Advanced Ultrasound and Photoacoustic Imaging in Cardiology
Abstract
1. Introduction
2. Advanced US Imaging in Cardiology and DL Techniques
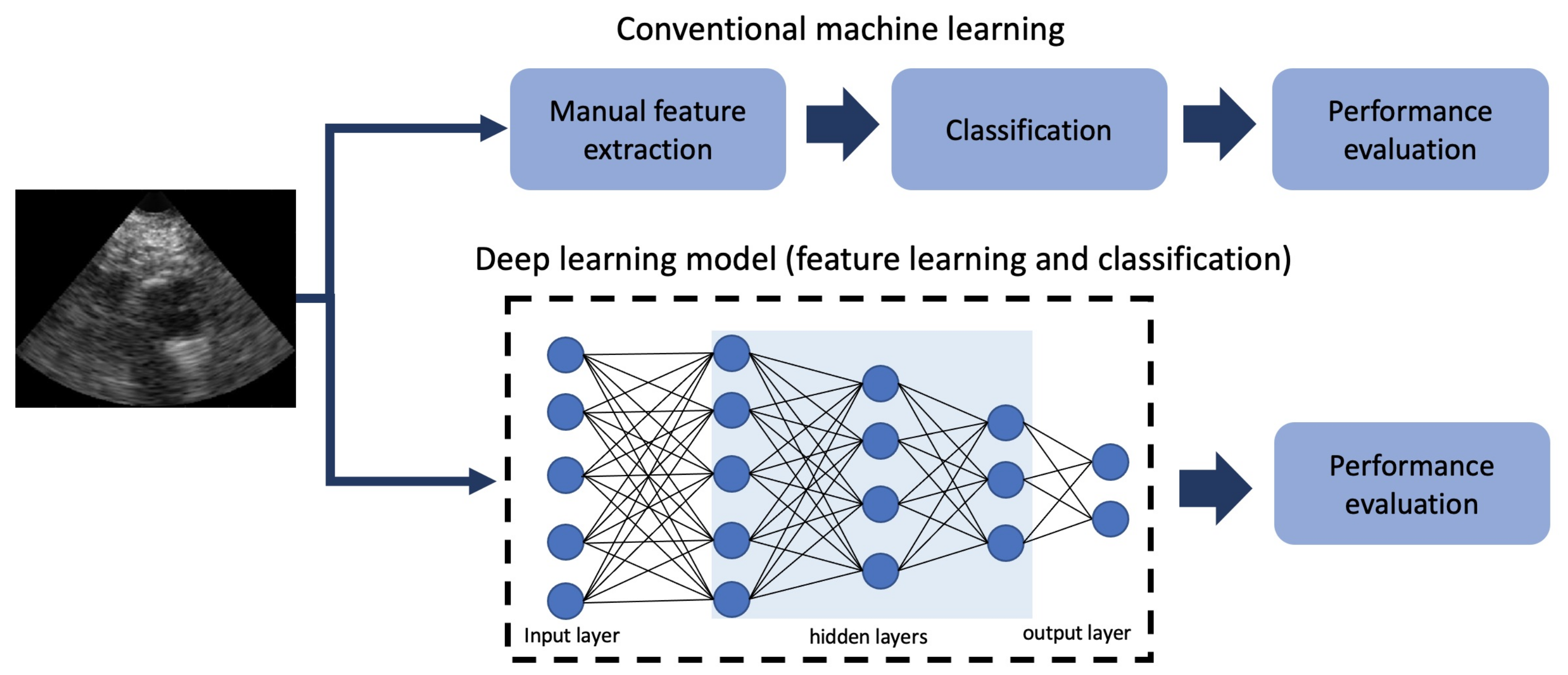
2.1. DL Techniques in US Imaging in Cardiology
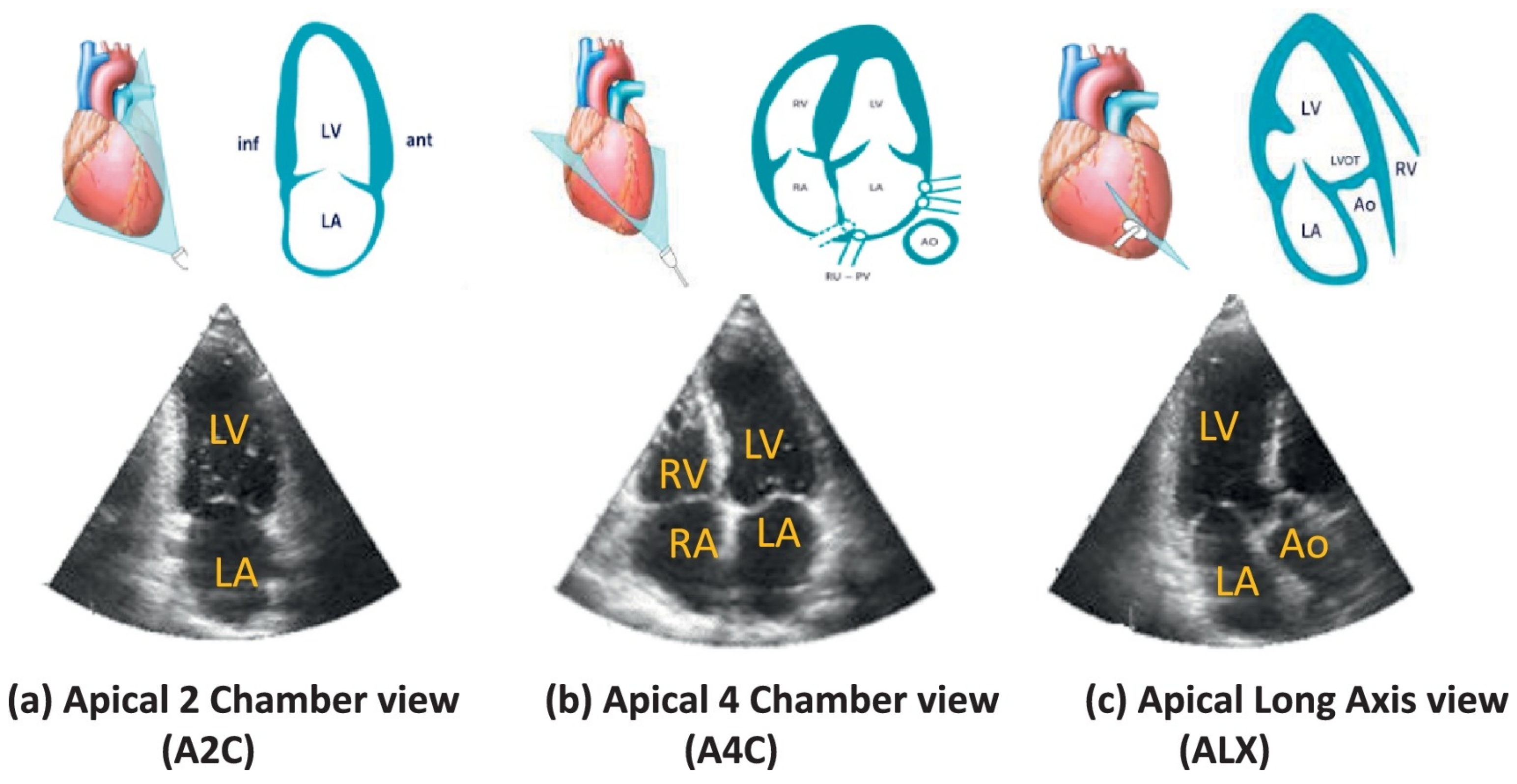
2.1.1. Advanced Techniques for Cardiac Viewpoint Classification
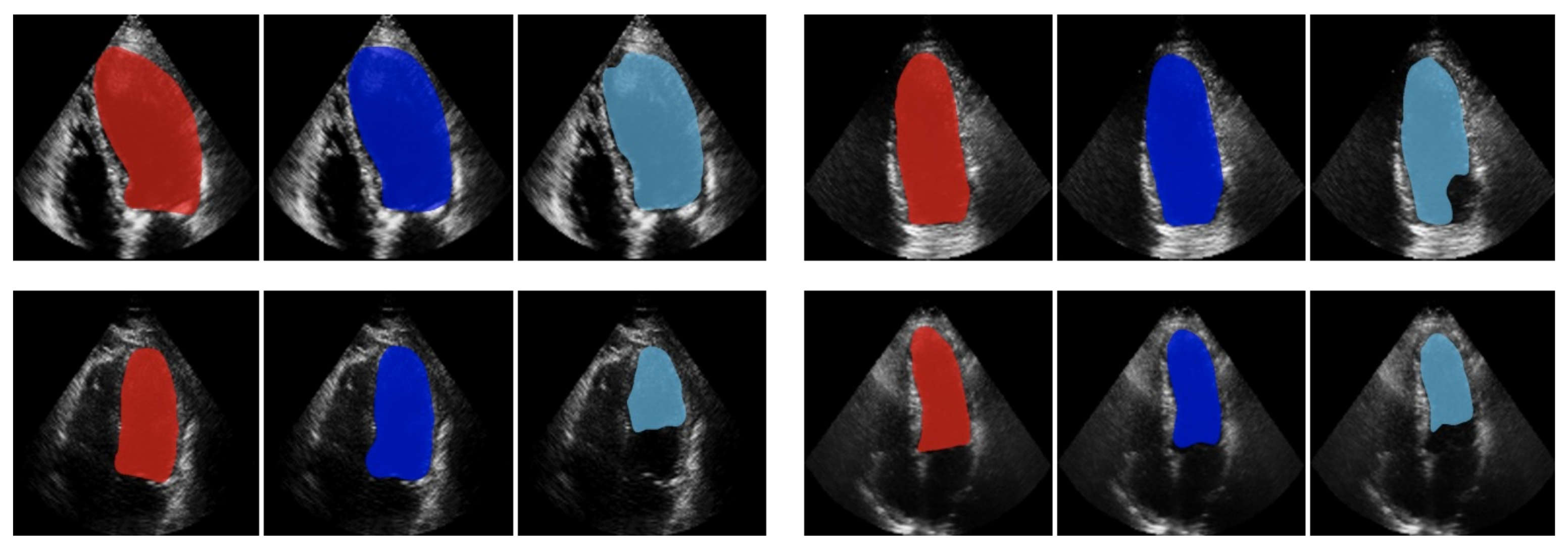
2.1.2. Advanced Techniques in US Imaging to Improve Left Ventricle Segmentation
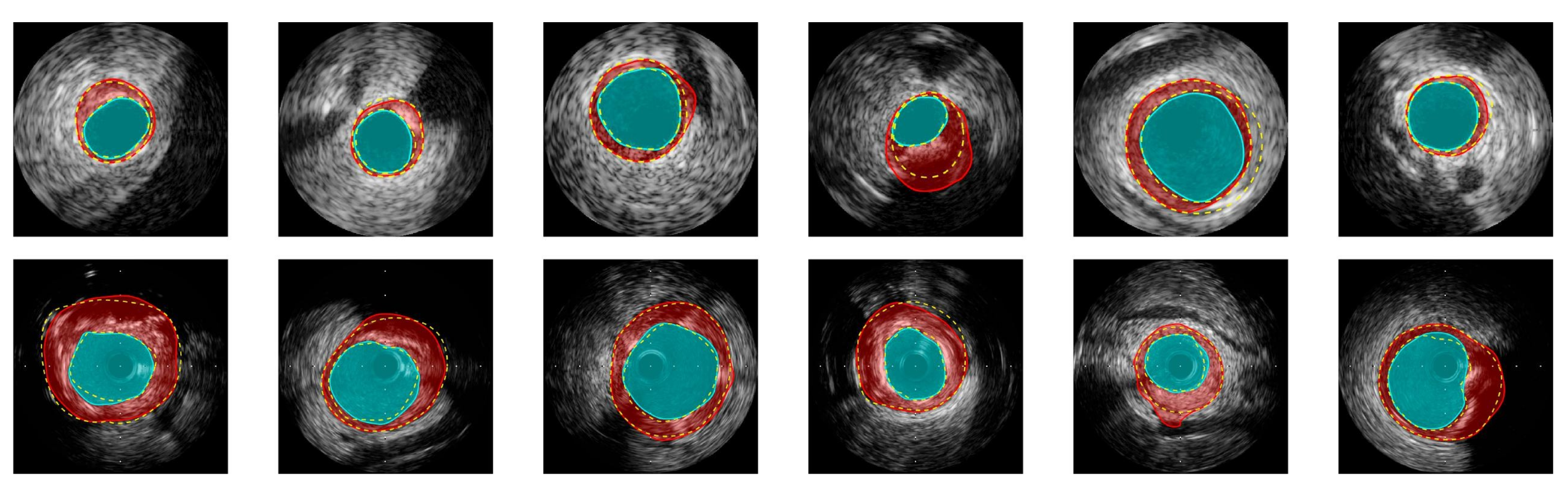
2.1.3. Advances in Intravascular Ultrasound (IVUS) Image Segmentation and Characterization
2.1.4. Advances of Point of Care Ultrasound (POCUS)
3. PA Imaging and DL Techniques in Cardiology
3.1. The Development of PA Imaging Techniques in Cardiology
3.2. Intravascular PA Imaging of Vulnerable Atherosclerotic Plaques
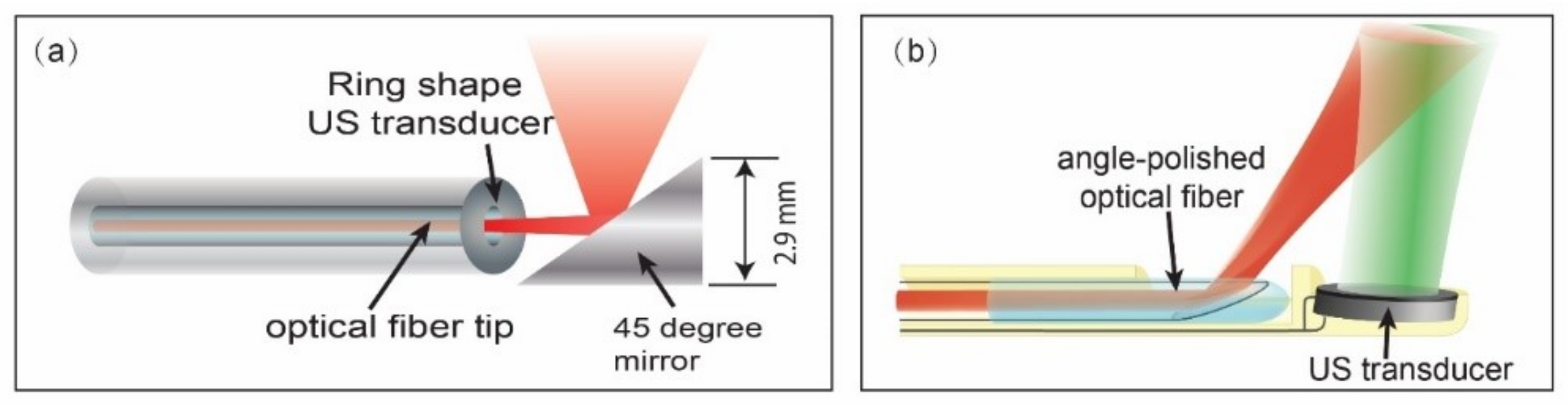
3.2.1. IVPA Imaging Catheter Development
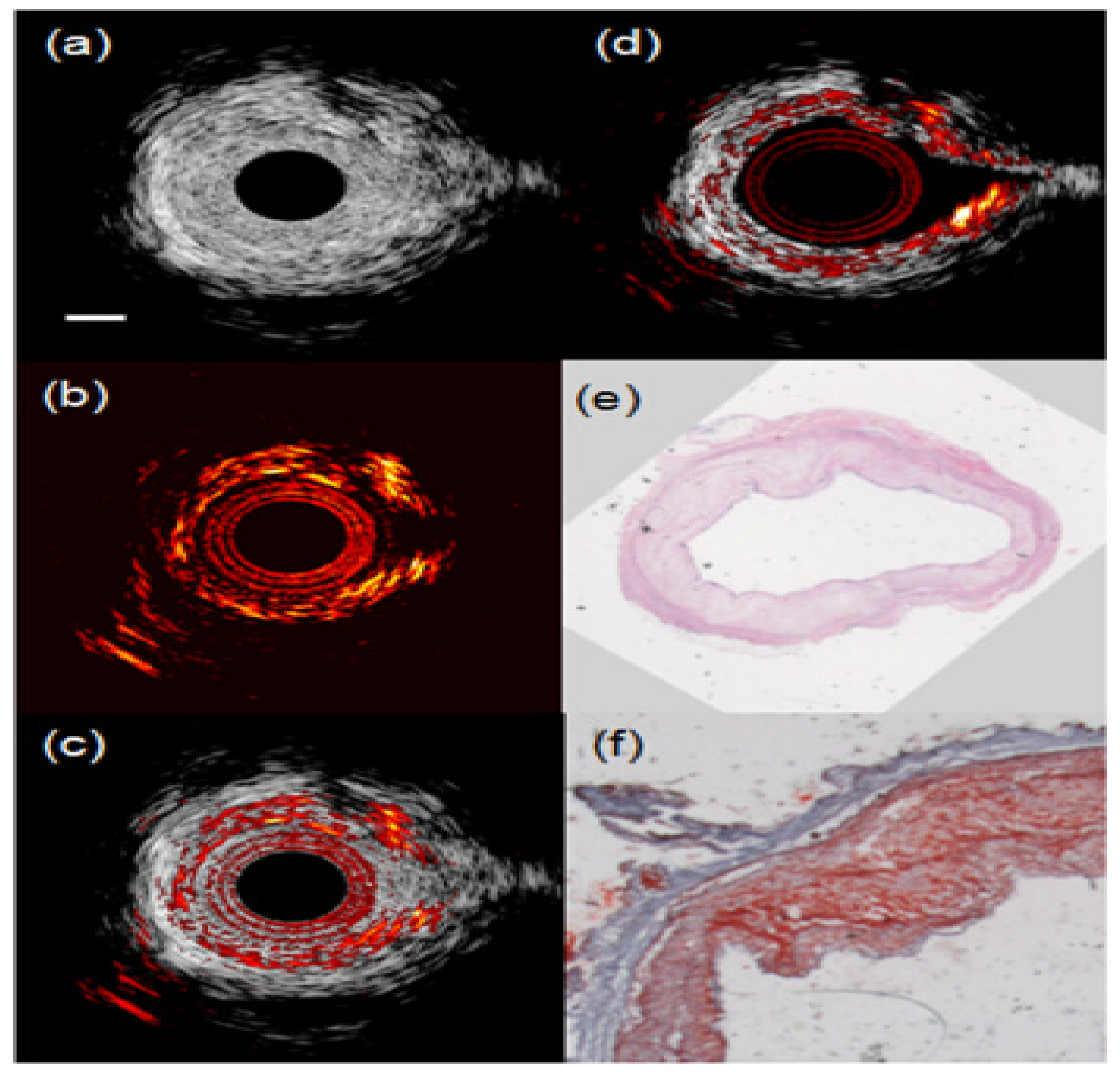
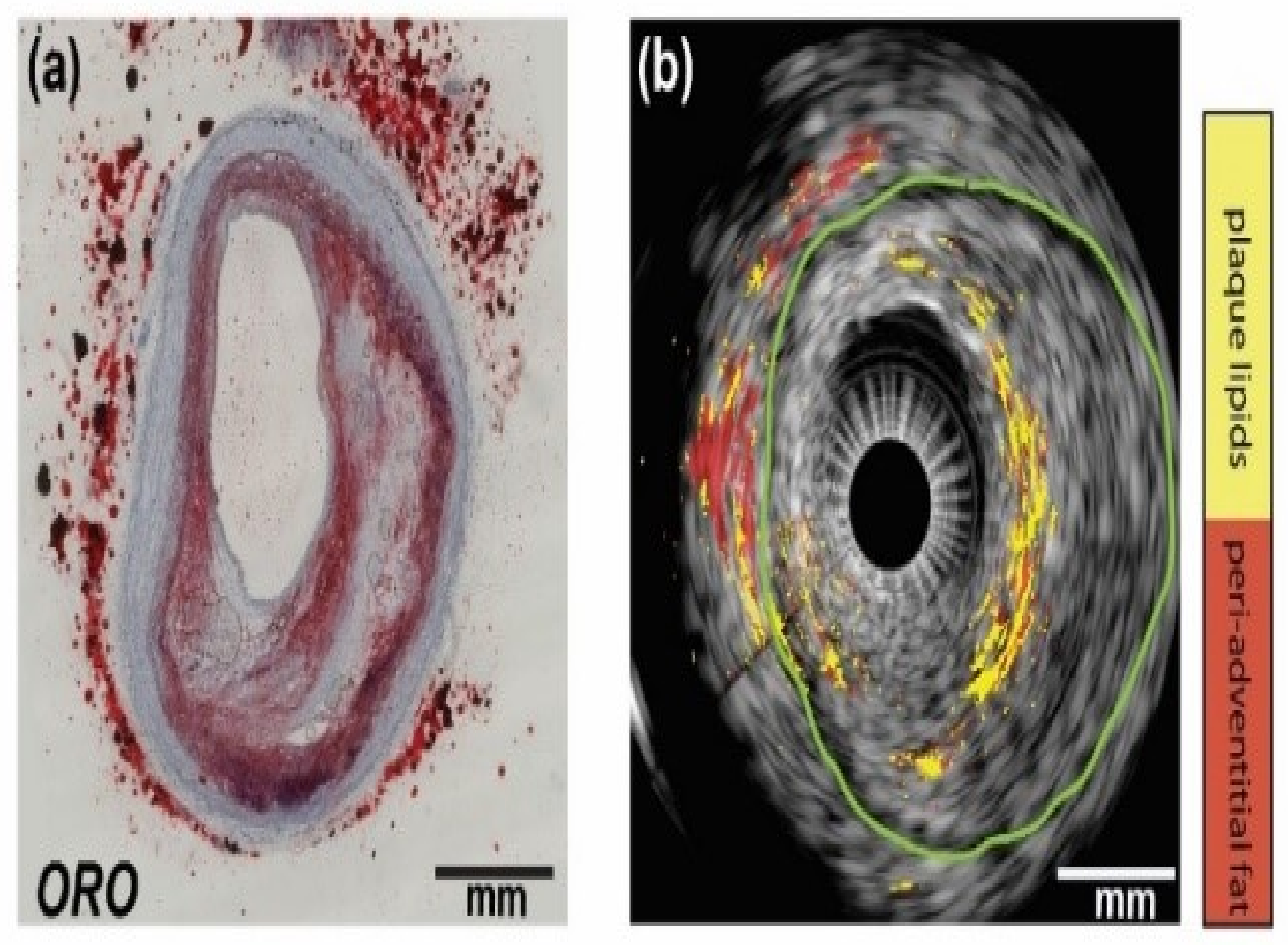
3.2.2. IVPA Imaging of Diverse Compositions in Vulnerable Plaques
3.2.3. Towards In Vivo IVPA Imaging of Vulnerable Atherosclerotic Plaques
3.3. Non-Invasive PA Imaging for Cardiovascular Applications
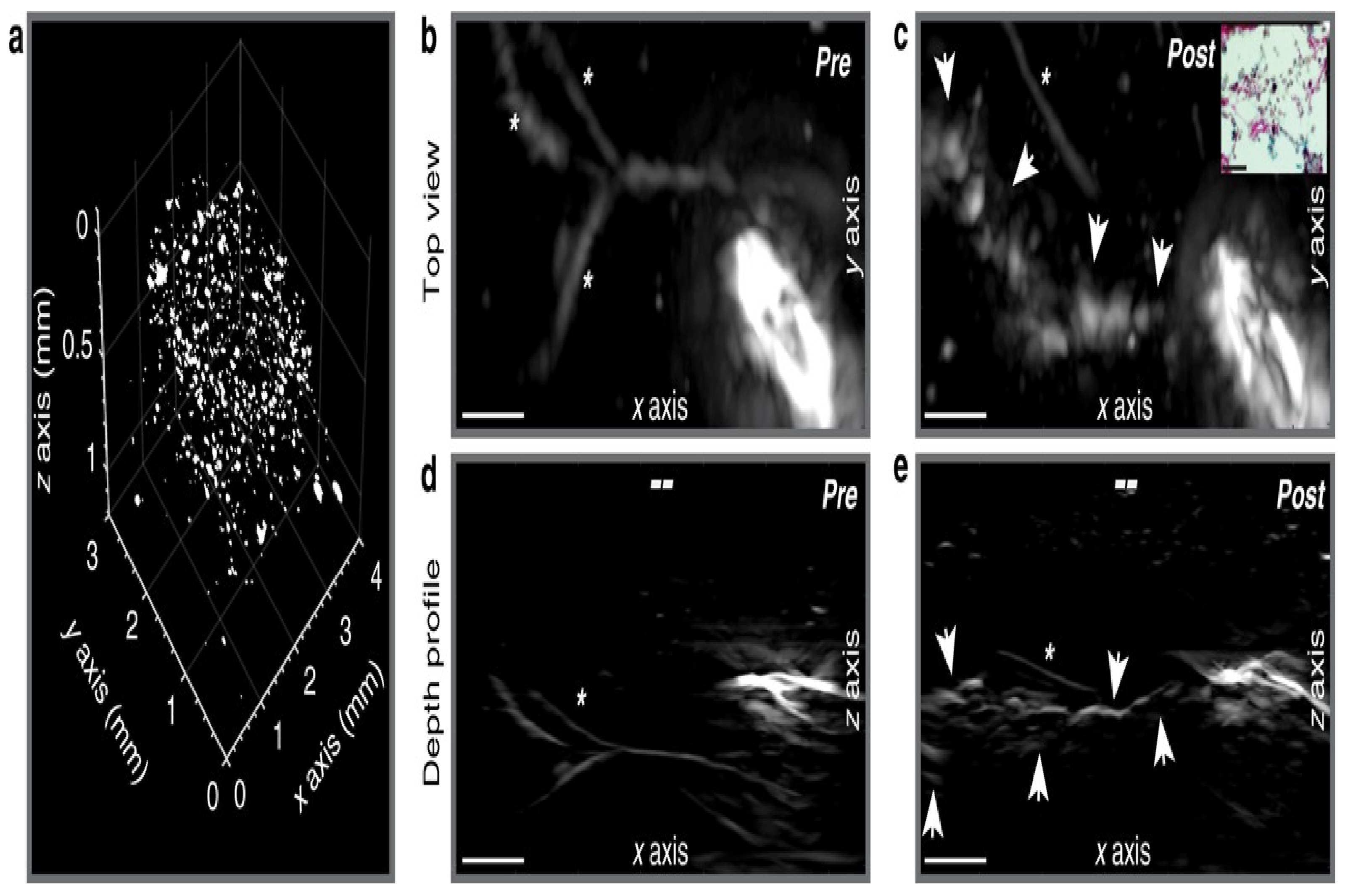
3.4. PA Imaging of Cardiac Arrhythmia
3.5. Application of DL in PA Imaging in Cardiology
4. Discussion and Future Opportunities
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Cardiovascular Disease Programme; Noncommunicable Disease and Mental Health Cluster. In Integrated Management of Cardiovascular Risk; World Health Organization: Geneva, Switzerland, 2002; ISBN 9241562242. [Google Scholar]
- Tarride, J.E.; Lim, M.; DesMeules, M.; Luo, W.; Burke, N.; O’Reilly, D.; Bowen, J.; Goeree, R. A review of the cost of cardiovascular disease. Can. J. Cardiol. 2009, 25, e195–e202. [Google Scholar] [CrossRef]
- Karlas, A.; Fasoula, N.A.; Paul-Yuan, K.; Reber, J.; Kallmayer, M.; Bozhko, D.; Seeger, M.; Eckstein, H.H.; Wildgruber, M.; Ntziachristos, V. Cardiovascular optoacoustics: From mice to men—A review. Photoacoustics 2019, 14, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Dave, J.K.; Mc Donald, M.E.; Mehrotra, P.; Kohut, A.R.; Eisenbrey, J.R.; Forsberg, F. Recent technological advancements in cardiac ultrasound imaging. Ultrasonics 2018, 84, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Beard, P. Biomedical photoacoustic imaging. Interface Focus 2011, 1, 602–631. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.T.; Laufer, J.G.; Beard, P.C.; Arridge, S.R. Quantitative spectroscopic photoacoustic imaging: A review. J. Biomed. Opt. 2012, 17, 061202. [Google Scholar] [CrossRef]
- Wang, L.V.; Hu, S. Photoacoustic tomography: In vivo imaging from organelles to organs. Science 2012, 335, 1458–1462. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Johnson, L.A.; Liu, S.; Moons, D.S.; Ma, T.; Zhou, Q.; Rice, M.D.; Ni, J.; Wang, X.; Higgins, P.D.; et al. Characterizing intestinal inflammation and fibrosis in Crohn’s disease by photoacoustic imaging: Feasibility study. Biomed. Opt. Express 2016, 7, 2837–2848. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Springeling, G.; Lovrak, M.; Mastik, F.; Iskander-Rizk, S.; Wang, T.; Van Beusekom, H.M.; Van Der Steen, A.; Van Soest, G. Real-time volumetric lipid imaging in vivo by intravascular photoacoustics at 20 frames per second. Biomed. Opt. Express 2017, 8, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Bengio, Y.; LeCun, Y. Scaling learning algorithms towards AI. Large-Scale Kernel Mach. 2007, 34, 1–41. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, UK, 2016; Volume 1. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Van Boxtel, J.; Vousten, V.R.; Pluim, J.; Rad, N.M. Hybrid Deep Neural Network for Brachial Plexus Nerve Segmentation in Ultrasound Images. arXiv 2021, arXiv:2106.00373. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Awasthi, N.; Jain, G.; Kalva, S.K.; Pramanik, M.; Yalavarthy, P.K. Deep Neural Network-Based Sinogram Super-Resolution and Bandwidth Enhancement for Limited-Data Photoacoustic Tomography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2020, 67, 2660–2673. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, N.; Prabhakar, K.R.; Kalva, S.K.; Pramanik, M.; Babu, R.V.; Yalavarthy, P.K. PA-Fuse: Deep supervised approach for the fusion of photoacoustic images with distinct reconstruction characteristics. Biomed. Opt. Express 2019, 10, 2227–2243. [Google Scholar] [CrossRef] [PubMed]
- Edler, I.; Hertz, C.H. The use of ultrasonic reflectoscope for the continuous recording of the movements of heart walls. Clin. Physiol. Funct. Imaging 2004, 24, 118–136. [Google Scholar] [CrossRef]
- Chesler, E. Ultrasound in cardiology. S. Afr. Med. J. 1973, 47, 1625–1637. [Google Scholar] [PubMed]
- Cobbold, R.S. Foundations of Biomedical Ultrasound; Oxford University Press: Oxford, UK, 2006. [Google Scholar]
- Provost, J.; Papadacci, C.; Arango, J.E.; Imbault, M.; Fink, M.; Gennisson, J.L.; Tanter, M.; Pernot, M. 3D ultrafast ultrasound imaging in vivo. Phys. Med. Biol. 2014, 59, L1. [Google Scholar] [CrossRef]
- Cikes, M.; Tong, L.; Sutherland, G.R.; D’hooge, J. Ultrafast cardiac ultrasound imaging: Technical principles, applications, and clinical benefits. JACC Cardiovasc. Imaging 2014, 7, 812–823. [Google Scholar] [CrossRef] [PubMed]
