Automated Processing and Phenotype Extraction of Ovine Medical Images Using a Combined Generative Adversarial Network and Computer Vision Pipeline
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ovine Ischium Scan Collection
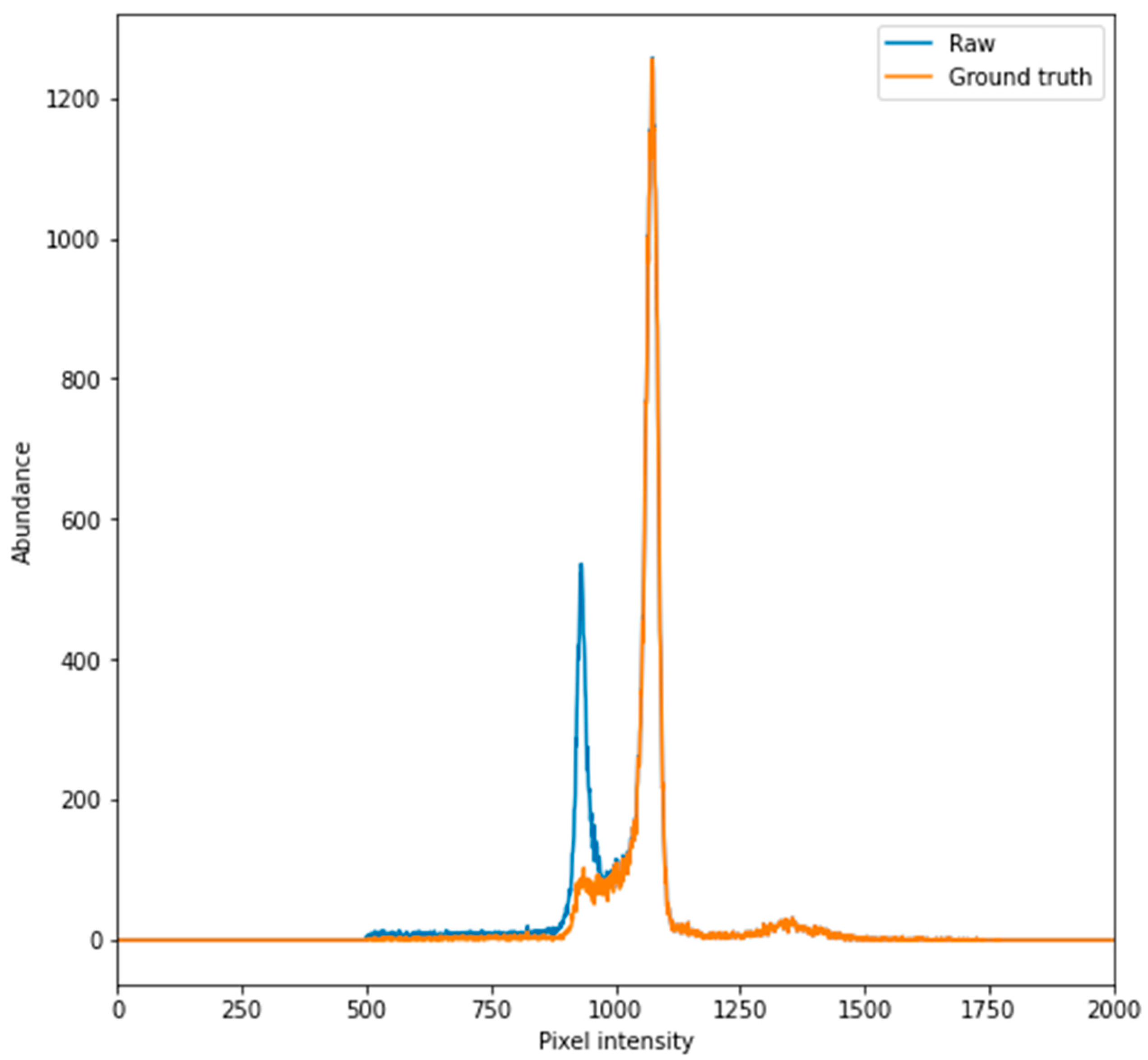
2.2. Determination of Tissue Pixel Intensities
2.3. Manual Image Processing and Phenotype Analysis
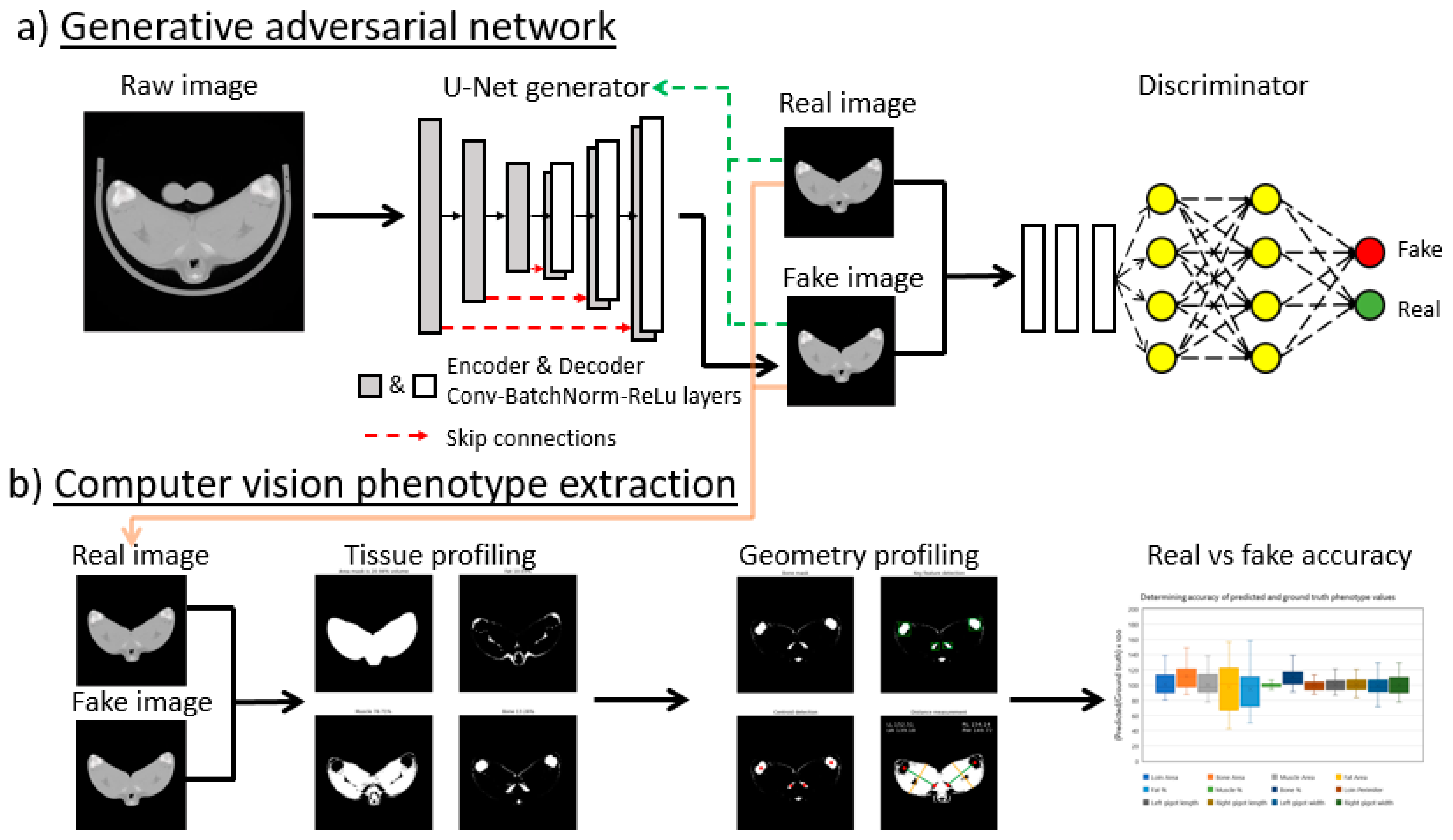
2.4. GAN Model
2.4.1. GAN Training
2.4.2. Image Processing Using Trained GAN on Unseen Data
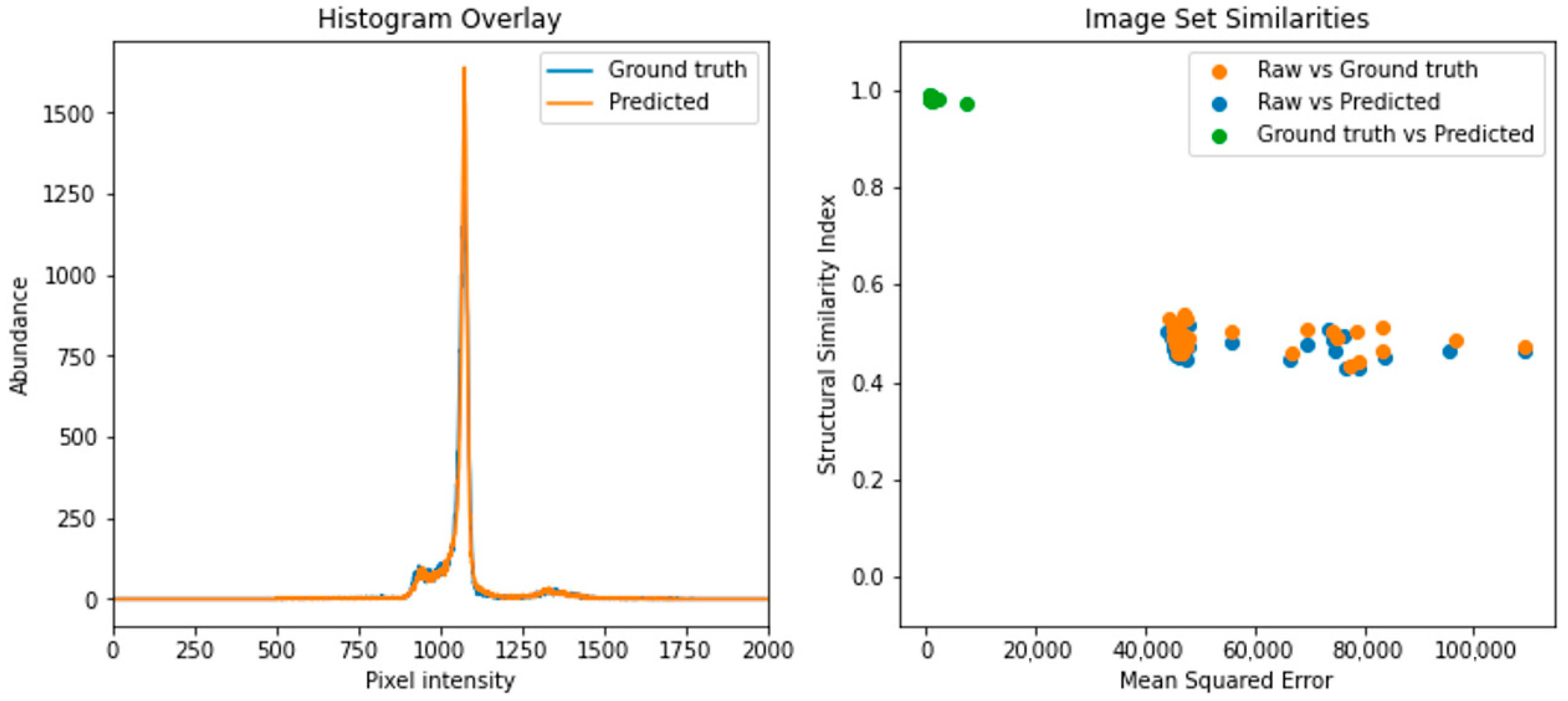
2.5. CT Scan Similarity Comparison
2.5.1. CT Scan Histogram Comparison
2.5.2. Calculation of Image Similarity
2.6. Phenotype Measurement Using Computer Vision
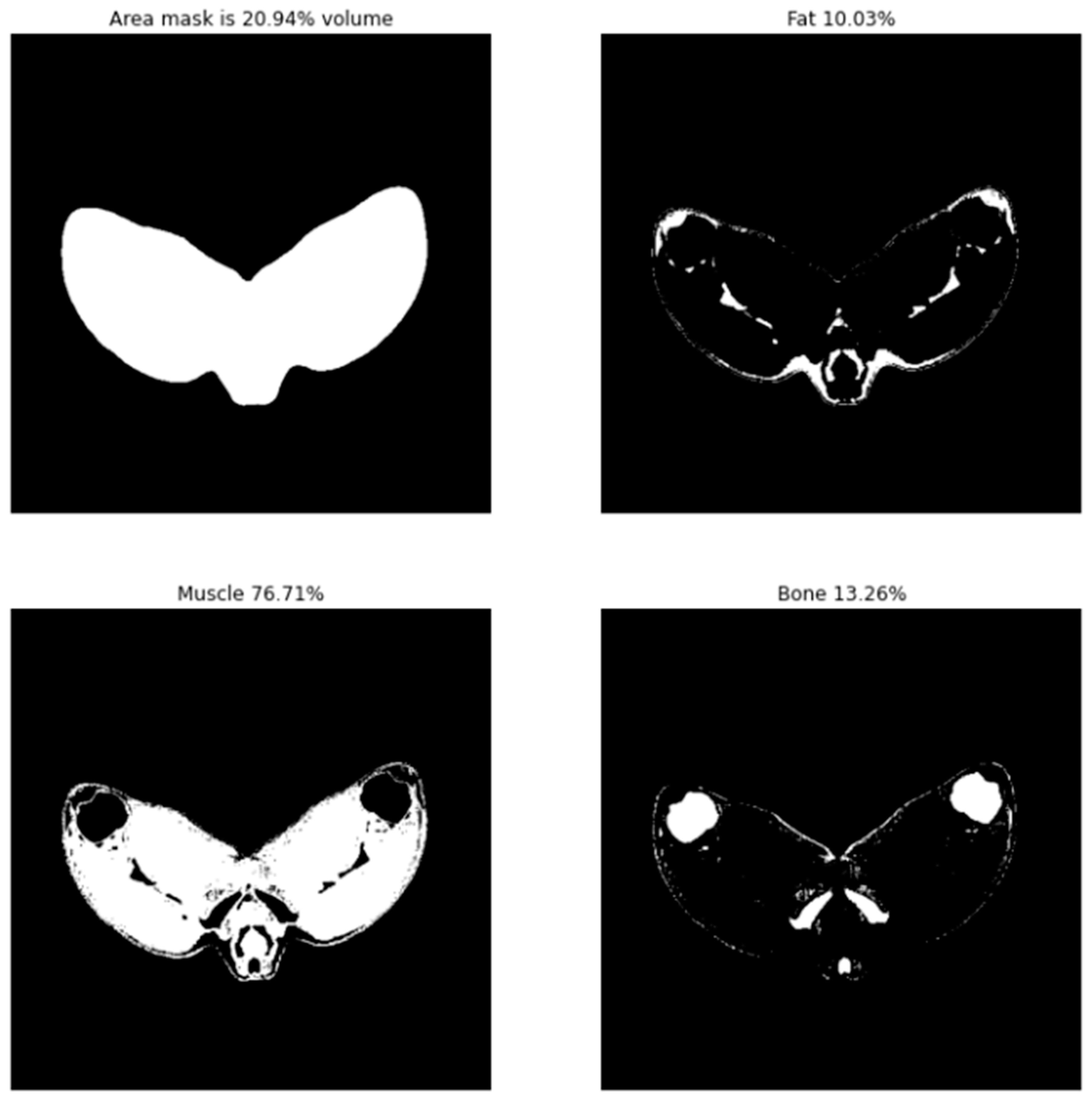
2.6.1. Tissue Distribution
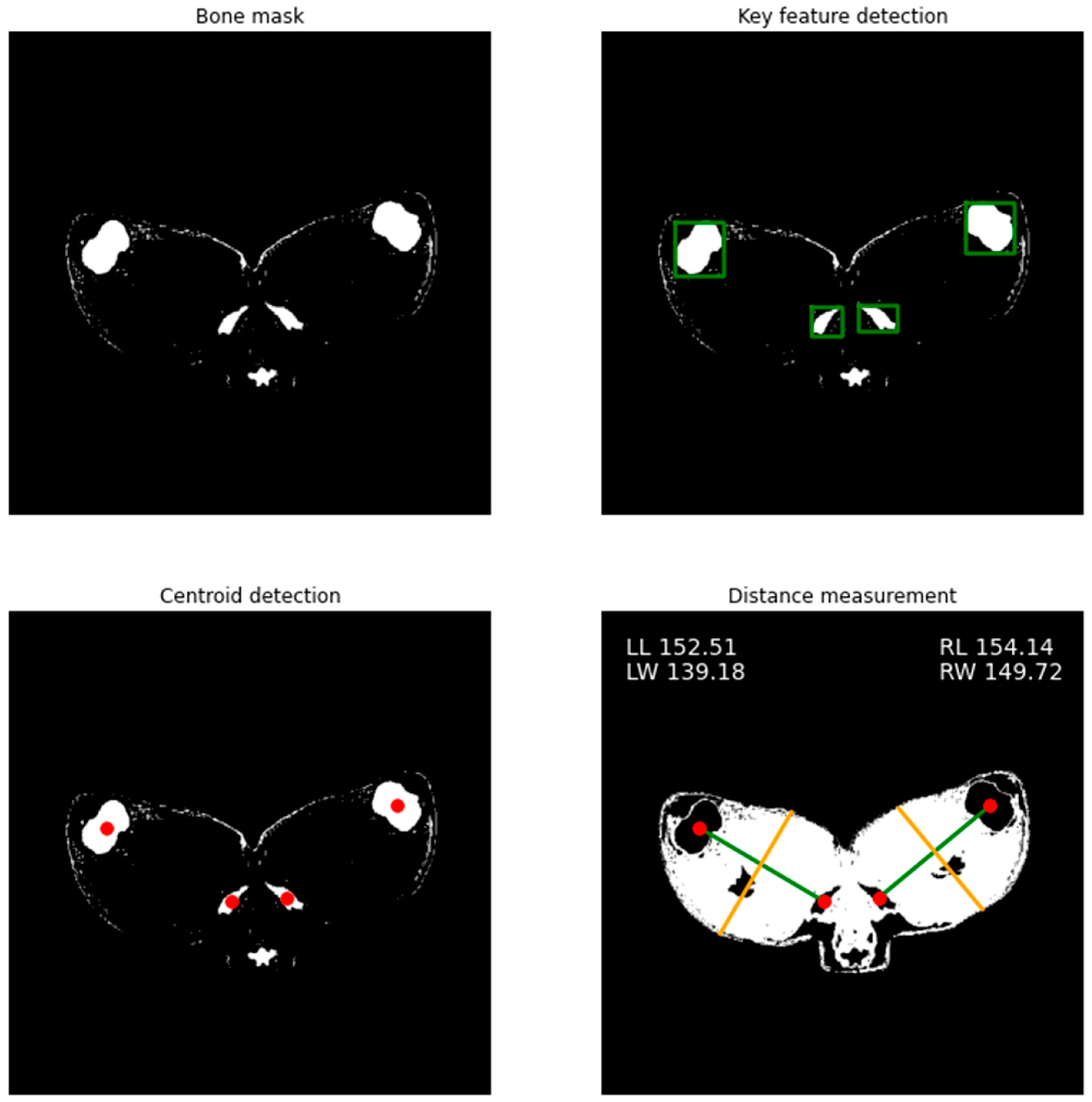
2.6.2. Skeleton Geometry
2.7. Computing Hardware and Software
3. Results
3.1. CT Scan Processing Using Trained GAN
3.1.1. Images Produced from Trained Model
3.1.2. Image Similarity Metrics Confirm a High Degree of Similarity
3.2. Automated Phenotype Extraction
3.2.1. Leg Tissue Composition
3.2.2. Gigot Length and Width Phenotype Extraction
3.2.3. Phenotype Extraction Accuracy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Shaping the future of livestock; Report No. I8384EN. In Proceedings of the 10th Global Forum for Food and Agriculture, Berlin, Germany, 20 January 2018; pp. 18–20. [Google Scholar]
