1. Introduction
Electrocardiogram (ECG) has always been among the most investigated signals due to multiple reasons. ECG monitoring allows preventing cardiovascular and many correlated diseases (such as diabetes and hypertension) [
1], and it is widely adopted in surgical interventions, sport activity monitoring, and daily home healthcare [
2]. As stated in [
3,
4], the standard 12-lead ECG recording is able to effectively reflect rich spatial information of the heart’s movements allowing the recognition of specific pathological features [
5]; thus, it is widely used in clinic and hospital applications. In a standard 12-lead monitoring system, heart activity is detected by employing up to 10 electrodes placed in standardized positions, which provide information regarding the following: three bipolar limb leads I, II, and III; three augmented limb leads, aVR, aVL, and aVF; and six precordial leads, V1, V2, V3, V4, V5, and V6. The cardiologist is able to make a diagnosis by comparing the ECG signals acquired from the different leads with reference ECG waveforms reported in the scientific literature. The minimum number and set of selected leads depend on the need of observing specific waveforms that could be then related to specific pathologies. As a consequence, an analysis aimed at determining the minimum number of sensors to be used, as reported in [
6], cannot be carried out unless referring to specific pathologies. Although providing complete ECG information, the acquisition of 12-lead ECG in people’s daily lives is still a challenge [
3]. In fact, several devices are occasionally used for at-home patient monitoring; nevertheless, they are still not convenient and comfortable enough for 24/7 usage due to the obtrusive 12-lead setting [
3].
Nowadays, continuous remote monitoring of physiological parameters is demanded for patients performing therapy at home, and it is currently allowed by Internet-of-Things technology, which in this case takes the term of Internet of Medical Things (IoMT). IoMT generally constitutes an elaborated paradigm, with medical things meaning wearable devices and smart sensors tied to human bodies (sometimes implanted in bodies), allowing the acquisition of biosignals and other vital parameters. Standard configurations of IoMT remote monitoring systems, where ECG signal is acquired at Nyquist rate, do not suit storage and transmission requirements as a huge amount of data need to be stored and transmitted [
7], especially when a high number of patients is monitored [
8]. Moreover, power consumption is often related to signal data rate, since data transmission is the main cause of energy dissipation in many interfaces (such as Wireless Local Area Network (WLAN) and Wireless Wide Area Network (WWAN) interfaces) [
9].
In Wavelet or Fourier domains, ECG signals are demonstrated to be sparse [
9], i.e., they can be represented by a reduced number of samples. Relying on sparsity property, the technique of Compressed Sensing (CS) has been proposed in order to reduce the number of samples representing the signals of interest and to reconstruct their digital version without compromising signal quality [
10]. The CS has been vastly investigated in the field of biomedical signal processing not only for ECG monitoring but also for electroencephalogram, electromyogram, electrooculogram, galvanic skin response, and heart sound signals [
9,
11,
12]. Adopting CS entails a reduction in data rate with respect to signal information content. Among the resulting benefits, it is possible to mention not only an increase in battery lifespan of sensor nodes but also an easier allocation of time slots for communications, e.g., when multiple systems are used sharing the same band or different physiological signals are acquired simultaneously.
ECG acquisition based on CS has been implemented both in hardware and software by means of analog or digital methods. The main advantage of analog methods, typically known as compressive sampling, consists in decreasing the sampling rate by employing architectures working below the Nyquist limit [
13]. On the other hand, digital methods are not intended to sample under the Nyquist rate but are intended to reduce the number of samples to be transmitted. Digital approaches have the main advantage of lower power consumption due to the lack of dissipating devices (such as mixers) in hardware implementations; thus, they are considered preferable in wireless applications [
9]. In reality, other compression algorithms for ECG signals are built on transforms in Fourier or Wavelet domains [
14,
15,
16,
17,
18] or on fractal-based transforms [
19]. These algorithms are potentially able to reconstruct the original ECG signal with good performance. Nevertheless, their high computational complexity and buffer requirements do not adapt to real time implementations of ECG monitoring [
14,
15]. Differently from the aforementioned compression algorithms, the CS reduces instead the computational complexity related to the compression phase such that it complies with the limited physical resources of IoMT sensor nodes. Instead, the most computationally heavy task which consists of the waveform reconstruction relies in the higher layers of the IoMT architecture that are usually deployed in the cloud where powerful resources are available. Research is also in progress in order to perform anomaly detection directly from compressed samples without the need of reconstructing the waveforms [
20].