- Villemain, O.; Baranger, J.; Friedberg, M.K.; Papadacci, C.; Dizeux, A.; Messas, E.; Tanter, M.; Pernot, M.; Mertens, L. Ultrafast ultrasound imaging in pediatric and adult cardiology: Techniques, applications, and perspectives. JACC Cardiovasc. Imaging 2020, 13, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Wells, P. Ultrasonic colour flow imaging. Phys. Med. Biol. 1994, 39, 2113. [Google Scholar] [CrossRef] [PubMed]
- Tee, M.; Noble, J.A.; Bluemke, D.A. Imaging techniques for cardiac strain and deformation: Comparison of echocardiography, cardiac magnetic resonance and cardiac computed tomography. Expert Rev. Cardiovasc. Ther. 2013, 11, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Bercoff, J.; Tanter, M.; Fink, M. Supersonic shear imaging: A new technique for soft tissue elasticity mapping. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2004, 51, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, A.F.; Kaspar, M.; Staub, D. Contrast-enhanced ultrasound: Clinical applications in patients with atherosclerosis. Int. J. Cardiovasc. Imaging 2016, 32, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Versluis, M.; Stride, E.; Lajoinie, G.; Dollet, B.; Segers, T. Ultrasound contrast agent modeling: A review. Ultrasound Med. Biol. 2020, 46, 2117–2144. [Google Scholar] [CrossRef]
- Raffort, J.; Adam, C.; Carrier, M.; Ballaith, A.; Coscas, R.; Jean-Baptiste, E.; Hassen-Khodja, R.; Chakfé, N.; Lareyre, F. Artificial intelligence in abdominal aortic aneurysm. J. Vasc. Surg. 2020, 72, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.C.; Then, P.H. Deep learning for cardiac computer-aided diagnosis: Benefits, issues & solutions. Mhealth 2017, 3, 45. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. Handb. Brain Theory Neural Netw. 1995, 3361, 1995. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Wharton, G.; Steeds, R.; Allen, J.; Phillips, H.; Jones, R.; Kanagala, P.; Lloyd, G.; Masani, N.; Mathew, T.; Oxborough, D.; et al. A minimum dataset for a standard adult transthoracic echocardiogram: A guideline protocol from the British Society of Echocardiography. Echo Res. Pract. 2015, 2, G9–G24. [Google Scholar] [CrossRef]
- Ebadollahi, S.; Chang, S.F.; Wu, H. Automatic view recognition in echocardiogram videos using parts-based representation. In Proceedings of the 2004 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 27 June–2 July 2004. [Google Scholar]
- Otey, M.; Bi, J.; Krishna, S.; Rao, B.; Stoeckel, J.; Katz, A.; Han, J.; Parthasarathy, S. Automatic view recognition for cardiac ultrasound images. In Proceedings of the 1st international workshop on computer vision for intravascular and intracardiac imaging, Copenhagen, Denmark, 6 October 2006; pp. 187–194. [Google Scholar]
- Agarwal, D.; Shriram, K.; Subramanian, N. Automatic view classification of echocardiograms using histogram of oriented gradients. In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging, San Francisco, CA, USA, 7–11 April 2013; pp. 1368–1371. [Google Scholar]
- Wu, H.; Bowers, D.M.; Huynh, T.T.; Souvenir, R. Echocardiogram view classification using low-level features. In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging, San Francisco, CA, USA, 7–11 April 2013; pp. 752–755. [Google Scholar]
- Qian, Y.; Wang, L.; Wang, C.; Gao, X. The synergy of 3D SIFT and sparse codes for classification of viewpoints from echocardiogram videos. In Proceedings of the MICCAI International Workshop on Medical Content-Based Retrieval for Clinical Decision Support, Nice, France, 1 October 2012; Springer: Berlin/Heidelberg, Germany; pp. 68–79. [Google Scholar]
- Aschkenasy, S.V.; Jansen, C.; Osterwalder, R.; Linka, A.; Unser, M.; Marsch, S.; Hunziker, P. Unsupervised image classification of medical ultrasound data by multiresolution elastic registration. Ultrasound Med. Biol. 2006, 32, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.K.; Park, J.; Georgescu, B.; Comaniciu, D.; Simopoulos, C.; Otsuki, J. Image-based multiclass boosting and echocardiographic view classification. In Proceedings of the 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’06), New York, NY, USA, 17–22 June 2006; Volume 2, pp. 1559–1565. [Google Scholar]
- Park, J.H.; Zhou, S.K.; Simopoulos, C.; Otsuki, J.; Comaniciu, D. Automatic cardiac view classification of echocardiogram. In Proceedings of the 2007 IEEE 11th International Conference on Computer Vision, Rio de Janeiro, Brazil, 14–21 October 2007; pp. 1–8. [Google Scholar]
- Khamis, H.; Zurakhov, G.; Azar, V.; Raz, A.; Friedman, Z.; Adam, D. Automatic apical view classification of echocardiograms using a discriminative learning dictionary. Med. Image Anal. 2017, 36, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Zhou, S.K.; Simopoulos, C.; Comaniciu, D. AutoGate: Fast and automatic Doppler gate localization in B-mode echocardiogram. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, New York, NY, USA, 6–10 September; Springer: Berlin/Heidelberg, Germany; pp. 230–237.
- Penatti, O.A.; Werneck, R.d.O.; de Almeida, W.R.; Stein, B.V.; Pazinato, D.V.; Júnior, P.R.M.; Torres, R.d.S.; Rocha, A. Mid-level image representations for real-time heart view plane classification of echocardiograms. Comput. Biol. Med. 2015, 66, 66–81. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Noble, J.A. Fetal ultrasound image classification using a bag-of-words model trained on sonographers’ eye movements. Procedia Comput. Sci. 2016, 90, 157–162. [Google Scholar] [CrossRef]