- Rexroad, C.; Vallet, J.; Matukumalli, L.K.; Reecy, J.; Bickhart, D.; Blackburn, H.; Boggess, M.; Cheng, H.; Clutter, A.; Cockett, N.; et al. Genome to phenome: Improving animal health, production, and well-being—A new USDA blueprint for animal genome research 2018–2027. Front. Genet. 2019, 10, 1–29. [Google Scholar] [CrossRef]
- Gonzalez-Recio, O.; Coffey, M.P.; Pryce, J.E. On the value of the phenotypes in the genomic era. J. Dairy Sci. 2014, 97, 7905–7915. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Molano, E.; Kapsona, V.V.; Ilska, J.J.; Desire, S.; Conington, J.; Mucha, S.; Banos, G. Genetic analysis of novel phenotypes for farm animal resilience to weather variability. BMC Genet. 2019, 20, 84. [Google Scholar] [CrossRef]
- Brito, L.F.; Oliveira, H.R.; McConn, B.R.; Schinckel, A.P.; Arrazola, A.; Marchant-Forde, J.N.; Johnson, J.S. Large-Scale Phenotyping of Livestock Welfare in Commercial Production Systems: A New Frontier in Animal Breeding. Front. Genet. 2020, 11, 793. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef] [PubMed]
- Santos, B.F.S.; Van Der Werf, J.H.J.; Gibson, J.P.; Byrne, T.J.; Amer, P.R. Genetic and economic benefits of selection based on performance recording and genotyping in lower tiers of multi-tiered sheep breeding schemes. Genet. Sel. Evol. 2017, 49, 10. [Google Scholar] [CrossRef]
- Duijvesteijn, N.; Bolormaa, S.; Daetwyler, H.D.; Van Der Werf, J.H.J. Genomic prediction of the polled and horned phenotypes in Merino sheep. Genet. Sel. Evol. 2018, 50, 28. [Google Scholar] [CrossRef] [Green Version]
