. https://doi.org/10.3390/s21248197
Chew KT, Raman V, Then PHH. Remote Arrhythmia Detection for Eldercare in Malaysia. Sensors. 2021; 21(24):8197. https://doi.org/10.3390/s21248197
Chicago/Turabian StyleChew, Kevin Thomas, Valliappan Raman, and Patrick Hang Hui Then. 2021. "Remote Arrhythmia Detection for Eldercare in Malaysia" Sensors 21, no. 24: 8197. https://doi.org/10.3390/s21248197
APA StyleChew, K. T., Raman, V., & Then, P. H. H. (2021). Remote Arrhythmia Detection for Eldercare in Malaysia. Sensors, 21(24), 8197. https://doi.org/10.3390/s21248197