- Gao, X.; Li, W.; Loomes, M.; Wang, L. A fused deep learning architecture for viewpoint classification of echocardiography. Inf. Fusion 2017, 36, 103–113. [Google Scholar] [CrossRef]
- Madani, A.; Arnaout, R.; Mofrad, M.; Arnaout, R. Fast and accurate view classification of echocardiograms using deep learning. NPJ Dig. Med. 2018, 1, 1–8. [Google Scholar] [CrossRef]
- Vaseli, H.; Liao, Z.; Abdi, A.H.; Girgis, H.; Behnami, D.; Luong, C.; Dezaki, F.T.; Dhungel, N.; Rohling, R.; Gin, K.; et al. Designing lightweight deep learning models for echocardiography view classification. In Proceedings of the Medical Imaging 2019: Image-Guided Procedures, Robotic Interventions, and Modeling, San Diego, CA, USA, 16–21 February 2019; Volume 10951, p. 109510F. [Google Scholar]
- Mignotte, M.; Meunier, J.; Tardif, J.C. Endocardial boundary e timation and tracking in echocardiographic images using deformable template and markov random fields. Pattern Anal. Appl. 2001, 4, 256–271. [Google Scholar] [CrossRef]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Khellaf, F.; Leclerc, S.; Voorneveld, J.D.; Bandaru, R.S.; Bosch, J.G.; Bernard, O. Left ventricle segmentation in 3D ultrasound by combining structured random forests with active shape models. In Proceedings of the Medical Imaging 2018: Image Processing, International Society for Optics and Photonics, Houston, TX, USA, 11–13 February 2018; Volume 10574, p. 105740J. [Google Scholar]
- Georgescu, B.; Zhou, X.S.; Comaniciu, D.; Gupta, A. Database-guided segmentation of anatomical structures with complex appearance. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–26 June 2005; Volume 2, pp. 429–436. [Google Scholar]
- Carneiro, G.; Nascimento, J.C. Incremental on-line semi-supervised learning for segmenting the left ventricle of the heart from ultrasound data. In Proceedings of the 2011 International Conference on Computer Vision, Barcelona, Spain, 6–13 November 2011; pp. 1700–1707. [Google Scholar]
- Martinez, H.P.; Bengio, Y.; Yannakakis, G.N. Learning deep physiological models of affect. IEEE Comput. Intell. Mag. 2013, 8, 20–33. [Google Scholar] [CrossRef]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep learning for cardiac image segmentation: A review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 12 June 2015; pp. 3431–3440. [Google Scholar]
- Smistad, E.; Østvik, A. 2D left ventricle segmentation using deep learning. In Proceedings of the 2017 IEEE International Ultrasonics Symposium (IUS), Washington, DC, USA, 6–9 September 2017; pp. 1–4. [Google Scholar]
- Zyuzin, V.; Sergey, P.; Mukhtarov, A.; Chumarnaya, T.; Solovyova, O.; Bobkova, A.; Myasnikov, V. Identification of the left ventricle endocardial border on two-dimensional ultrasound images using the convolutional neural network Unet. In Proceedings of the 2018 Ural Symposium on Biomedical Engineering, Radioelectronics and Information Technology (USBEREIT), Yekaterinburg, Russia, 7–8 May 2018; pp. 76–78. [Google Scholar]
- Yu, L.; Guo, Y.; Wang, Y.; Yu, J.; Chen, P. Segmentation of fetal left ventricle in echocardiographic sequences based on dynamic convolutional neural networks. IEEE Trans. Biomed. Eng. 2016, 64, 1886–1895. [Google Scholar] [CrossRef]
- Zyuzin, V.; Chumarnaya, T. Comparison of Unet architectures for segmentation of the left ventricle endocardial border on two-dimensional ultrasound images. In Proceedings of the 2019 Ural Symposium on Biomedical Engineering, Radioelectronics and Information Technology (USBEREIT), Yekaterinburg, Russia, 25–26 April 2019; pp. 110–113. [Google Scholar]
- Ahn, S.S.; Ta, K.; Thorn, S.; Langdon, J.; Sinusas, A.J.; Duncan, J.S. Multi-frame Attention Network for Left Ventricle Segmentation in 3D Echocardiography. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; pp. 348–357. [Google Scholar]
- Smistad, E.; Salte, I.M.; Dalen, H.; Lovstakken, L. Real-time temporal coherent left ventricle segmentation using convolutional LSTMs. In Proceedings of the IEEE International Ultrasonics Symposium, Virtual Symposium, 11–16 September 2021. [Google Scholar]
- Carneiro, G.; Nascimento, J.C.; Freitas, A. The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Trans. Image Process. 2011, 21, 968–982. [Google Scholar] [CrossRef] [PubMed]
- Osher, S.; Sethian, J.A. Fronts propagating with curvature-dependent speed: Algorithms based on Hamilton-Jacobi formulations. J. Comput. Phys. 1988, 79, 12–49. [Google Scholar] [CrossRef]
- Jafari, M.H.; Girgis, H.; Liao, Z.; Behnami, D.; Abdi, A.; Vaseli, H.; Luong, C.; Rohling, R.; Gin, K.; Tsang, T.; et al. A unified framework integrating recurrent fully-convolutional networks and optical flow for segmentation of the left ventricle in echocardiography data. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; pp. 29–37. [Google Scholar]
- Carneiro, G.; Nascimento, J.; Freitas, A. Robust left ventricle segmentation from ultrasound data using deep neural networks and efficient search methods. In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, The Netherlands, 14–17 April 2010; pp. 1085–1088. [Google Scholar]
- Nascimento, J.C.; Carneiro, G. Non-rigid segmentation using sparse low dimensional manifolds and deep belief networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 24–27 June 2014; pp. 288–295. [Google Scholar]