- Seidel, A.; Krattenmacher, N.; Thaller, G. Dealing with complexity of new phenotypes in modern dairy cattle breeding. Anim. Front. 2020, 10, 23–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroy, G.; Besbes, B.; Boettcher, P.; Hoffmann, I.; Capitan, A.; Baumung, R. Rare phenotypes in domestic animals: Unique resources for multiple applications. Anim. Genet. 2016, 47, 141–153. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Lehmann, K.; Krauss, G. SSO1450—A CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 2009, 583, 1928–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunger, L.; Macfarlane, J.M.; Lambe, N.R.; Conington, J.; McLean, K.A.; Moore, K.; Glasbey, C.A.; Simm, G. Use of X-ray Computed Tomography (CT) in UK Sheep Production and Breeding. CT Scanning-Tech. Appl. 2011, 329–348. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lohumi, S.; Lim, H.S.; Gotoh, T.; Cho, B.K.; Jung, S. Determination of intramuscular fat content in beef using magnetic resonance imaging. J. Fac. Agric. Kyushu Univ. 2015, 60, 157–162. [Google Scholar] [CrossRef]
- McLaren, A.; Kaseja, K.; McLean, K.A.; Boon, S.; Lambe, N.R. Genetic analyses of novel traits derived from CT scanning for implementation in terminal sire sheep breeding programmes. Livest. Sci. 2021, 250, 104555. [Google Scholar] [CrossRef]
- Savage, N. How-ai-is-improving-cancer-diagnost. Nature 2020, 579, S14. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology 2020, 296, E65–E71. [Google Scholar] [CrossRef]