Although multiple electrodes and leads are actually adopted by measurement systems in biomedical field, ECG monitoring through CS has been addressed in the literature mainly in the case of one electrode with one lead [
9,
13,
21,
22,
23,
24,
25,
26]. The aim of this paper is to propose a digital CS-based method for ECG monitoring of multi-lead signals. Obviously, a significant part of the information content sensed by various electrodes is common to all the signals on the different leads. Such property is exploited by the proposed method by jointly reconstructing the multi-lead signals from the same support. The multi-lead proposed method implements a dynamic deterministic approach [
26] in the sense that the compression mechanism is adapted to the acquired signals to include more information on cardiac features and improve reconstruction quality. The method is designed to be implemented in the Ambient-Intelligent Tele-monitoring and Telemetry for Incepting and Catering Over hUman Sustainability (ATTICUS) system described in [
27]. This system consists of a smart T-shirt, called S-WEAR, capable of monitoring from one up to six ECG leads. S-WEAR has a modular architecture which can be adapted for multi-leads monitoring without substantially increasing overall costs of the entire system and without compromising its comfort. The ATTICUS system is characterized by a three-level Decision Support System (DSS). The first level is installed in the S-WEAR and allows automatically detecting a limited set of anomalies, such as when the heart rate is too high. The second level is installed on an electronic device, called S-BOX, that is installed at the patient’s home, and is able to detect a broader range of anomalies, such as irregular heartbeat. Finally, the third level is installed on the server, and it makes use of advanced machine-learning techniques to decide if it is necessary to notify the alarm of the physician at the monitoring center. They allow automatic detection: atrial fibrillation, ventricular tachycardia, congestive heart failure, and four types of arrhythmia conditions. Some of these machine learning techniques work on multi-lead acquisitions for assuring high accuracy [
27]. If an alarm is sent to the physician, long recordings of the acquired multi-lead signals before and after the detected event are sent to her/him in order to make a diagnosis. This requires the storage of a large amount of data that need to be compressed.
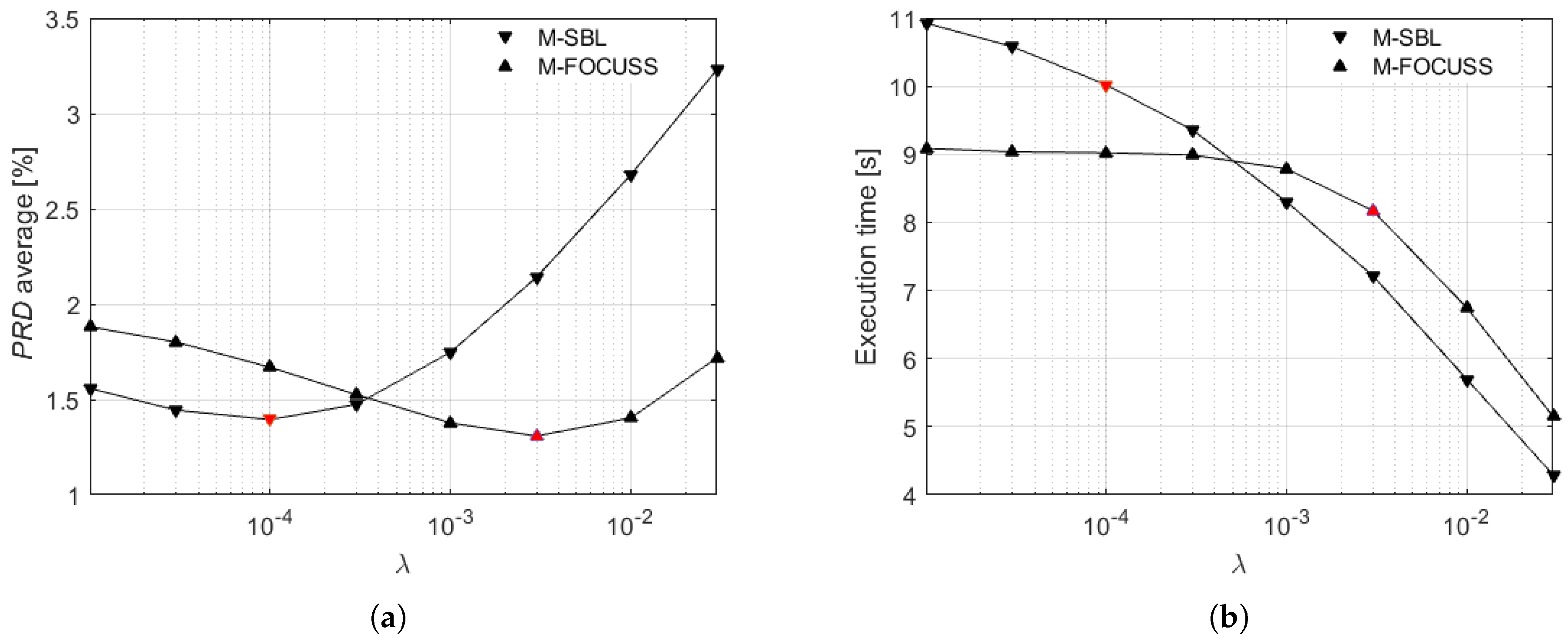
A preliminary version of the method has been introduced in [
28], where the multi-lead reconstruction was carried out by dynamically evaluating the signal frame acquired on the first lead through a threshold set by a percentile. In the work presented here, the following innovations have been added: (i) the dynamic sensing matrix is built from a combination of the most significant leads in terms of information content, i.e., lead II and aVF, instead of the first lead; (ii) for the reconstruction, a comparison in terms of signal quality and execution time between Multiple Sparse Bayesian Learning (M-SBL) and the Multiple FOCal Underdetermined System Solver (M-FOCUSS) is performed; (iii) experimental validation is extended with a set of signals of subjects affected by specific pathologies; and (iv) an experimental comparison with other four relevant literature methods proposing CS for multi-lead ECG monitoring has been added.