- Nascimento, J.C.; Carneiro, G. One shot segmentation: Unifying rigid detection and non-rigid segmentation using elastic regularization. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 42, 3054–3070. [Google Scholar] [CrossRef]
- Veni, G.; Moradi, M.; Bulu, H.; Narayan, G.; Syeda-Mahmood, T. Echocardiography segmentation based on a shape-guided deformable model driven by a fully convolutional network prior. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 898–902. [Google Scholar]
- Oktay, O.; Ferrante, E.; Kamnitsas, K.; Heinrich, M.; Bai, W.; Caballero, J.; Cook, S.A.; De Marvao, A.; Dawes, T.; O‘Regan, D.P.; et al. Anatomically constrained neural networks (ACNNs): Application to cardiac image enhancement and segmentation. IEEE Trans. Med. Imaging 2017, 37, 384–395. [Google Scholar] [CrossRef]
- Bernard, O.; Bosch, J.G.; Heyde, B.; Alessandrini, M.; Barbosa, D.; Camarasu-Pop, S.; Cervenansky, F.; Valette, S.; Mirea, O.; Bernier, M.; et al. Standardized evaluation system for left ventricular segmentation algorithms in 3D echocardiography. IEEE Trans. Med. Imaging 2015, 35, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, G.; Nascimento, J.C. The use of on-line co-training to reduce the training set size in pattern recognition methods: Application to left ventricle segmentation in ultrasound. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 948–955. [Google Scholar]
- Ta, K.; Ahn, S.S.; Lu, A.; Stendahl, J.C.; Sinusas, A.J.; Duncan, J.S. A semi-supervised joint learning approach to left ventricular segmentation and motion tracking in echocardiography. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1734–1737. [Google Scholar]
- Ta, K.; Ahn, S.S.; Stendahl, J.C.; Sinusas, A.J.; Duncan, J.S. A Semi-supervised Joint Network for Simultaneous Left Ventricular Motion Tracking and Segmentation in 4D Echocardiography. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 468–477. [Google Scholar]
- Jafari, M.H.; Girgis, H.; Abdi, A.H.; Liao, Z.; Pesteie, M.; Rohling, R.; Gin, K.; Tsang, T.; Abolmaesumi, P. Semi-supervised learning for cardiac left ventricle segmentation using conditional deep generative models as prior. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 649–652. [Google Scholar]
- Parrillo, J.E.; Dellinger, R.P. Critical Care Medicine e-Book: Principles of Diagnosis and Management in the Adult; Elsevier Health Sciences: Amsterdam, The Netherlands, 2018; ISBN 978-0-323-44676-1. [Google Scholar]
- Garcìa-Garcìa, H.M.; Gogas, B.D.; Serruys, P.W.; Bruining, N. IVUS-based imaging modalities for tissue characterization: Similarities and differences. Int. J. Cardiovasc. Imaging 2011, 27, 215–224. [Google Scholar] [CrossRef]
- Yang, J.; Tong, L.; Faraji, M.; Basu, A. IVUS-Net: An intravascular ultrasound segmentation network. In Proceedings of the International Conference on Smart Multimedia, Toulon, France, 24–26 August 2018; pp. 367–377. [Google Scholar]
- Yang, J.; Faraji, M.; Basu, A. Robust segmentation of arterial walls in intravascular ultrasound images using Dual Path U-Net. Ultrasonics 2019, 96, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Hu, Z.; Lin, Q.; Hau, W.K.; Gao, Z.; Zhang, H. An artificial neural network method for lumen and media-adventitia border detection in IVUS. Comput. Med. Imaging Graph. 2017, 57, 29–39. [Google Scholar] [CrossRef]
- Balakrishna, C.; Dadashzadeh, S.; Soltaninejad, S. Automatic detection of lumen and media in the IVUS images using U-Net with VGG16 Encoder. arXiv 2018, arXiv:1806.07554. [Google Scholar]
- Balocco, S.; Gatta, C.; Ciompi, F.; Wahle, A.; Radeva, P.; Carlier, S.; Unal, G.; Sanidas, E.; Mauri, J.; Carillo, X.; et al. Standardized evaluation methodology and reference database for evaluating IVUS image segmentation. Comput. Med. Imaging Graph. 2014, 38, 70–90. [Google Scholar] [CrossRef] [PubMed]
- Rockafellar, R.T.; Wets, R.J.B. Variational Analysis; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; Volume 317. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Bargsten, L.; Riedl, K.A.; Wissel, T.; Brunner, F.J.; Schaefers, K.; Sprenger, J.; Grass, M.; Seiffert, M.; Blankenberg, S.; Schlaefer, A. Tailored methods for segmentation of intravascular ultrasound images via convolutional neural networks. In Proceedings of the Medical Imaging 2021: Ultrasonic Imaging and Tomography, San Diego, CA, USA, 15–20 February 2021; Volume 11602, p. 1160204. [Google Scholar]
- Olender, M.L.; Athanasiou, L.S.; Michalis, L.K.; Fotiadis, D.I.; Edelman, E.R. A Domain Enriched Deep Learning Approach to Classify Atherosclerosis Using Intravascular Ultrasound Imaging. IEEE J. Sel. Top. Signal Process. 2020, 14, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Shen, T.Y.; Chen, C.C.; Chang, W.T.; Lee, P.Y.; Huang, C.C.J. Automatic detection of atherosclerotic plaque and calcification from intravascular ultrasound images by using deep convolutional neural networks. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2021, 68, 1762–1772. [Google Scholar] [CrossRef]
- Junker, R.; Schlebusch, H.; Luppa, P.B. Point-of-care testing in hospitals and primary care. Dtsch. Ärztebl. Int. 2010, 107, 561. [Google Scholar] [CrossRef]