- Lim, L.J.; Tison, G.H.; Delling, F.N. Artificial Intelligence in Cardiovascular Imaging. Methodist Debakey Cardiovasc. J. 2020, 16, 138–145. [Google Scholar] [CrossRef]
- Denholm, S.J.; Brand, W.; Mitchell, A.P.; Wells, A.T.; Krzyzelewski, T.; Smith, S.L.; Wall, E.; Coffey, M.P. Predicting bovine tuberculosis status of dairy cows from mid-infrared spectral data of milk using deep learning. J. Dairy Sci. 2020, 103, 9355–9367. [Google Scholar] [CrossRef]
- Brand, W.; Wells, A.T.; Smith, S.L.; Denholm, S.J.; Wall, E.; Coffey, M.P. Predicting pregnancy status from mid-infrared spectroscopy in dairy cow milk using deep learning. J. Dairy Sci. 2021, 104, 4980–4990. [Google Scholar] [CrossRef]
- Soffer, S.; Ben-Cohen, A.; Shimon, O.; Amitai, M.M.; Greenspan, H.; Klang, E. Convolutional Neural Networks for Radiologic Images: A Radiologist’s Guide. Radiology 2019, 290, 590–606. [Google Scholar] [CrossRef]
- Hou, X.; Gong, Y.; Liu, B.; Sun, K.; Liu, J.; Xu, B.; Duan, J.; Qiu, G. Learning based image transformation using convolutional neural networks. IEEE Access 2018, 6, 49779–49792. [Google Scholar] [CrossRef]
- Bod, M. A guide to recurrent neural networks and backpropagation. Rnn Dan Bpnn 2001, 2, 1–10. [Google Scholar]
- Benos, L.; Tagarakis, A.C.; Dolias, G.; Berruto, R.; Kateris, D.; Bochtis, D. Machine learning in agriculture: A comprehensive updated review. Sensors 2021, 21, 3758. [Google Scholar] [CrossRef]