The rest of the paper is organized as follows. An overview of the CS-based methods available in the literature for the compression of multi-lead ECG signals is proposed in
Section 2.
Section 3 presents the proposed method by detailing the two phases of compression and reconstruction. In
Section 4, the implementation of the proposed method is described.
Section 5 illustrates the experimental results of the proposed method for several sets of signals. Specifically, in
Section 5.1, an analysis versus the regularization parameter used in the reconstruction algorithm is presented. In
Section 5.2, the performance of the proposed method versus the compression ratio and the number of leads is analyzed. In
Section 5.3, an experimental comparison of the results of the proposed method with those achieved other four relevant literature methods is reported. Lastly,
Section 6 is devoted to conclusions.
2. Related Works
As stated above, in the literature, few studies are focused on the use of CS methods for multi-lead monitoring [
7,
29,
30,
31]. Those methods outperform the single lead CS methods because they rely on the fact that the ECG signals from multi-lead channels are not independent but they have the electrical heart vector as a common source of information. In particular, the ECG signals on the different leads correspond to the projections in different directions of the electrical heart vector.
In [
31], the adopted multi-lead CS method is based on Filtered Modulated-Multiplexer (FM-Mux) architecture. In this case, firstly, each ECG signal is modulated with a pseudo-random sequence having a rate higher than the analyzed bandwidth. Secondly, the signals are convoluted with low-pass linear time-invariant (LTI) filters having a bandwidth equal to the rate of the sequence. Finally, the signals are added together onto a single channel and uniformly sampled by an Analog-to-Digital Converter (ADC) at a rate lower than the Nyquist one and equal to the rate of the pseudo-random sequence. The results of [
31] demonstrated that the reconstruction quality in terms of
is lower than 9% by considering a Compression Ratio (
) of 5.
In [
30], the ECG signals are acquired by ADCs working at the Nyquist rate and then compressed by multiplying the acquired samples with a sparse binary matrix. The results demonstrate that the adopted method achieves good reconstruction quality, i.e.,
lower than 9%, for
around 3.
A multi-lead CS method, based on the same sparse binary matrix as before is proposed in [
7]. In this case, the performance of several Weighted mixed-norm minimization (WMNM)-based joint sparse recovery algorithms was assessed, and a
around 7% was achieved with a
of 7 by means of the Prior Weighted MNM (PWMNM) algorithm.
In [
29], a sparse binary matrix was adopted as a sensing matrix together with a sparsity matrix based on the Daubechies 6 wavelet. In this case, a
lower than 9% was achieved with a
around 4 [
29].
The above-mentioned methods are based on sensing matrices that are randomly built; on the other hand, in the work here presented, a deterministic matrix is adopted. This deterministic matrix was already tested in [
26] for single lead monitoring. In this paper, its use is analyzed in the case of multi-lead monitoring with the aim of outperforming the performance in terms of reconstruction quality of random-based approaches.
