Detection of Mild Cognitive Impairment with MEG Functional Connectivity Using Wavelet-Based Neuromarkers
Abstract
:1. Introduction
- (1)
- A regional analysis of the effectiveness of different cortical regions for MCI detection is undertaken to examine the phenomena of hyper- and hypo-connections between regions of interest (ROIs) which may have a significant impact on MCI vs. HC classification with respect to ROI selections.
- (2)
- Based on the findings from the regional analysis, a system for MCI/HC classification is developed. In particular, the effectiveness of the novel wavelet-based neuromarkers for MCI detection is evaluated. The results show that the proposed method can achieve an average classification accuracy of about 84%, using only MEG data from the selected cortical regions.
- (3)
- A comparative analysis using two different evaluation procedures is undertaken. One is based on the well-known Monte Carlo repetition of random splits, i.e., into 95/5% of training/test subsets. The other test procedure is a subject-based 20-fold leave-one-out (LOO) cross-validation, in order to better mimic the actual clinical scenario with individual participants (each class contained 20 subjects).
- (4)
- The region selection and recognition problems are addressed from a practical perspective by exploring the statistical significance of the results, which is a crucial aspect that is often ignored in the performance evaluation for such recognition systems.
2. Region of Interest Analysis
2.1. Connectivity Indicator Computation
2.2. Pair-Wise ROI Analysis
- (1)
- Each ROI contained hundreds of MEG values in the source space. First, the values of each ROI were averaged: this resulted in ten mean MEG values for the ten selected ROIs.
- (2)
- For each averaged MEG value per ROI, the coherences with respect to the remaining nine mean ROI values were computed, which produced nine coherences for each ROI.
- (3)