- Parmar, N.; Vaswani, A.; Uszkoreit, J.; Kaiser, L.; Shazeer, N.; Ku, A.; Tran, D. Image transformer. In Proceedings of the 35th International Conference on Machine Learning ICML 2018, Stockholm, Sweden, 10–15 July 2018; pp. 6453–6462. [Google Scholar]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar] [CrossRef] [Green Version]
- Zhong, G.; Gao, W.; Liu, Y.; Yang, Y.; Wang, D.H.; Huang, K. Generative adversarial networks with decoder–encoder output noises. Neural Networks 2020, 127, 19–28. [Google Scholar] [CrossRef]
- Singh, N.K.; Raza, K. Medical Image Generation using Generative Adversarial Networks. arXiv 2020, arXiv:2005.10687. [Google Scholar]
- Armanious, K.; Jiang, C.; Fischer, M.; Küstner, T.; Hepp, T.; Nikolaou, K.; Gatidis, S.; Yang, B. MedGAN: Medical image translation using GANs. Comput. Med. Imaging Graph. 2020, 79, 101684. [Google Scholar] [CrossRef] [PubMed]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep Learning for Computer Vision: A Brief Review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef]
- Meer, P.; Mintz, D.; Rosenfeld, A.; Kim, D.Y. Robust regression methods for computer vision: A review. Int. J. Comput. Vis. 1991, 6, 59–70. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.W. Colour measurements by computer vision for food quality control—A review. Trends Food Sci. Technol. 2013, 29, 5–20. [Google Scholar] [CrossRef]
- Brosnan, T.; Sun, D.W. Improving quality inspection of food products by computer vision—A review. J. Food Eng. 2004, 61, 3–16. [Google Scholar] [CrossRef]