3. The Proposed Method
IoMT networks allow constantly assessing the health status of subjects monitored through biomedical measurement systems [
26]. The working principle of a generic IoMT-enabled system is based on the transfer of information from several sensor nodes to a cloud server that constitute a physical layer and the information integration layer of the IoMT model [
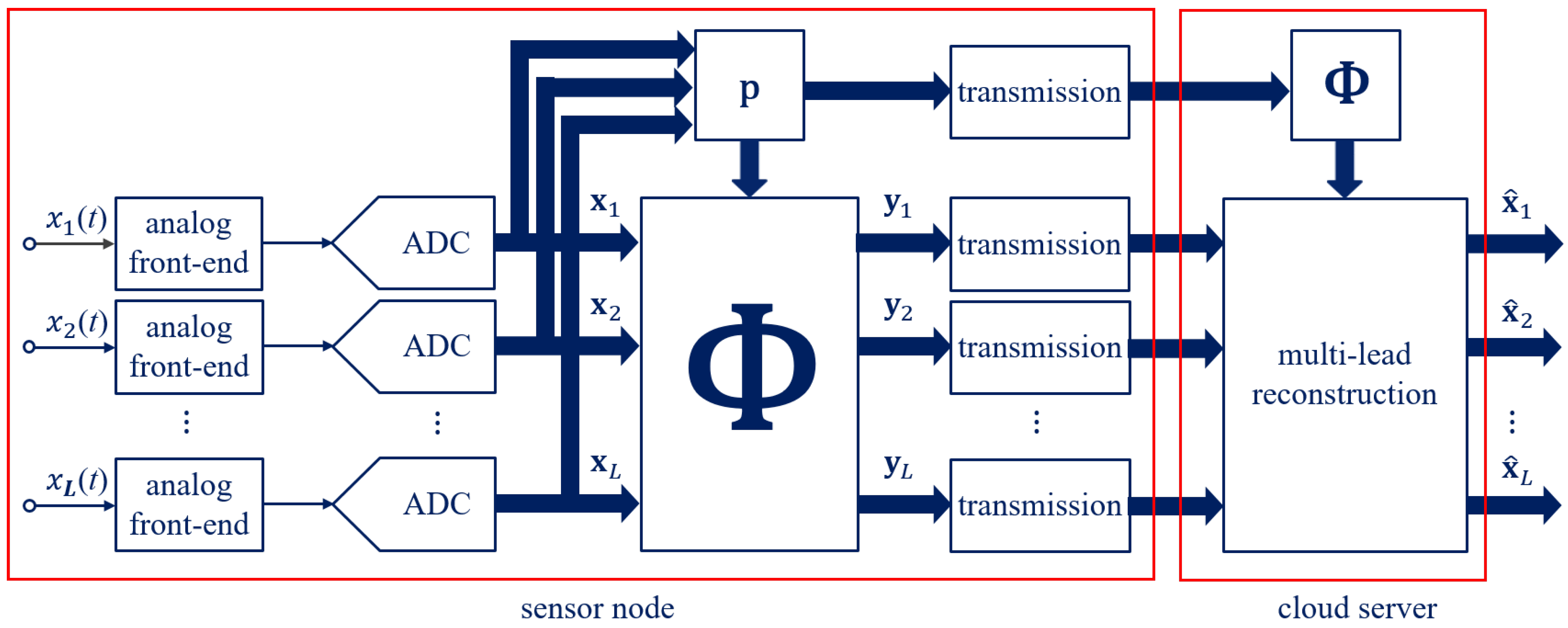
32], respectively. Each sensor node acquires and transmits the biosignal samples. The cloud server stores the received samples, which are later employed to recover original information about the health status. Therefore, in an IoMT network—which can be shared also among thousands of nodes—considerable data rates have to be handled.
Generically, the sensor node of the ECG monitoring system consists of multiple electrodes that form
L leads. The ECG signal on each lead
, with
, is acquired through an analog front-end and an Analog-to-Digital Converter (ADC) working at the Nyquist rate. A record of
N acquired samples is here represented as the vector
. Overall, in the time frame of each record, the ECG monitoring system acquires and transmits a matrix of
samples.
Let each vector acquired at the Nyquist rate, i.e., each column
of the matrix
, be sparse, i.e., represented by a few non-null coefficients in a given transform domain described by a
sparsity matrix . In this case, each vector
can be expressed as follows:
where
is a vector of the signal coefficients with few nonzero elements. Overal, the matrix
can be expressed as follows:
with
.
The ECG signal is usually sparse in several Wavelet and Fourier domains. As reported in [
9], one of the sparsity matrices achieving the highest reconstruction performance is obtained by means of the Mexican hat wavelet kernel. In this paper, a Mexican hat wavelet matrix
defined according to [
33] is used:
where
is a vector that describes the Mexican hat wavelet function having
, and
and
denote scale and translation parameters, respectively:
with
.
The multi-lead method proposed in this paper for ECG monitoring is intended to optimize, through CS [
10], the transfer of information in IoMT networks. In particular, as depicted in
Figure 1, the sensor node sub-samples the multi-lead ECG signals coming from electrodes, while the cloud server jointly reconstructs them. The sensor node can be implemented by simple hardware consisting of electrodes and leads along with a microcontroller and a radio frequency interface, since the ECG signals are firstly acquired at the Nyquist rate and then compressed. The joint reconstruction is instead relied on by the cloud server and is typically characterized by unconstrained resources. The multi-lead proposed method consisting in the two phases of compression and reconstruction is detailed in the following.
In the sensor node, the vector acquired on each lead,
, is compressed in an
M-dimensional measurement space, with
. The core idea behind the proposed method is the formulation of a dynamic
sensing matrix of size
that sub-samples every frame of
N samples of the multi-lead signals and that is effective for a joint reconstruction. In particular, data reduction in sub-sampling systems is typically expressed by the
Compression Ratio (
).
In order to build the dynamic sensing matrix
, a vector
of
N binary digits must be preliminarily introduced. The vector
is defined from a vector
that constitutes a proper combination of the leads with the most significant contribution. Specifically, the vector
is built as the root mean square of the bipolar limb lead II and the augmented limb lead aVF.