- The nine coherence values for each ROI were summed together and the resulting summation is defined as the degree of connectivity for each ROI.
- (4)
- Considering that there are 20 participants per class and 180 s per participant, 20 × 180 = 3600 epoch per ROI for each class are produced.
- (5)
- The max, min, mean, and SD for these 3600 epochs, with respect to the degree of connectivity for each ROI, are computed and shown in Table 1.
2.3. Statistical Analysis for the Connectivity Indicator
3. Methodology for MCI Detection
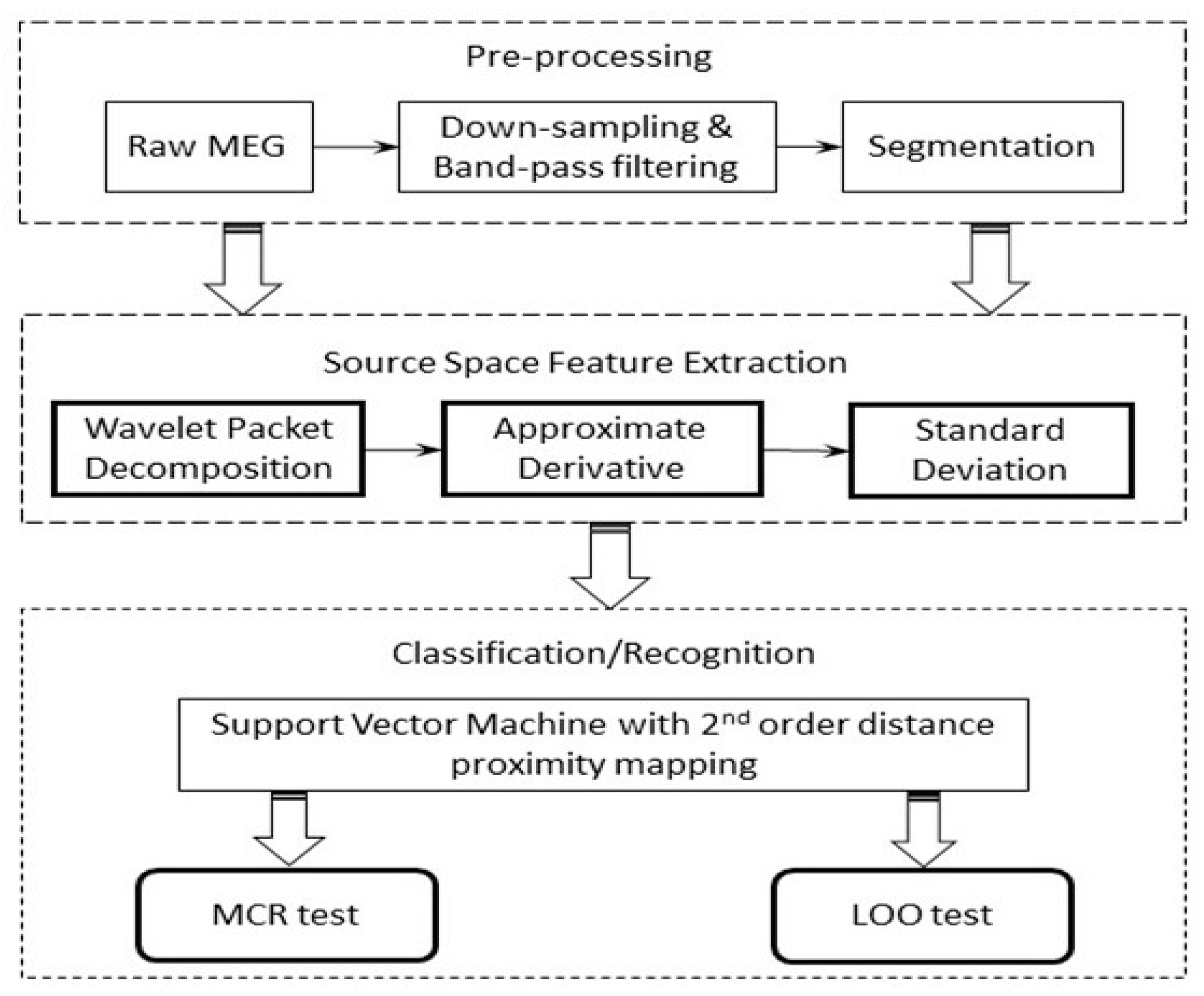
3.1. Pipeline of the MCI Detection System
- (1)
- For the source space MEG data, in total, 180 s of the recording was preserved, which was then further segmented into a number of windows in the time domain.
- (2)
- The epochs (10 s per epoch) were then down-sampled to 200 Hz, followed by a band-pass filtering process extracting the signal between 0.1 Hz to 100 Hz, which cover all the typical bands (Delta, Theta, Alpha, Beta, and Gamma) using a 4th-order Butterworth filter. Here, we purposely included the frequency content that was higher than 60 Hz, as in principle the MEG modality may retain useful information in the high-frequency range (high gamma band).
- (3)
- The wavelet packet decomposition was performed on the resulting time-domain signals. WPD was performed up to level 3; each level covers the selected full frequency range, which facilitates the multi-resolution analysis of the signal in question.
- (4)
- (5)
- Finally, the multi-dimensional feature vector was constructed by concatenating the standard deviations (SDs) of the obtained approximate derivative coefficients from each band.
- (6)
- A range of classifiers that have shown to be effective for MEG/EEG signal classifications were employed to investigate the effectiveness of the features, including k-nearest neighbor (k-NN), linear discriminate analysis (LDA), and support vector machine (SVM).