- Deva Koresh, J. Computer Vision Based Traffic Sign Sensing for Smart Transport. J. Innov. Image Process. 2019, 1, 11–19. [Google Scholar] [CrossRef]
- Glasbey, C.A.; Young, M.J. Maximum a posteriori estimation of image boundaries by dynamic programming. J. R. Stat. Soc. Ser. C Appl. Stat. 2002, 51, 209–221. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Zhu, B.; Liu, J.Z.; Cauley, S.F.; Rosen, B.R.; Rosen, M.S. Image reconstruction by domain-transform manifold learning. Nature 2018, 555, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Kaji, S.; Kida, S. Overview of image-to-image translation by use of deep neural networks: Denoising, super-resolution, modality conversion, and reconstruction in medical imaging. Radiol. Phys. Technol. 2019, 12, 235–248. [Google Scholar] [CrossRef] [Green Version]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. In Proceedings of the 4th International Conference on Learning Representations, ICLR 2016, San Juan, Puerto Rico, 2–4 May 2016; pp. 1–16. [Google Scholar]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. Proc. IEEE Int. Conf. Comput. Vis. 2017, 2017, 2242–2251. [Google Scholar] [CrossRef] [Green Version]
- Sara, U.; Akter, M.; Uddin, M.S. Image Quality Assessment through FSIM, SSIM, MSE and PSNR—A Comparative Study. J. Comput. Commun. 2019, 7, 8–18. [Google Scholar] [CrossRef] [Green Version]
- AGandhi, S.; Kulkarni, C.V. MSE vs. SSIM. Int. J. Sci. Eng. Res. 2013, 4, 930–934. [Google Scholar]