Actually, different lead combinations can also be considered. For example, another combination allowing to sub-sample with high
values is the root mean square of the leads II, aVR and V6, where one lead for each lead group is selected. Generally, lead II is the most commonly used one for accurately assessing cardiac rhythm as it usually provides a good view of the P wave [
34], while including precordial leads may help to follow R wave progression. Another vector, called
, is obtained from the vector
, and the average
of
is defined as follows.
Then, a threshold value
is determined starting from the vector
. The threshold
is given by the 60th percentile computed on the
N samples of
. The percentile value is fixed on the basis of an experimental analysis. The vector
is evaluated by comparing
with
. In particular, it contains 1 when the corresponding sample of
is higher than or equal to
; otherwise, it is 0:
where
is the
n-th element of the vector
, with
. In this manner, the vector
presents ones where the ECG acquired in the selected leads has higher amplitude. Finally, the sensing matrix
is built by the vector
circularly shifting on each row by an amount equal to
.
The compression consists of the multiplication of the the vector
acquired on each lead by the sensing matrix
, thus obtaining
M compressed samples or overall the matrix
of size
.
The compression process can be interpreted as the cross-correlation of each vector acquired on the different leads with the vector. The matrix and the vector are then transmitted to the cloud server.
In the cloud server, the ECG received in compressed form can be reconstructed at the Nyquist rate. First of all, the sensing matrix
is rebuilt within the cloud server in the same manner as in the sensor node, thanks to the vector
. Moreover, the use of a sparsity matrix is required for the ECG reconstruction. In addition to the columns reported in (
4), a further
N-size column
is employed to take into account a possible offset in the signal.
It is worth noting that the signals on the different leads are highly correlated, since they have the heart as common source and are synchronous. Therefore, they will share the same support, and the nonzero elements of each column
will lie mostly in the same positions. This means that the matrix
is row-sparse; that is, it has few nonzero rows. Exploiting this consideration, the matrix
can be reconstructed by starting from the matrix of compressed samples
(
11), the dynamic sensing matrix
(
10), and the Mexican hat wavelet matrix
(
12) and by solving a joint sparse recovery problem that can be expressed as the following [
35]:
where
denotes the Frobenius norm,
is a regularization parameter that must be chosen to balance quality in the estimation of the cost function with sparsity [
35,
36], and
J is a cost function that uses a
p-norm, with
, in order to lead towards a sparse solution:
with
the
n-th row of
. The cost function (
14) counts the number of non-null rows in
, and it is the extension to matrices of the
-norm that counts the non-null elements in a vector [
35,
36]. Once the
has been obtained, the matrix
is evaluated as follows.
The reconstruction is performed on each frame of
transmitted samples that are compressed in the sensor node from the
acquired samples. Fundamentally, through the proposed method, only the
matrix
of the compressed samples is transmitted together with the
N-size vector
in place of the
matrix
of the samples acquired at the Nyquist rate. The main advantage of the proposed method is the dynamic ECG evaluation. In other CS implementations for ECG monitoring, the sensing matrix
is usually randomly constructed according to a probability distribution [
9,
13,
21,
25,
29]. As an example, in [
21], the reconstruction performance of different CS methods is compared by considering several distributions, such as Bernoulli or Gaussian. However, adopting random matrices entails that the reconstruction performance may significantly vary depending on the correlation between the entries of the sensing matrix and the acquired samples. Such limitation is overcome in the proposed ECG monitoring system, since the sensing matrix is not randomly generated. Deterministic matrices have been already proposed in [
22,
23] as a viable alternative for ECG compression that does not require random number generation on the chip [
22]. Indeed, random number generation on chip can be a heavy computational load for sensor nodes equipped with simple microcontrollers, while deterministic matrices could easily be employed also in wearable devices. The substantial difference of the proposed method with the other deterministic methods [
22,
23] is that the dynamic sensing matrix is built as a circulant matrix that depends on a combination of the leads with the most significant contribution. Thus, being adapted to the distribution of the acquired ECG, it contains more information on the signal features, guaranteeing better reconstruction quality. Finally, it is important to point out that the proposed method exploits the common information content that all the multi-lead signals share. In fact, only one sensing matrix and one sparsity matrix are adopted regardless the number of leads.
4. Implementation of the Proposed Method
The implementation of the proposed method for multi-lead ECG monitoring is presented in this Section. Both the phases of compression in the sensor node and reconstruction in the cloud server, described in previous
Section 3, were implemented in the MATLAB environment. ECG signals from the Physikalisch-Technische Bundesanstalt (PTB) Diagnostic ECG Database, available online at the PhysioNet website [
37], were examined. For each monitored patient, the signals related to the monitoring of 12 leads are available. For reducing the signal distortions due to the power line disturbances, the signals were filtered by a notch filter at the harmonics
, with
50 Hz and
. Each signal of the PTB database is sampled at 1
Samples/
. The duration of the signal frame was chosen equal to 1
; therefore, the size of each frame results in
. The size of
M depends, instead, on the adopted
.