- Killu, K.; Coba, V.; Mendez, M.; Reddy, S.; Adrzejewski, T.; Huang, Y.; Ede, J.; Horst, M. Model point-of-care ultrasound curriculum in an intensive care unit fellowship program and its impact on patient management. Crit. Care Res. Pract. 2014, 2014, 934796. [Google Scholar] [CrossRef]
- Marin, J.R.; Lewiss, R.E.; American Academy of Pediatrics; Committee on Pediatric Emergency Medicine; American College of Emergency Physicians; Pediatric Emergency Medicine Committee. Point-of-care ultrasonography by pediatric emergency medicine physicians. Pediatrics 2015, 135, e1113–e1122. [Google Scholar] [CrossRef]
- Solomon, S.D.; Saldana, F. Point-of-care ultrasound in medical education–stop listening and look. N. Engl. J. Med. 2014, 370, 1083–1085. [Google Scholar] [CrossRef]
- Singh, M.R.; Jackson, J.S.; Newberry, M.A.; Riopelle, C.; Tran, V.H.; PoSaw, L.L. Barriers to point-of-care ultrasound utilization during cardiac arrest in the emergency department: A regional survey of emergency physicians. Am. J. Emerg. Med. 2021, 41, 28–34. [Google Scholar] [CrossRef]
- Kimura, B.J. Point-of-care cardiac ultrasound techniques in the physical examination: Better at the bedside. Heart 2017, 103, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Montinari, M.R.; Minelli, S. The first 200 years of cardiac auscultation and future perspectives. J. Multidiscip. Healthc. 2019, 12, 183. [Google Scholar] [CrossRef]
- Di Bello, V.; La Carrubba, S.; Conte, L.; Fabiani, I.; Posteraro, A.; Antonini-Canterin, F.; Barletta, V.; Nicastro, I.; Mariotti, E.; Severino, S.; et al. Incremental value of pocket-sized echocardiography in addition to physical examination during inpatient cardiology evaluation: A multicenter Italian study (SIEC). Echocardiography 2015, 32, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.C.; Lahham, S.; Maldonado, G.; Klaus, S.; Aish, B.; Sylwanowicz, L.V.; Yanuck, J.; Wilson, S.P.; Shieh, M.; Anderson, C.L.; et al. Hypertrophic cardiomyopathy in youth athletes: Successful screening with point-of-care ultrasound by medical students. J. Ultrasound Med. 2017, 36, 1109–1115. [Google Scholar] [CrossRef]
- Kalagara, H.; Coker, B.; Gerstein, N.S.; Kukreja, P.; Deriy, L.; Pierce, A.; Townsley, M.M. Point of Care Ultrasound (POCUS) for the Cardiothoracic Anesthesiologist. J. Cardiothorac. Vasc. Anesth. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Gaspari, R.; Weekes, A.; Adhikari, S.; Noble, V.E.; Nomura, J.T.; Theodoro, D.; Woo, M.; Atkinson, P.; Blehar, D.; Brown, S.M.; et al. Emergency department point-of-care ultrasound in out-of-hospital and in-ED cardiac arrest. Resuscitation 2016, 109, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.; Stawicki, S.; Evans, D.C.; Bahner, D.; Sparks, S.; Sharpe, R.; Cipolla, J. From FAST to E-FAST: An overview of the evolution of ultrasound-based traumatic injury assessment. Eur. J. Trauma Emerg. Surg. 2016, 42, 119–126. [Google Scholar] [CrossRef]
- Blaivas, M.; Blaivas, L. Are all deep learning architectures alike for point-of-care ultrasound? Evidence from a cardiac image classification model suggests otherwise. J. Ultrasound Med. 2020, 39, 1187–1194. [Google Scholar] [CrossRef]
- Blaivas, M.; Blaivas, L.; Philips, G.; Merchant, R.; Levy, M.; Abbasi, A.; Eickhoff, C.; Shapiro, N.; Corl, K. Development of a deep learning network to classify inferior vena cava collapse to predict fluid responsiveness. J. Ultrasound Med. 2021, 40, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Huh, J.; Ye, J.C. Contrast and Resolution Improvement of POCUS Using Self-consistent CycleGAN. In Domain Adaptation and Representation Transfer, and Affordable Healthcare and AI for Resource Diverse Global Health; Springer: Berlin/Heidelberg, Germany, 2021; pp. 158–167. [Google Scholar]
- Shokoohi, H.; LeSaux, M.A.; Roohani, Y.H.; Liteplo, A.; Huang, C.; Blaivas, M. Enhanced point-of-care ultrasound applications by integrating automated feature-learning systems using deep learning. J. Ultrasound Med. 2019, 38, 1887–1897. [Google Scholar] [CrossRef]
- Blaivas, M.; Arntfield, R.; White, M. DIY AI, deep learning network development for automated image classification in a point-of-care ultrasound quality assurance program. J. Am. Coll. Emerg. Physicians Open 2020, 1, 124–131. [Google Scholar] [CrossRef]
- Cheema, B.S.; Walter, J.; Narang, A.; Thomas, J.D. Artificial intelligence–enabled POCUS in the COVID-19 ICU: A new spin on cardiac ultrasound. Case Rep. 2021, 3, 258–263. [Google Scholar]
- Naghavi, M.; Libby, P.; Falk, E.; Casscells, S.W.; Litovsky, S.; Rumberger, J.; Badimon, J.J.; Stefanadis, C.; Moreno, P.; Pasterkamp, G.; et al. From vulnerable plaque to vulnerable patient: A call for new definitions and risk assessment strategies: Part I. Circulation 2003, 108, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Chen, Z.Q.; Bao, Y.H.; Jiao, L.Q.; Ling, F. Correlation between carotid intraplaque hemorrhage and clinical symptoms: Systematic review of observational studies. Stroke 2007, 38, 2382–2390. [Google Scholar] [CrossRef]
- Cao, Y.; Hui, J.; Kole, A.; Wang, P.; Yu, Q.; Chen, W.; Sturek, M.; Cheng, J.X. High-sensitivity intravascular photoacoustic imaging of lipid–laden plaque with a collinear catheter design. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Jansen, K.; Springeling, G.; van der Steen, A.F.; van Soest, G. Impact of device geometry on the imaging characteristics of an intravascular photoacoustic catheter. Appl. Opt. 2014, 53, 8131–8139. [Google Scholar] [CrossRef] [PubMed]
