Molecular Beacon Assay Development for Severe Acute Respiratory Syndrome Coronavirus 2 Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis of SARS-CoV-2 Genome
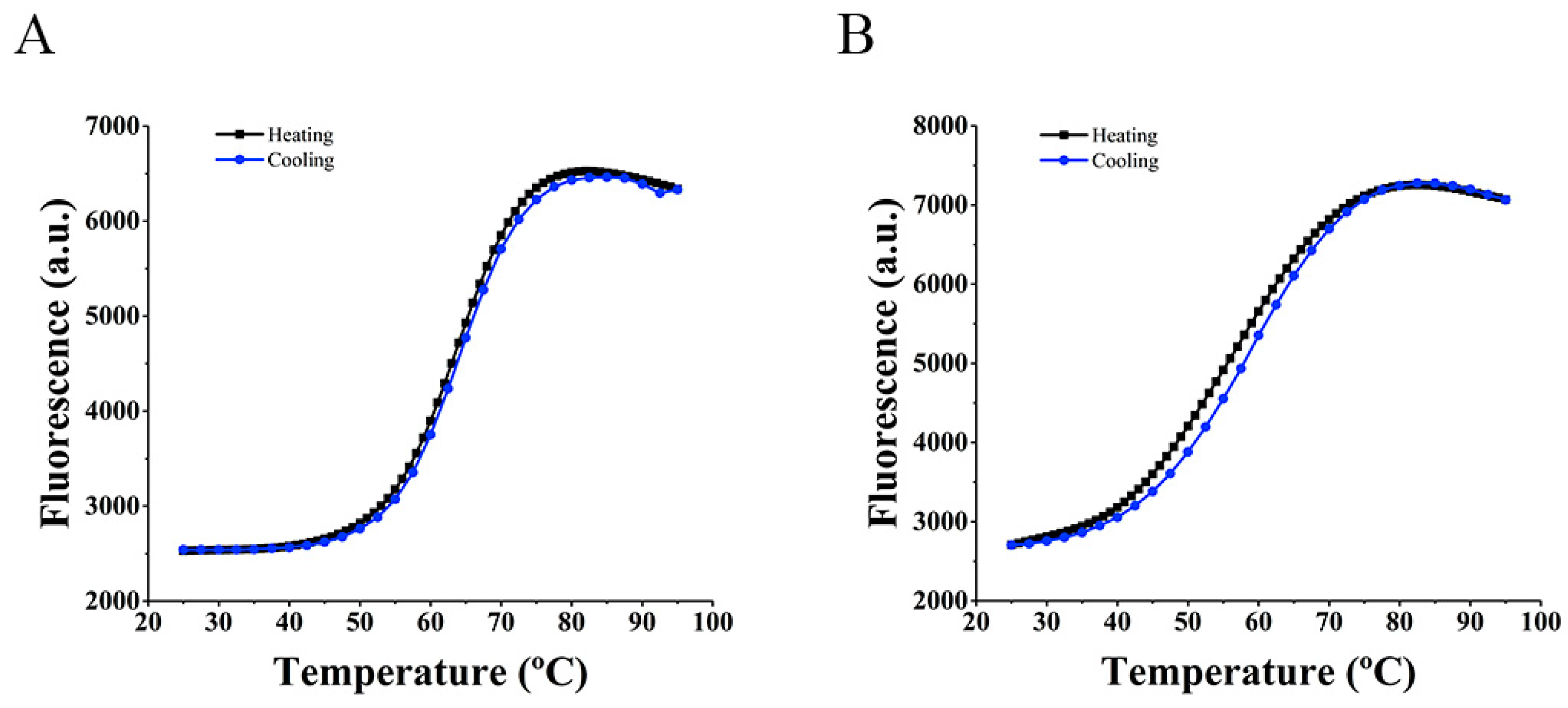
2.2. FRET Melting Assay
2.2.1. Circular Dichroism (CD) Spectroscopy