When proposing a compression method, a good practice consists in verifying that the compression does not alter significantly the clinical information contained in the signal. The performance of a compression method for ECG signals and other biosignals is typically evaluated by the percentage of root-mean-squared difference (
) [
9,
12,
13,
21,
23,
24,
25,
26,
28,
29]. In this paper, the
is computed for the ECG signal related to each lead
l:
where
is the vector of the ECG samples acquired at the Nyquist rate, and
is the reconstructed vector, with
indicating the
-norm. In ECG monitoring, clinical information contained in the original signal acquired at the Nyquist rate is considered generally preserved if the reconstructed signal exhibits a
lower than 9% [
21]. Therefore, in analyzing the proposed method, particular attention will be paid to such value as upper bound for good reconstruction and monitoring.
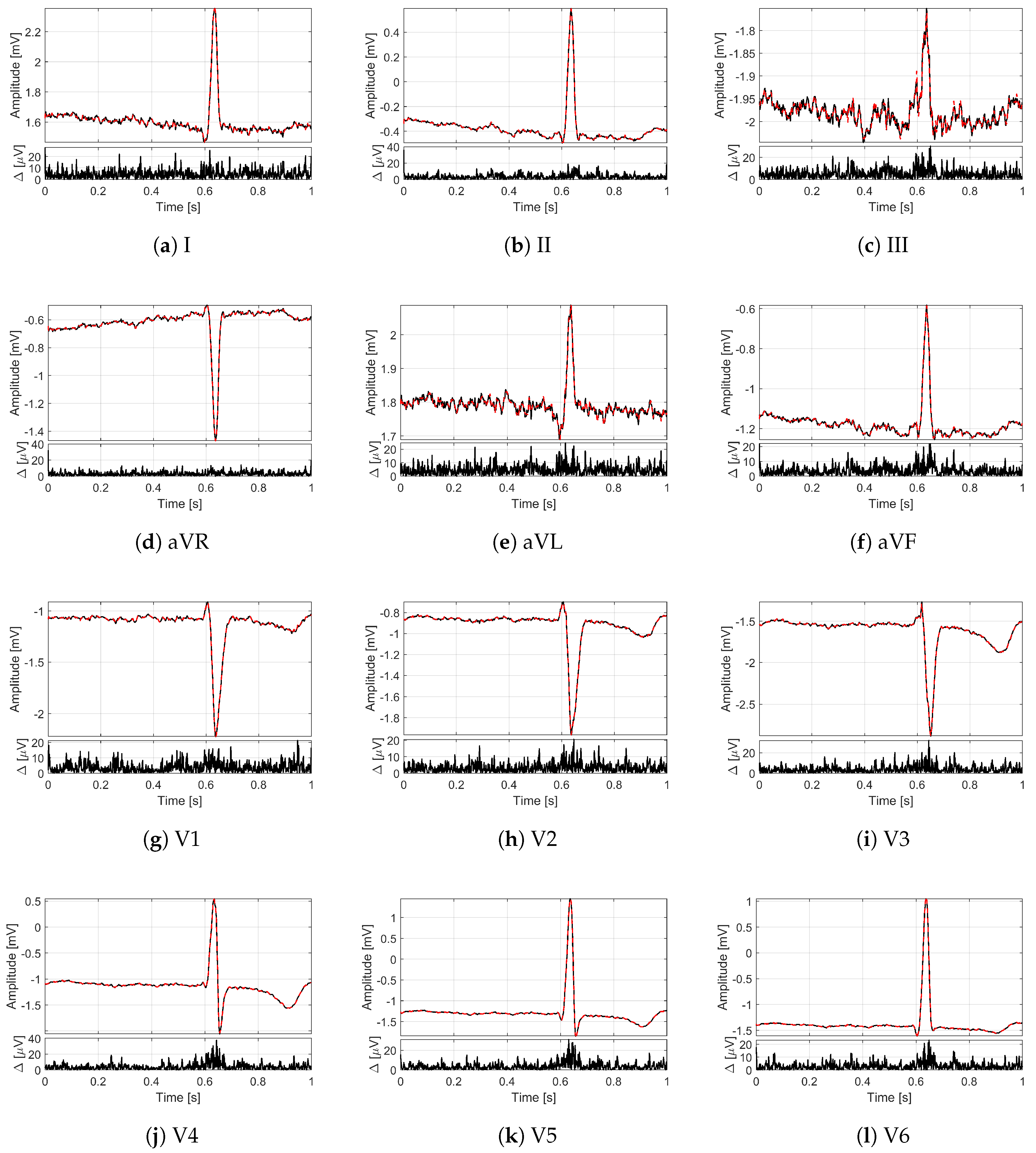
In order to provide an idea of the reconstruction quality achieved by the proposed method for multi-lead ECG monitoring,
Figure 2 and
Figure 3 illustrate some recordings from the PTB database labeled as myocardial infarction. The minimization problem (
13) with 12 measurement vectors was solved by using the M-FOCUSS algorithm [
35], with regularization parameters fixed to
. The original signals are drawn with black lines, while the signals reconstructed from
compressed samples (i.e., with
) are represented by red dashed lines. In particular, in
Figure 2, the lead I of the signal
is shown for a time window of 10
where different deflections of the QRS complex can be appreciated.