- Iskander-Rizk, S.; Wu, M.; Springeling, G.; Mastik, F.; Beurskens, R.H.; van der Steen, A.F.; van Soest, G. Catheter design optimization for practical intravascular photoacoustic imaging (IVPA) of vulnerable plaques. In Proceedings of the Diagnostic and Therapeutic Applications of Light in Cardiology 2018, San Francisco, CA, USA, 27–28 January 2018; Volume 10471, p. 1047111. [Google Scholar]
- Li, Y.; Gong, X.; Liu, C.; Lin, R.; Hau, W.; Bai, X.; Song, L. High-speed intravascular spectroscopic photoacoustic imaging at 1000 A-lines per second with a 0.9-mm diameter catheter. J. Biomed. Opt. 2015, 20, 065006. [Google Scholar] [CrossRef]
- Wu, M.; van der Steen, A.F.; Regar, E.; van Soest, G. Emerging technology update intravascular photoacoustic imaging of vulnerable atherosclerotic plaque. Interv. Cardiol. Rev. 2016, 11, 120. [Google Scholar] [CrossRef]
- Jansen, K.; Wu, M.; van der Steen, A.F.; van Soest, G. Lipid detection in atherosclerotic human coronaries by spectroscopic intravascular photoacoustic imaging. Opt. Express 2013, 21, 21472–21484. [Google Scholar] [CrossRef]
- Jansen, K.; Wu, M.; van der Steen, A.F.; van Soest, G. Photoacoustic imaging of human coronary atherosclerosis in two spectral bands. Photoacoustics 2014, 2, 12–20. [Google Scholar] [CrossRef]
- Piao, Z.; Ma, T.; Li, J.; Wiedmann, M.T.; Huang, S.; Yu, M.; Kirk Shung, K.; Zhou, Q.; Kim, C.S.; Chen, Z. High speed intravascular photoacoustic imaging with fast optical parametric oscillator laser at 1.7 μm. Appl. Phys. Lett. 2015, 107, 083701. [Google Scholar] [CrossRef]
- Sethuraman, S.; Amirian, J.H.; Litovsky, S.H.; Smalling, R.W.; Emelianov, S.Y. Ex vivo characterization of atherosclerosis using intravascular photoacoustic imaging. Opt. Express 2007, 15, 16657–16666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, S.; Ji, X.; Zhou, Q.; Xing, D. Characterization of lipid-rich aortic plaques by intravascular photoacoustic tomography: Ex vivo and in vivo validation in a rabbit atherosclerosis model with histologic correlation. J. Am. Coll. Cardiol. 2014, 64, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Su, J.L.; Amirian, J.; Litovsky, S.H.; Smalling, R.; Emelianov, S. Detection of lipid in atherosclerotic vessels using ultrasound-guided spectroscopic intravascular photoacoustic imaging. Opt. Express 2010, 18, 4889–4897. [Google Scholar] [CrossRef] [PubMed]
- Iskander-Rizk, S.; van der Steen, A.F.W.; van Soest, G. Photoacoustic imaging for guidance of interventions in cardiovascular medicine. Phys. Med. Biol. 2019, 64, 16TR01. [Google Scholar] [CrossRef]
- Wang, B.; Karpiouk, A.; Yeager, D.; Amirian, J.; Litovsky, S.; Smalling, R.; Emelianov, S. Intravascular photoacoustic imaging of lipid in atherosclerotic plaques in the presence of luminal blood. Opt. Lett. 2012, 37, 1244–1246. [Google Scholar] [CrossRef]
- Wu, M.; Jansen, K.; van der Steen, A.F.; van Soest, G. Specific imaging of atherosclerotic plaque lipids with two-wavelength intravascular photoacoustics. Biomed. Opt. Express 2015, 6, 3276–3286. [Google Scholar] [CrossRef] [PubMed]
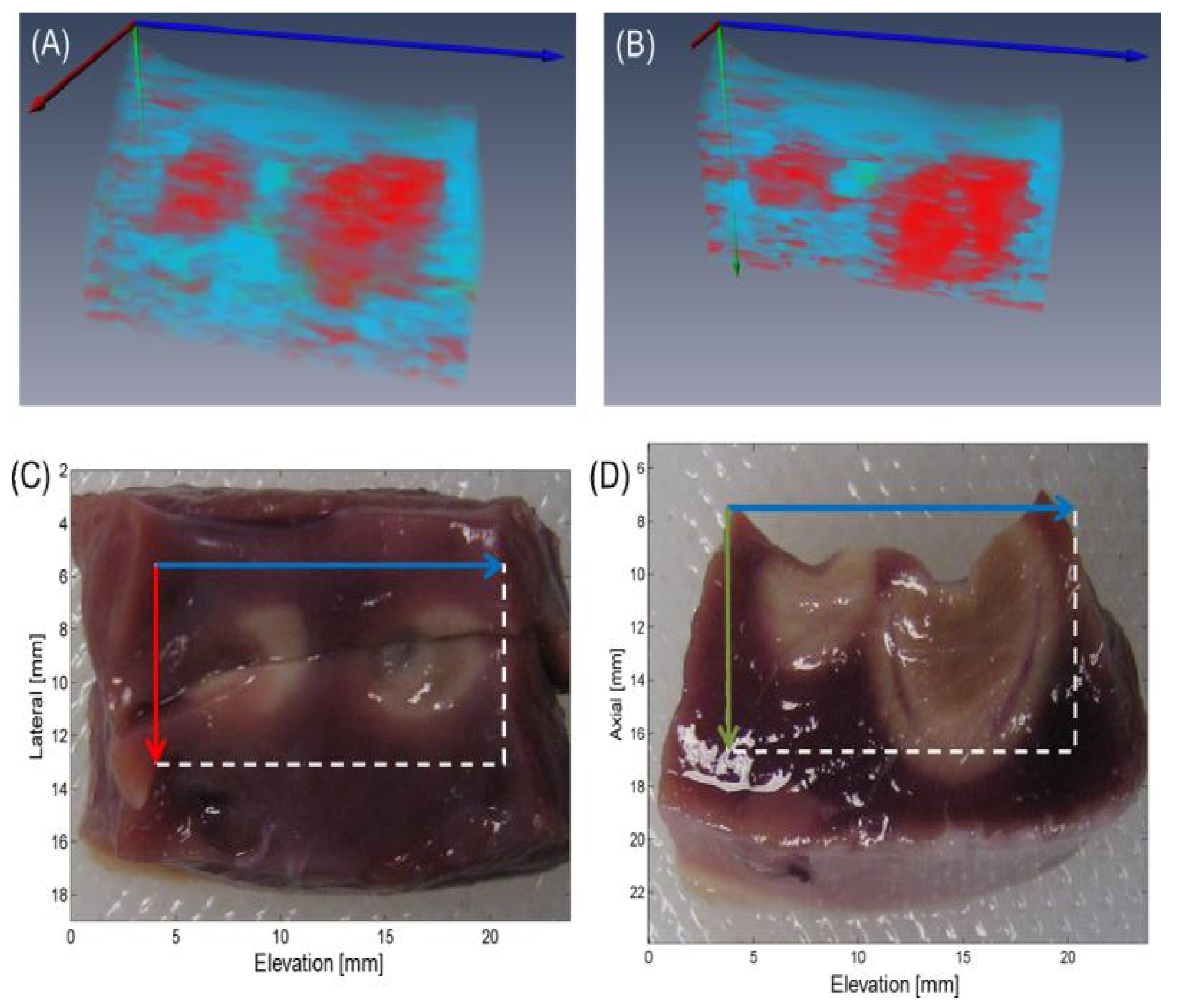
- Iskander-Rizk, S.; Visscher, M.; Moerman, A.M.; Korteland, S.A.; Van der Heiden, K.; Van der Steen, A.F.; Van Soest, G. Micro Spectroscopic Photoacoustic (μsPA) imaging of advanced carotid atherosclerosis. Photoacoustics 2021, 22, 100261. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yantsen, E.; Larson, T.; Karpiouk, A.B.; Sethuraman, S.; Su, J.L.; Sokolov, K.; Emelianov, S.Y. Plasmonic intravascular photoacoustic imaging for detection of macrophages in atherosclerotic plaques. Nano Lett. 2009, 9, 2212–2217. [Google Scholar] [CrossRef]
- Bui, N.Q.; Hlaing, K.K.; Lee, Y.W.; Kang, H.W.; Oh, J. Ex vivo detection of macrophages in atherosclerotic plaques using intravascular ultrasonic-photoacoustic imaging. Phys. Med. Biol. 2016, 62, 501. [Google Scholar]
- Qin, H.; Zhao, Y.; Zhang, J.; Pan, X.; Yang, S.; Xing, D. Inflammation-targeted gold nanorods for intravascular photoacoustic imaging detection of matrix metalloproteinase-2 (MMP2) in atherosclerotic plaques. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1765–1774. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, Y.; Li, Z.; Li, C.; Wang, Q. A novel photoacoustic nanoprobe of ICG@ PEG-Ag 2 S for atherosclerosis targeting and imaging in vivo. Nanoscale 2016, 8, 12531–12539. [Google Scholar] [CrossRef] [PubMed]
- Weidenfeld, I.; Zakian, C.; Duewell, P.; Chmyrov, A.; Klemm, U.; Aguirre, J.; Ntziachristos, V.; Stiel, A.C. Homogentisic acid-derived pigment as a biocompatible label for optoacoustic imaging of macrophages. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Xie, Z.; Shu, C.; Yang, D.; Chen, H.; Chen, C.; Dai, G.; Lam, K.H.; Zhang, J.; Wang, X.; Sheng, Z.; et al. In vivo intravascular photoacoustic imaging at a high speed of 100 frames per second. Biomed. Opt. Express 2020, 11, 6721–6731. [Google Scholar] [CrossRef] [PubMed]
- Arabul, M.U.; Heres, M.; Rutten, M.C.; van Sambeek, M.R.; van de Vosse, F.N.; Lopata, R.G. Toward the detection of intraplaque hemorrhage in carotid artery lesions using photoacoustic imaging. J. Biomed. Opt. 2016, 22, 041010. [Google Scholar] [CrossRef]