2.2.2. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.2.3. Hybridization Temperature Determination
2.3. Clinical Samples
2.3.1. cDNA Synthesis and Amplification
2.3.2. cDNA Detection Using Molecular Beacon
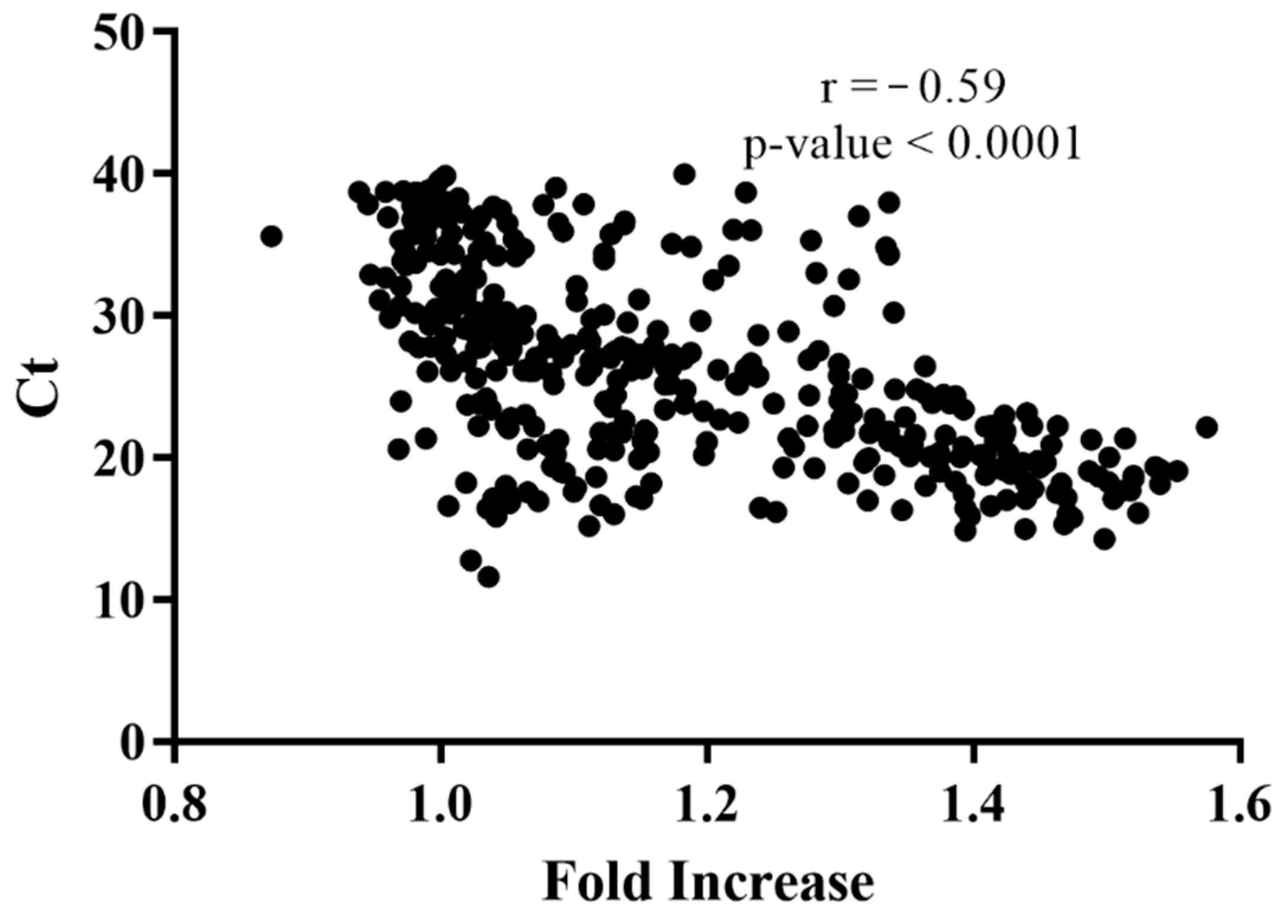
2.4. Real-Time PCR for Clinical Samples
3. Results
4. Discussion
- i.