- Van Der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. Scikit-image: Image processing in python. PeerJ 2014, 2014, e453. [Google Scholar] [CrossRef]
- Séverine, R. Analyse D’image Géométrique et Morphométrique par Diagrammes de Forme et Voisinages Adaptatifs Généraux. Ph.D. Thesis, ENSMSE, Saint-Etienne, France, 2011. [Google Scholar]
- NVIDIA NVIDIA DGX Station: AI Workstation for Data Science Teams. Available online: https://www.nvidia.com/en-gb/data-center/dgx-station-a100/ (accessed on 21 May 2021).
- Tokui, S.; Okuta, R.; Akiba, T.; Niitani, Y.; Ogawa, T.; Saito, S.; Suzuki, S.; Uenishi, K.; Vogel, B.; Vincent, H.Y. Chainer: A deep learning framework for accelerating the research cycle. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 2002–2011. [Google Scholar] [CrossRef]
- Lassau, N.; Ammari, S.; Chouzenoux, E.; Gortais, H.; Herent, P.; Devilder, M.; Soliman, S.; Meyrignac, O.; Talabard, M.P.; Lamarque, J.P.; et al. Integrating deep learning CT-scan model, biological and clinical variables to predict severity of COVID-19 patients. Nat. Commun. 2021, 12, 634. [Google Scholar] [CrossRef] [PubMed]
- Saood, A.; Hatem, I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Med. Imaging 2021, 21, 19. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Phuoc, T.; Li, C.; Theis, L.; Richardt, C.; Yang, Y.L. HoloGAN: Unsupervised learning of 3D representations from natural images. In Proceedings of the 2019 International Conference on Computer Vision Workshop, ICCVW 2019, Seoul, Korea, 27–28 October 2019; pp. 2037–2040. [Google Scholar] [CrossRef] [Green Version]