- Muller, J.W.; van Hees, R.; van Sambeek, M.; Boutouyrie, P.; Rutten, M.; Brands, P.; Wu, M.; Lopata, R. Towards in vivo photoacoustic imaging of vulnerable plaques in the carotid artery. Biomed. Opt. Express 2021, 12, 4207–4218. [Google Scholar] [CrossRef]
- Neuschmelting, V.; Burton, N.C.; Lockau, H.; Urich, A.; Harmsen, S.; Ntziachristos, V.; Kircher, M.F. Performance of a multispectral optoacoustic tomography (MSOT) system equipped with 2D vs. 3D handheld probes for potential clinical translation. Photoacoustics 2016, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Merčep, E.; Deán-Ben, X.L.; Razansky, D. Imaging of blood flow and oxygen state with a multi-segment optoacoustic ultrasound array. Photoacoustics 2018, 10, 48–53. [Google Scholar] [CrossRef]
- Taruttis, A.; Herzog, E.; Razansky, D.; Ntziachristos, V. Real-time imaging of cardiovascular dynamics and circulating gold nanorods with multispectral optoacoustic tomography. Opt. Express 2010, 18, 19592–19602. [Google Scholar] [CrossRef]
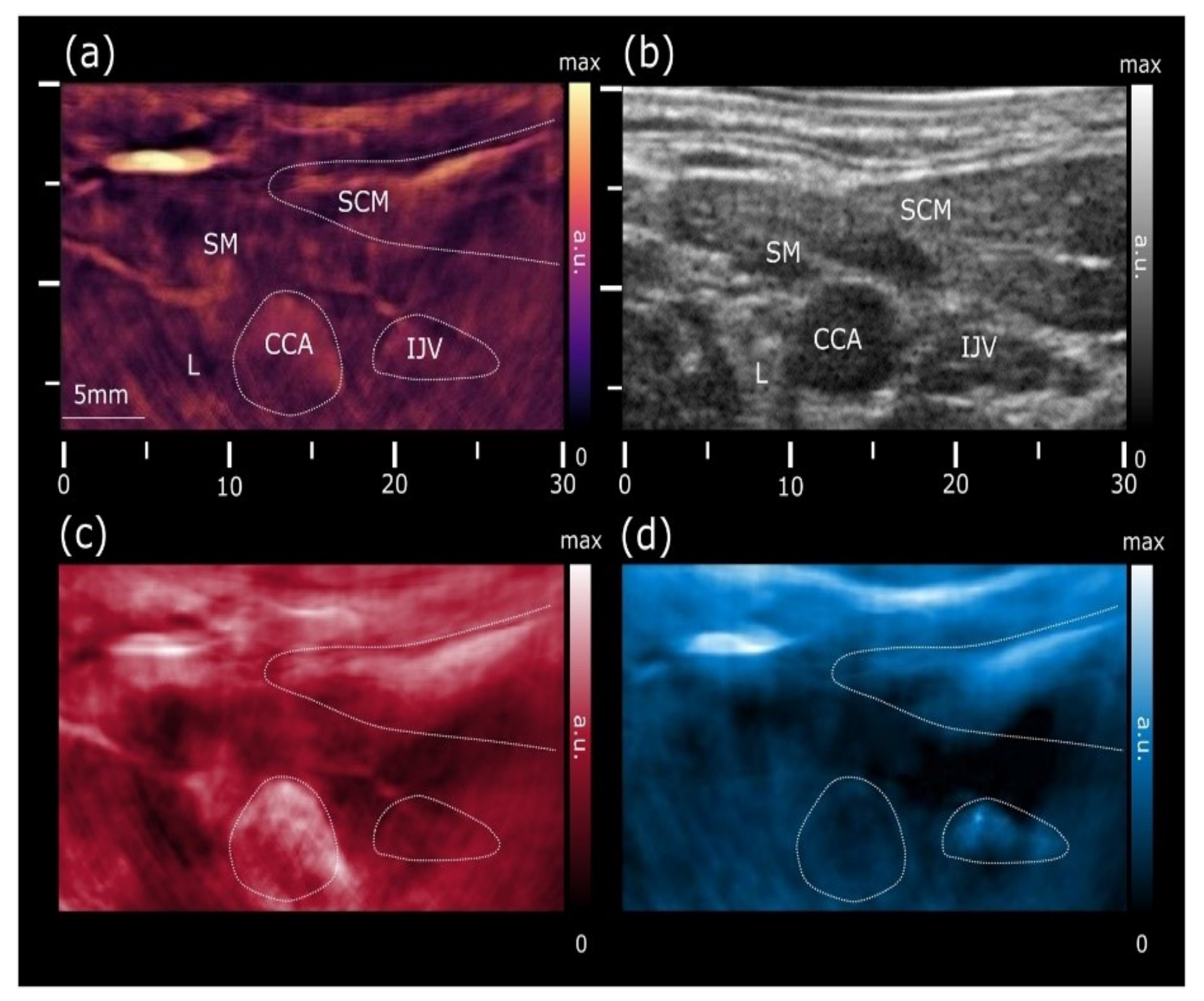
- Deán-Ben, X.L.; Razansky, D. Functional optoacoustic human angiography with handheld video rate three dimensional scanner. Photoacoustics 2013, 1, 68–73. [Google Scholar] [CrossRef]
- Ivankovic, I.; Merčep, E.; Schmedt, C.G.; Deán-Ben, X.L.; Razansky, D. Real-time volumetric assessment of the human carotid artery: Handheld multispectral optoacoustic tomography. Radiology 2019, 291, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Karlas, A.; Reber, J.; Diot, G.; Bozhko, D.; Anastasopoulou, M.; Ibrahim, T.; Schwaiger, M.; Hyafil, F.; Ntziachristos, V. Flow-mediated dilatation test using optoacoustic imaging: A proof-of-concept. Biomed. Opt. Express 2017, 8, 3395–3403. [Google Scholar] [PubMed]
- Taruttis, A.; Timmermans, A.C.; Wouters, P.C.; Kacprowicz, M.; van Dam, G.M.; Ntziachristos, V. Optoacoustic imaging of human vasculature: Feasibility by using a handheld probe. Radiology 2016, 281, 256–263. [Google Scholar] [CrossRef]
- Karlas, A.; Kallmayer, M.; Bariotakis, M.; Fasoula, N.A.; Liapis, E.; Hyafil, F.; Pelisek, J.; Wildgruber, M.; Eckstein, H.H.; Ntziachristos, V. Multispectral optoacoustic tomography of lipid and hemoglobin contrast in human carotid atherosclerosis. Photoacoustics 2021, 23, 100283. [Google Scholar] [CrossRef]
- Steinkamp, P.J.; Vonk, J.; Huisman, L.A.; Meersma, G.J.; Diercks, G.F.; Hillebrands, J.L.; Nagengast, W.B.; Zeebregts, C.J.; Slart, R.H.; Boersma, H.H.; et al. VEGF-Targeted Multispectral Optoacoustic Tomography and Fluorescence Molecular Imaging in Human Carotid Atherosclerotic Plaques. Res. Square 2021. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Huang, Q.; Li, Y. Measurement of cardiac output by use of noninvasively measured transient hemodilution curves with photoacoustic technology. Biomed. Opt. Express 2014, 5, 1445–1452. [Google Scholar] [CrossRef]
- Kang, D.; Huang, Q.; Li, Y. Noninvasive photoacoustic measurement of the composite indicator dilution curve for cardiac output estimation. Biomed. Opt. Express 2015, 6, 536–543. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stewart, S.; Hart, C.L.; Hole, D.J.; McMurray, J.J. A population-based study of the long-term risks associated with atrial fibrillation: 20-year follow-up of the Renfrew/Paisley study. Am. J. Med. 2002, 113, 359–364. [Google Scholar] [CrossRef]
- Bouchard, R.; Dana, N.; Di Biase, L.; Natale, A.; Emelianov, S. Photoacoustic characterization of radiofrequency ablation lesions. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2012, San Francisco, CA, USA, 22–24 January 2012; Volume 8223, p. 82233K. [Google Scholar]
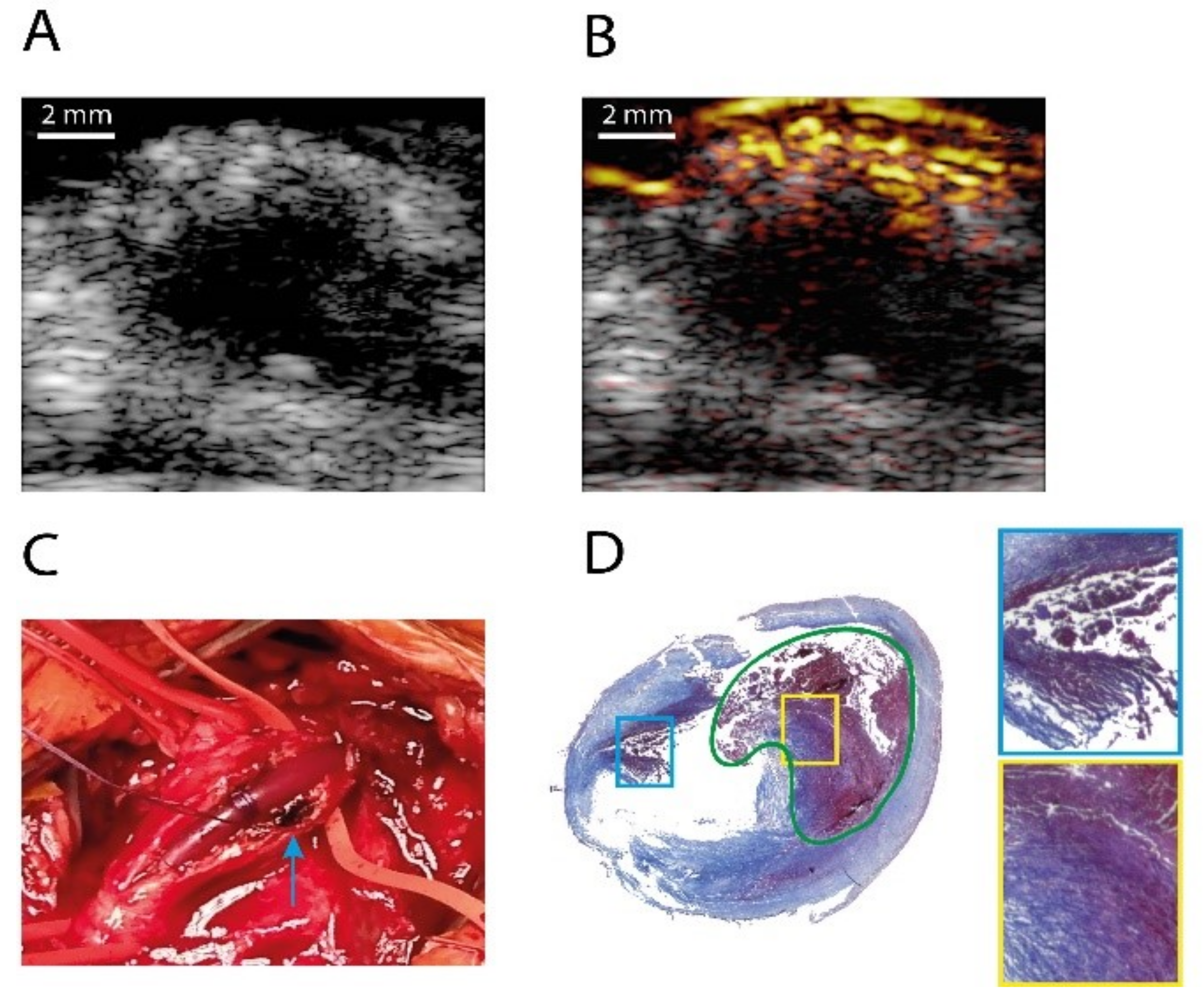
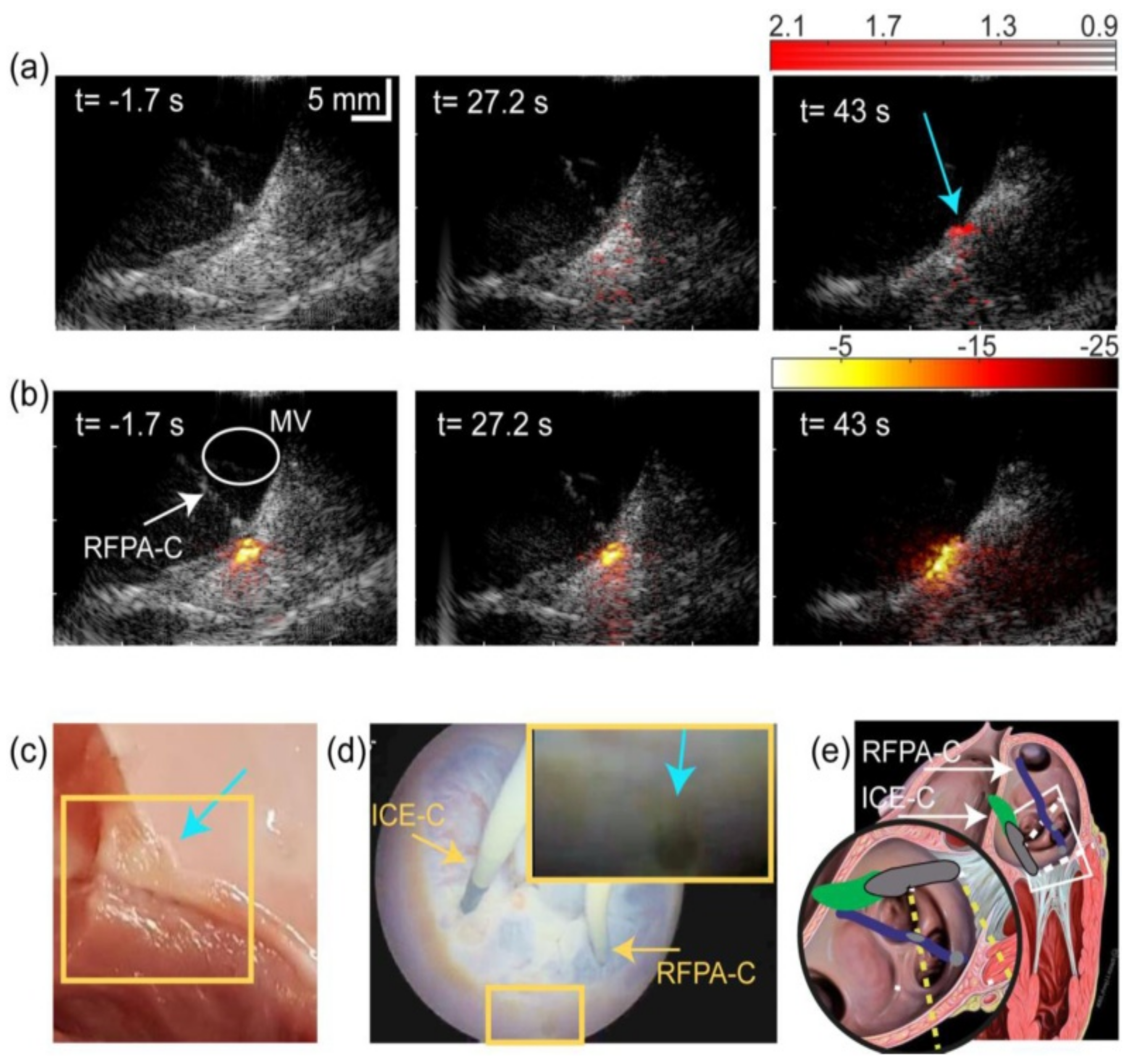
- Iskander-Rizk, S.; Kruizinga, P.; Beurskens, R.; Springeling, G.; Mastik, F.; de Groot, N.M.; Knops, P.; van der Steen, A.F.; van Soest, G. Real-time photoacoustic assessment of radiofrequency ablation lesion formation in the left atrium. Photoacoustics 2019, 16, 100150. [Google Scholar] [CrossRef]
- Dana, N.; Di Biase, L.; Natale, A.; Emelianov, S.; Bouchard, R. In vitro photoacoustic visualization of myocardial ablation lesions. Heart Rhythm 2014, 11, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Özsoy, Ç.; Floryan, M.; Deán-Ben, X.L.; Razansky, D. Endocardial irrigated catheter for volumetric optoacoustic mapping of radio-frequency ablation lesion progression. Opt. Lett. 2019, 44, 5808–5811. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Vu, T.; Sankin, G.; Winship, B.; Boydston, K.; Terry, R.; Zhong, P.; Yao, J. Internal-illumination photoacoustic tomography enhanced by a graded-scattering fiber diffuser. IEEE Trans. Med. Imaging 2020, 40, 346–356. [Google Scholar] [CrossRef]
- Özsoy, Ç.; Özbek, A.; Reiss, M.; Deán-Ben, X.L.; Razansky, D. Ultrafast four-dimensional imaging of cardiac mechanical wave propagation with sparse optoacoustic sensing. Proc. Natl. Acad. Sci. USA 2021, 118, 45. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Qiao, H.; Dai, Q.; Ma, C. Deep learning in photoacoustic imaging: A review. J. Biomed. Opt. 2021, 26, 040901. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lan, H.; Gao, F.; Gao, F. Review of deep learning for photoacoustic imaging. Photoacoustics 2021, 21, 100215. [Google Scholar] [CrossRef]
- Gröhl, J.; Schellenberg, M.; Dreher, K.; Maier-Hein, L. Deep learning for biomedical photoacoustic imaging: A review. Photoacoustics 2021, 22, 100241. [Google Scholar] [CrossRef] [PubMed]