- Ease of result interpretation, requiring less training and enabling health care staff to use the test correctly. The current RT-qPCR tests employ multiplex reactions that render a multitude of amplification plots for two to four different genes, which are not readily interpreted by health care staff with limited training in molecular biology techniques or molecular diagnosis. Furthermore, despite OMS recommendations, the cycle threshold (Ct) and copy number values provided by RT-qPCR are not being used to drive the routine clinical practice. Therefore, the extra unused information provided by RT-qPCR is creating a needless and expensive bottleneck. End-point RT-PCR provides the same Positive/Negative result for the presence/absence of SARS-CoV-2 RNA at a potentially lower cost and higher throughput, with easier interpretation.
- ii.
- Less expensive lab apparatus and reagents needed. The proposed test does not require expensive real-time thermocyclers, being possible to execute using a simple end-point thermocycler or programmable heating block, and a plate reader or fluorescence reading apparatus. This enables the laboratories of developing countries with poor access to heavy lab machinery to implement the method in their facilities using existent equipment as end-point PCR is already used to diagnose other infectious diseases such as Hepatitis B, Ebola, and HIV. Furthermore, as the reactions are performed using a single set of primers and a single molecular beacon, the users can easily implement the protocol using existent enzyme master mixes and reagents, without the need to purchase expensive RT-qPCR SARS-CoV-2 detection kits.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 Infection: Origin, Transmission, and Characteristics of Human Coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- World Health Organization. Diagnostic Testing for SARS-CoV-2: Interim Guidance, 11 September 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Surkova, E.; Nikolayevskyy, V.; Drobniewski, F. False-Positive COVID-19 Results: Hidden Problems and Costs. Lancet Respir. Med. 2020, 8, 1167–1168. [Google Scholar] [CrossRef]
- Yuan, X.; Yang, C.; He, Q.; Chen, J.; Yu, D.; Li, J.; Zhai, S.; Qin, Z.; Du, K.; Chu, Z.; et al. Current and Perspective Diagnostic Techniques for COVID-19. ACS Infect. Dis. 2020, 6, 1998–2016. [Google Scholar] [CrossRef] [PubMed]
- Scarlata, S.; Yerramilli, V.S. Design of a Rapid and Reversible Fluorescence Assay to Detect COVID-19 and other Pathogens. medRxiv 2020. [Google Scholar] [CrossRef]
- Da Silva, S.J.R.; Pardee, K.; Pena, L. Loop-Mediated Isothermal Amplification (LAMP) for the Diagnosis of Zika Virus: A Review. Viruses 2019, 12, 19. [Google Scholar] [CrossRef] [Green Version]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [Green Version]
- Goel, G.; Kumar, A.; Puniya, A.K.; Chen, W.; Singh, K. Molecular Beacon: A Multitask Probe. J. Appl. Microbiol. 2005, 99, 435–442. [Google Scholar] [CrossRef]