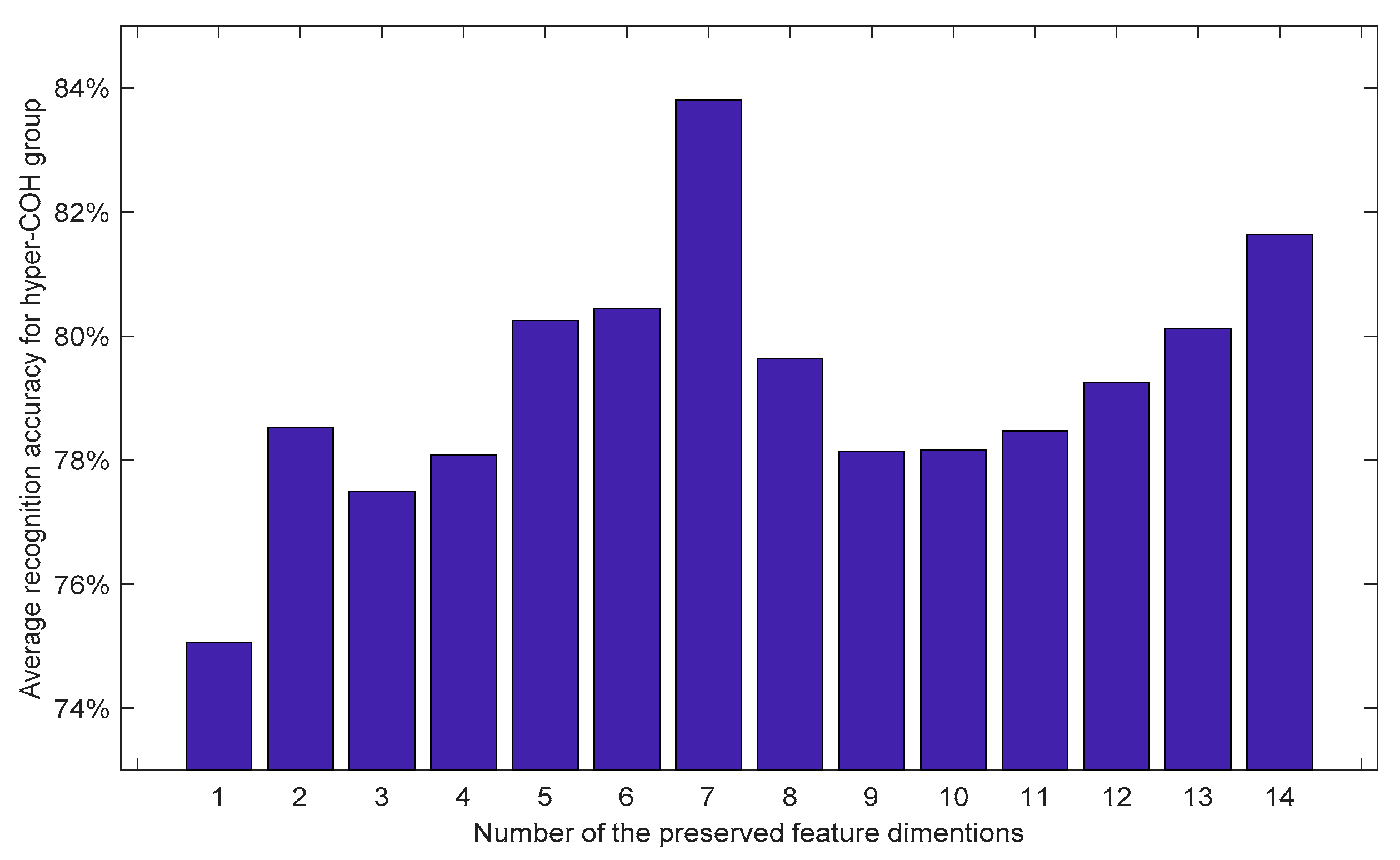
3.2. Experimental Analysis on Classification
3.3. Statistical Analysis of the Neuromarkers for Classification
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deng, J.; Xu, X.; Zhang, Z.; Fruhholz, S.; Schuller, B. Universum Autoencoder-Based Domain Adaptation for Speech Emotion Recognition. IEEE Signal Process. Lett. 2017, 24, 500–504. [Google Scholar] [CrossRef]
- Ciresan, D.; Meier, U.; Schmidhuber, J. Multi-column deep neural networks for image classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 3642–3649. [Google Scholar]
- Medvedeva, A.V.; Yahno, N.N. Functional Connectivity as a Neurophysiological Biomarker of Alzheimer’s Disease. J. Alzheimer’s Park. Dement. 2018, 3, 1–8. [Google Scholar]
- Gross, C.G. Brain, Vision, Memory: Tales in the History of Neuroscience; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Yang, S.; Bornot, J.M.S.; Wong-lin, K.; Prasad, G. M/EEG-based Bio-markers to predict the Mild Cognitive Impairment and Alzheimer’s disease: A Review from the Machine Learning Perspective. IEEE Trans. Biomed. Eng. 2019, 66, 2924–2935. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Cuesta, P.; Fernández, A.; Arahata, Y.; Iwata, K.; Kuratsubo, I.; Bundo, M.; Hattori, H.; Sakurai, T.; Fukuda, K.; et al. Electromagnetic sig-natures of the preclinical and prodromal stages of Alzheimer’s disease. Brain 2018, 141, 1470–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacini, F.; Sorrentino, P.; Lardone, A.; Rucco, R.; Baselice, F.; Cavaliere, C.; Aiello, M.; Orsini, M.; Iavarone, A.; Manzo, V.; et al. Amnestic mild cognitive impairment is associated with frequency-specific brain network alterations in temporal poles. Front. Aging Neurosci. 2018, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitriadis, S.I.; López, M.E.; Bruña, R.; Cuesta, P.; Marcos, A.; Maestú, F.; Pereda, E. How to build a functional con-nectomic biomarker for mild cognitive impairment from source reconstructed MEG Resting-state activity: The combination of ROI representation and connectivity estimator matters. Front. Neurosci. 2018, 12, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Elekta Neuromag Vector View 306 Channel Meg|Mindset. Available online: https://www.mindsetconsultinggroup.com/index.php/what-we-do/medical-imaging/elekta-neuromag-vector-view-306-channel-meg (accessed on 18 June 2020).