Figure 3 depicts, instead, all of the 12 leads of the signal
for a time window of 1
corresponding to a single frame. Moreover, for each lead, the absolute value of the difference
between original and reconstructed signals is reported too. What is worth noting is that all the 12 leads are correctly reconstructed. In fact, a good overlapping with the original signals can be appreciated not only for leads II and aVF (
Figure 3b,f), which are employed to build the sensing matrix but also for all the other leads. The performance in terms of
confirms the graphical results since the obtained value for the signal of
Figure 2 is 1.09%, while the obtained values for the signal of
Figure 3 vary from a minimum of 1.99% for lead V2 to a maximum of 5.08% for lead I. In any case, the
for
is much lower than the limit of 9%.
6. Conclusions
In this paper, a CS-based method for multi-lead ECG signal monitoring has been presented. In detail, the proposed method employs a deterministic sensing matrix dynamically built from a vector obtained by a proper combination of ECG signals of two different leads. According to such vectors, for each ECG frame, a compressed version of the signal is obtained and then transmitted to the cloud server by the sensor node, together with the vector determining the sensing matrix. Thus, in the cloud server, the sensing matrix can be rebuilt, and all the ECG leads can be recovered. Specifically, the sparsity matrix is based on a Mexican hat wavelet kernel.
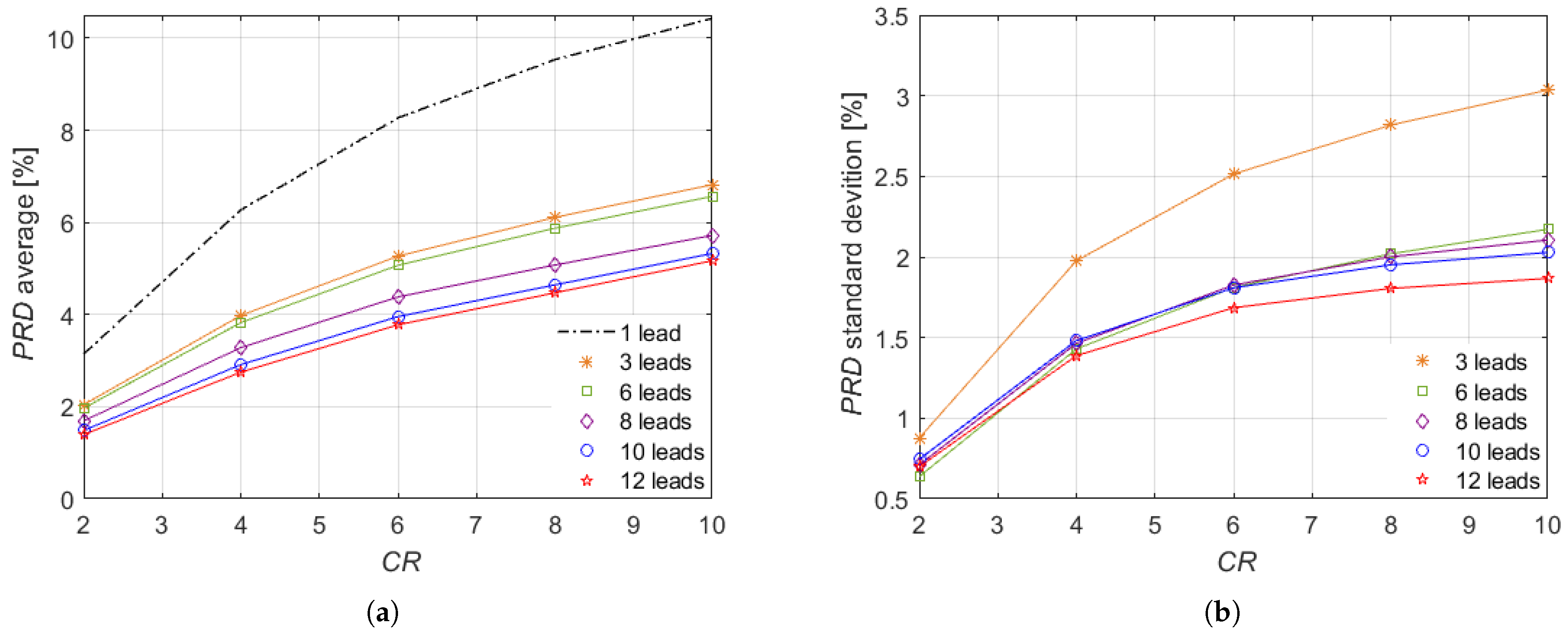
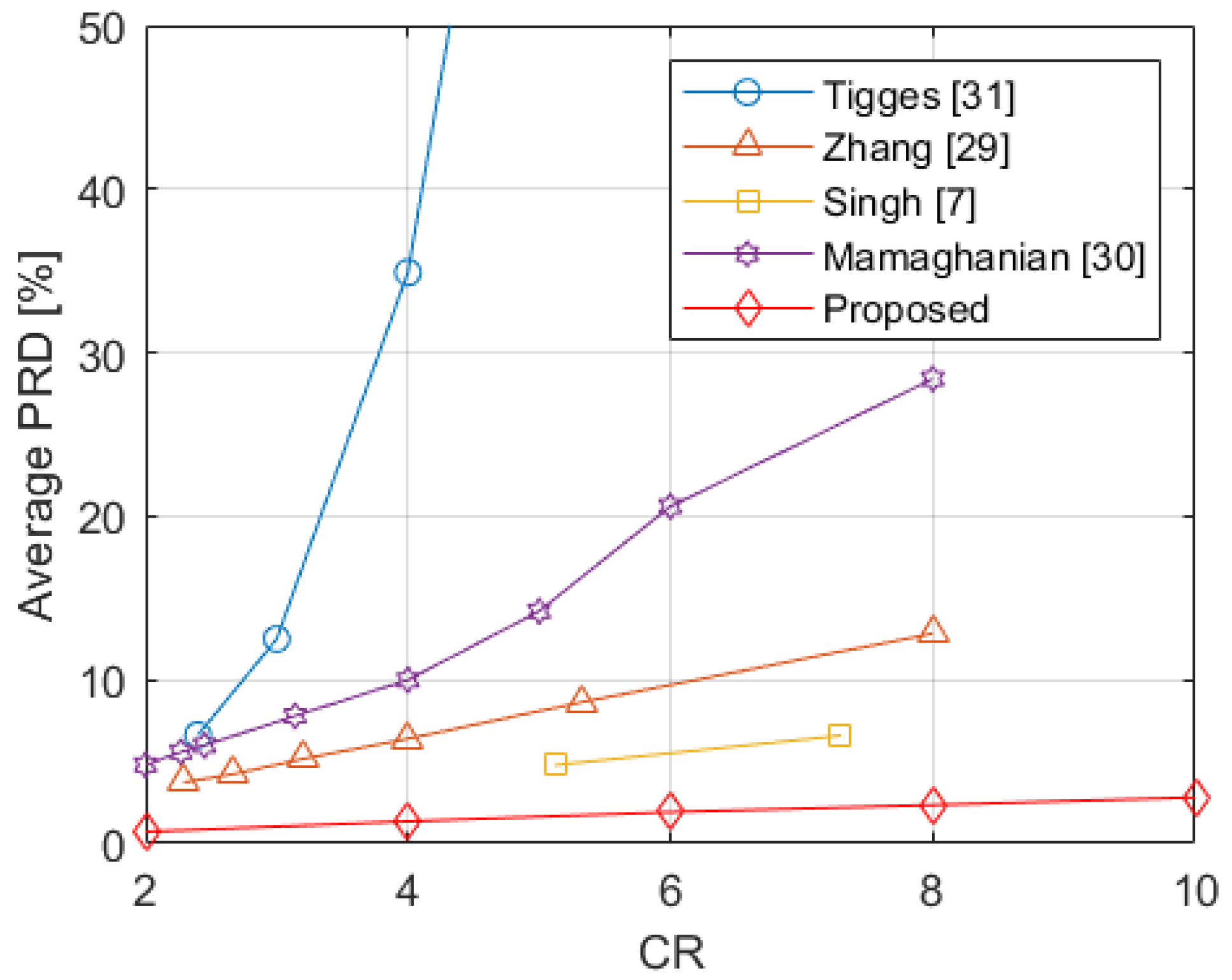
The method was evaluated through several investigations on a wide set of signals. The values obtained from the proposed method were analyzed against the number of considered leads and . The experimental results show better performance in case of ECG monitoring system with all the conventional 12 leads. In any case, the values are always lower than the bound of 9%, which has been indicated for the preservation of ECG information up to and reveals the proposed method suitability for ECG monitoring of subjects considered healthy as well as affected by pathologies such as myocardial infarction, cardiomyopathy, and bundle branch block. Furthermore, the method was compared with other four relevant literature papers proposing CS for multi-lead ECG monitoring. The proposed method improves significantly the signal reconstruction quality, as demonstrated by the lowest obtained in the experimental results among all the considered CS methods.
As future work, the implementations and testing on the hardware of the proposed method have been planned with the aim of demonstrating its suitability to be implemented on wearable devices in IoMT applications.