- Hadjinicolaou, A.V.; Farcas, G.A.; Demetriou, V.L.; Mazzulli, T.; Poutanen, S.M.; Willey, B.M.; Low, D.E.; Butany, J.; Asa, S.L.; Kain, K.C.; et al. Development of a Molecular-Beacon-Based Multi-Allelic Real-Time RT-PCR Assay for the Detection of Human Coronavirus Causing Severe Acute Respiratory Syndrome (SARS-CoV): A General Methodology for Detecting Rapidly Mutating Viruses. Arch. Virol. 2011, 156, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Sherrill-Mix, S.; van Duyne, G.D.; Bushman, F.D. Molecular Beacons Allow Specific RT-LAMP Detection of B.1.1.7 Variant SARS-CoV-2. medRxiv 2021. [Google Scholar] [CrossRef]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-Quadruplex, Friend or Foe: The Role of the G-Quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Juhas, M.; Tsang, C.M.; Kwok, C.K.; Li, Y.; Zhang, Y. Discovery of G-Quadruplex-Forming Sequences in SARS-CoV-2. Brief. Bioinform. 2021, 22, 1150–1160. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A Web-Based Server for Predicting G-Quadruplexes in Nucleotide Sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ke, X.; Gu, Y.; Liu, H.; Sun, X. Whole Genome Identification of Potential G-Quadruplexes and Analysis of the G-Quadruplex Binding Domain for SARS-CoV-2. Front. Genet. 2020, 11, 1430. [Google Scholar] [CrossRef]
- Panera, N.; Tozzi, A.E.; Alisi, A. The G-Quadruplex/Helicase World as a Potential Antiviral Approach against COVID-19. Drugs 2020, 80, 941–946. [Google Scholar] [CrossRef]
- Cui, H.; Zhang, L. G-Quadruplexes Are Present in Human Coronaviruses Including SARS-CoV-2. Front. Microbiol. 2020, 11, 567317. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem.-Int. Ed. 2020, 60, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Metifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.-L. G-Quadruplexes in Viruses: Function and Potential Therapeutic Applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, P.K.; Cha, J.; Barton, J.K. 1H NMR Determination of Base-Pair Lifetimes in Oligonucleotides Containing Single Base Mismatches. Nucleic Acids Res. 2002, 30, 4740–4750. [Google Scholar] [CrossRef] [Green Version]
- Božič, T.; Zalar, M.; Rogelj, B.; Plavec, J.; Šket, P. Structural Diversity of Sense and Antisense RNA Hexanucleotide Repeats Associated with ALS and FTLD. Molecules 2020, 25, 525. [Google Scholar] [CrossRef] [Green Version]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 Mutation Hot Spots Include a Novel RNA-Dependent-RNA Polymerase Variant. J. Transl. Med. 2020, 18, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamroskovic, J.; Obi, I.; Movahedi, A.; Chand, K.; Chorell, E.; Sabouri, N. Identification of Putative G-Quadruplex DNA Structures in S. Pombe Genome by Quantitative PCR Stop Assay. DNA Repair 2019, 82, 102678. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Arons, M.M.; Hatfield, K.M.; Reddy, S.C.; Kimball, A.; James, A.; Jacobs, J.R.; Taylor, J.; Spicer, K.; Bardossy, A.C.; Oakley, L.P.; et al. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N. Engl. J. Med. 2020, 382, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- la Scola, B.; le Bideau, M.; Andreani, J.; Hoang, V.T.; Grimaldier, C.; Colson, P.; Gautret, P.; Raoult, D. Viral RNA Load as Determined by Cell Culture as a Management Tool for Discharge of SARS-CoV-2 Patients from Infectious Disease Wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1059–1061. [Google Scholar] [CrossRef] [PubMed]



| Genome Region | Position | Length | QGRS 1 | G-Score | Total of Genomes Analyzed | Genomes with PQS | Conservation Level (%) |
|---|---|---|---|---|---|---|---|