- Öngün, C.; Temizel, A. Paired 3D model generation with conditional generative adversarial networks. In Proceedings of the European Conference on Computer Vision, ECCV 2018 Workshops, Munich, Germany, 8–14 September 2018; Springer: Cham, Switherland, 2019; Volume 11129, pp. 473–487. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robson, J.F.; Denholm, S.J.; Coffey, M. Automated Processing and Phenotype Extraction of Ovine Medical Images Using a Combined Generative Adversarial Network and Computer Vision Pipeline. Sensors 2021, 21, 7268. https://doi.org/10.3390/s21217268
Robson JF, Denholm SJ, Coffey M. Automated Processing and Phenotype Extraction of Ovine Medical Images Using a Combined Generative Adversarial Network and Computer Vision Pipeline. Sensors. 2021; 21(21):7268. https://doi.org/10.3390/s21217268
Chicago/Turabian StyleRobson, James Francis, Scott John Denholm, and Mike Coffey. 2021. "Automated Processing and Phenotype Extraction of Ovine Medical Images Using a Combined Generative Adversarial Network and Computer Vision Pipeline" Sensors 21, no. 21: 7268. https://doi.org/10.3390/s21217268
APA StyleRobson, J. F., Denholm, S. J., & Coffey, M. (2021). Automated Processing and Phenotype Extraction of Ovine Medical Images Using a Combined Generative Adversarial Network and Computer Vision Pipeline. Sensors, 21(21), 7268. https://doi.org/10.3390/s21217268