- Waibel, D.; Gröhl, J.; Isensee, F.; Kirchner, T.; Maier-Hein, K.; Maier-Hein, L. Reconstruction of initial pressure from limited view photoacoustic images using deep learning. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2018, San Francisco, CA, USA, 28 January–1 February 2018; Volume 10494, p. 104942S. [Google Scholar]
- Lan, H.; Yang, C.; Jiang, D.; Gao, F. Reconstruct the photoacoustic image based on deep learning with multi-frequency ring-shape transducer array. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 7115–7118. [Google Scholar]
- Antholzer, S.; Haltmeier, M.; Schwab, J. Deep learning for photoacoustic tomography from sparse data. Inverse Probl. Sci. Eng. 2019, 27, 987–1005. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.T.; Guan, S.; Chitnis, P.V. Comparing deep learning frameworks for photoacoustic tomography image reconstruction. Photoacoustics 2021, 23, 100271. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jeng, G.S.; Pelivanov, I.; O’Donnell, M. Deep-learning image reconstruction for real-time photoacoustic system. IEEE Trans. Med. Imaging 2020, 39, 3379–3390. [Google Scholar] [CrossRef]
- Lan, H.; Jiang, D.; Yang, C.; Gao, F.; Gao, F. Y-Net: Hybrid deep learning image reconstruction for photoacoustic tomography in vivo. Photoacoustics 2020, 20, 100197. [Google Scholar]
- Cai, C.; Deng, K.; Ma, C.; Luo, J. End-to-end deep neural network for optical inversion in quantitative photoacoustic imaging. Opt. Lett. 2018, 43, 2752–2755. [Google Scholar] [CrossRef] [PubMed]
- Chlis, N.K.; Karlas, A.; Fasoula, N.A.; Kallmayer, M.; Eckstein, H.H.; Theis, F.J.; Ntziachristos, V.; Marr, C. A sparse deep learning approach for automatic segmentation of human vasculature in multispectral optoacoustic tomography. Photoacoustics 2020, 20, 100203. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.Y.; Gao, Y.; Peng, L.; Zhou, L.; Liu, J.; Zhu, S.; Song, W. Hybrid deep learning network for vascular segmentation in photoacoustic imaging. Biomed. Opt. Express 2020, 11, 6445–6457. [Google Scholar] [CrossRef] [PubMed]
- Gröhl, J.; Schellenberg, M.; Dreher, K.K.; Holzwarth, N.; Tizabi, M.D.; Seitel, A.; Maier-Hein, L. Semantic segmentation of multispectral photoacoustic images using deep learning. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2021, Online, 6–11 March 2021; Volume 11642, p. 116423F. [Google Scholar]
- Yang, H.; Jüstel, D.; Prakash, J.; Karlas, A.; Helfen, A.; Masthoff, M.; Wildgruber, M.; Ntziachristos, V. Soft ultrasound priors in optoacoustic reconstruction: Improving clinical vascular imaging. Photoacoustics 2020, 19, 100172. [Google Scholar]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef] [PubMed]
- Kouw, W.M.; Loog, M. A review of single-source unsupervised domain adaptation. arXiv 2019, arXiv:1901.05335. [Google Scholar]
- Arabul, M.; Rutten, M.; Bruneval, P.; van Sambeek, M.; van de Vosse, F.; Lopata, R. Unmixing multi-spectral photoacoustic sources in human carotid plaques using non-negative independent component analysis. Photoacoustics 2019, 15, 100140. [Google Scholar] [CrossRef]
- An, L.; Cox, B. Independent component analysis for unmixing multi-wavelength photoacoustic images. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2016, San Francisco, CA, USA, 14–17 February 2016; Volume 9708, p. 970851. [Google Scholar]
- Ding, L.; Deán-Ben, X.L.; Burton, N.C.; Sobol, R.W.; Ntziachristos, V.; Razansky, D. Constrained inversion and spectral unmixing in multispectral optoacoustic tomography. IEEE Trans. Med. Imaging 2017, 36, 1676–1685. [Google Scholar] [CrossRef]
- Cao, Y.; Kole, A.; Lan, L.; Wang, P.; Hui, J.; Sturek, M.; Cheng, J.X. Spectral analysis assisted photoacoustic imaging for lipid composition differentiation. Photoacoustics 2017, 7, 12–19. [Google Scholar] [CrossRef] [PubMed]
| Application | Popular Deep Learning Models |
|---|---|
| Cardiac viewpoint classification | Custom architecture based on VGG, ResNet, DenseNet [50]; Custom architecture based on CNNs [49]; Custom architecture fusing spatial and temporal information using CNNs [48] |
| LV segmentation | U-net-based architectures [59,60,62,63,71]; CNN [61]; Deep belief network (DBN) [55,68,69,70,74]; U-net combined with RNNs [64,67,75]; U-net with TL-net [72,77] |
| IVUS image segmentation | U-net-based architectures [80,81,83,87,89], Autoencoder [82], CNN [88] |
| Point of care ultrasound (POCUS) | AlexNet, VGG-16, VGG-19, ResNet50, DenseNet201 [102]; LSTM [103]; CycleGAN [104]; |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Awasthi, N.; Rad, N.M.; Pluim, J.P.W.; Lopata, R.G.P. Advanced Ultrasound and Photoacoustic Imaging in Cardiology. Sensors 2021, 21, 7947. https://doi.org/10.3390/s21237947
Wu M, Awasthi N, Rad NM, Pluim JPW, Lopata RGP. Advanced Ultrasound and Photoacoustic Imaging in Cardiology. Sensors. 2021; 21(23):7947. https://doi.org/10.3390/s21237947
Chicago/Turabian StyleWu, Min, Navchetan Awasthi, Nastaran Mohammadian Rad, Josien P. W. Pluim, and Richard G. P. Lopata. 2021. "Advanced Ultrasound and Photoacoustic Imaging in Cardiology" Sensors 21, no. 23: 7947. https://doi.org/10.3390/s21237947
APA StyleWu, M., Awasthi, N., Rad, N. M., Pluim, J. P. W., & Lopata, R. G. P. (2021). Advanced Ultrasound and Photoacoustic Imaging in Cardiology. Sensors, 21(23), 7947. https://doi.org/10.3390/s21237947