| ORF1ab | 1,574 | 26 | GGTGTTGTTGGAGAAGGTTCCGAAGG | 19 | 211,072 | 207,006 | 98.07 |
| 13,385 | 20 | GGTATGTGGAAAGGTTATGG | 18 | 210,888 | 99.91 | ||
| S | 24,215 | 20 | GGTTGGACCTTTGGTGCAGG | 17 | 211,072 | 210,803 | 99.87 |
| 24,268 | 24 | GGCTTATAGGTTTAATGGTATTGG | 19 | 210,999 | 99.97 | ||
| 25,197 | 22 | GGCCATGGTACATTTGGCTAGG | 17 | 207,837 | 98.47 | ||
| N | 28,903 | 15 | GGCTGGCAATGGCGG | 18 | 211,072 | 191,696 | 90.82 |
| Variant | SARS-CoV-2 Gene | SARS-CoV-2 G4 Sequence | Total of Sequences Analyzed | Number of Sequences with PQS | Conservation (%) |
|---|---|---|---|---|---|
| Alpha | ORF1ab | GGTGTTGTTGGAGAAGGTTCCGAAGG | 153,813 | 151,813 | 99.02 |
| S | GGCTTATAGGTTTAATGGTATTGG | 153,259 | 99.96 | ||
| Delta | ORF1ab | GGTGTTGTTGGAGAAGGTTCCGAAGG | 184,757 | 180,642 | 97.77 |
| S | GGCTTATAGGTTTAATGGTATTGG | 184,691 | 99.96 | ||
| Beta | ORF1ab | GGTGTTGTTGGAGAAGGTTCCGAAGG | 13,185 | 12,942 | 98.16 |
| S | GGCTTATAGGTTTAATGGTATTGG | 13,175 | 99.92 | ||
| Gamma | ORF1ab | GGTGTTGTTGGAGAAGGTTCCGAAGG | 36,027 | 35,930 | 99.73 |
| S | GGCTTATAGGTTTAATGGTATTGG | 36,012 | 99.96 | ||
| Lambda | ORF1ab | GGTGTTGTTGGAGAAGGTTCCGAAGG | 211 | 211 | 100 |
| S | GGCTTATAGGTTTAATGGTATTGG | 211 | 100 | ||
| Mu | ORF1ab | GGTGTTGTTGGAGAAGGTTCCGAAGG | 3,266 | 3,247 | 99.42 |
| S | GGCTTATAGGTTTAATGGTATTGG | 3,263 | 99.91 |
| MB1 | MB2 | |||||
|---|---|---|---|---|---|---|
| Temperature (°C) | t = 0 Min | t = 5 Min | t = 21 Min | t = 0 Min | t = 5 Min | t = 21 Min |
| 65 | 51.05 | 45.09 | 42.31 | 61.29 | 66.81 | 65.94 |
| 63.8 | 58.65 | 57.64 | 56.51 | 60.09 | 62.01 | 60.86 |
| 62 | 59.68 | 57.81 | 56.75 | 78.69 | 72.64 | 64.07 |
| 59.1 | 64.42 | 63.26 | 62.18 | 82.31 | 73.63 | 65.91 |
| 55.7 | 59.58 | 57.75 | 55.69 | 49.42 | 48.48 | 45.86 |
| 52.9 | 72.69 | 68.76 | 63.37 | 77.06 | 68.49 | 62.01 |
| 51 | 68.03 | 66.23 | 61.45 | 76.58 | 69.99 | 66.78 |
| 50 | 60.36 | 62.92 | 60.34 | 70.81 | 65.35 | 60.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, J.; Lopes-Nunes, J.; Figueiredo, J.; Santos, T.; Miranda, A.; Riscado, M.; Sousa, F.; Duarte, A.P.; Socorro, S.; Tomaz, C.T.; et al. Molecular Beacon Assay Development for Severe Acute Respiratory Syndrome Coronavirus 2 Detection. Sensors 2021, 21, 7015. https://doi.org/10.3390/s21217015
Carvalho J, Lopes-Nunes J, Figueiredo J, Santos T, Miranda A, Riscado M, Sousa F, Duarte AP, Socorro S, Tomaz CT, et al. Molecular Beacon Assay Development for Severe Acute Respiratory Syndrome Coronavirus 2 Detection. Sensors. 2021; 21(21):7015. https://doi.org/10.3390/s21217015
Chicago/Turabian StyleCarvalho, Josué, Jéssica Lopes-Nunes, Joana Figueiredo, Tiago Santos, André Miranda, Micaela Riscado, Fani Sousa, Ana Paula Duarte, Sílvia Socorro, Cândida Teixeira Tomaz, and et al. 2021. "Molecular Beacon Assay Development for Severe Acute Respiratory Syndrome Coronavirus 2 Detection" Sensors 21, no. 21: 7015. https://doi.org/10.3390/s21217015
APA StyleCarvalho, J., Lopes-Nunes, J., Figueiredo, J., Santos, T., Miranda, A., Riscado, M., Sousa, F., Duarte, A. P., Socorro, S., Tomaz, C. T., Felgueiras, M., Teixeira, R., Faria, C., & Cruz, C. (2021). Molecular Beacon Assay Development for Severe Acute Respiratory Syndrome Coronavirus 2 Detection. Sensors, 21(21), 7015. https://doi.org/10.3390/s21217015