- Poza, J.; Gómez, C.; García, M.; Corralejo, R.; Fernández, A.; Hornero, R. Analysis of neural dynamics in mild cognitive impairment and Alzheimer’s disease using wavelet turbulence. J. Neural Eng. 2014, 11, 026010. [Google Scholar] [CrossRef]
- Klonowski, W. Everything you wanted to ask about EEG but were afraid to get the right answer. Nonlinear Biomed. Phys. 2009, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Fiscon, G.; Weitschek, E.; Cialini, A.; Felici, G.; Bertolazzi, P.; De Salvo, S.; Bramanti, A.; Bramanti, P.; De Cola, M.C. Combining EEG signal processing with supervised methods for Alzheimer’s patients classification. BMC Med. Inform. Decis. Mak. 2018, 18, 1–10. [Google Scholar] [CrossRef]
- López, M.E.; Turrero, A.; Cuesta, P.; Lopez-Sanz, D.; Bruna, R.; Marcos, A.; Gil, P.; Yus, M.; Barabash, A.; Cabranes, J.A.; et al. Searching for Primary Predictors of Conversion from Mild Cognitive Impairment to Alz-heimer’s Disease: A Multivariate Follow-Up Study. J. Alzheimer’s Dis. 2016, 52, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Taulu, S.; Simola, J. Spatiotemporal signal space separation method for rejecting nearby interference in MEG measurements. Phys. Med. Biol. 2006, 51, 1759–1768. [Google Scholar] [CrossRef]
- Jatoi, M.A.; Kamel, N.; Malik, A.S.; Faye, I.; Begum, T. A survey of methods used for source localization using EEG signals. Biomed. Signal Process. Control 2014, 11, 42–52. [Google Scholar] [CrossRef]
- Hincapié, A.S.; Kujala, J.; Mattout, J.; Pascarella, A.; Daligault, S.; Delpuech, C.; Mery, D.; Cosmelli, D.; Jerbi, K. The impact of MEG source reconstruction method on source-space connectivity estimation: A comparison between mini-mum-norm solution and beamforming. Neuroimage 2017, 156, 29–42. [Google Scholar] [CrossRef]
- Vicente, J.M.F.; Álvarez-Sánchez, J.R.; De la Paz López, F.; Moreo, J.T.; Adeli, H. (Eds.) Natural and Artificial Computation for Biomedicine and Neuroscience: International Work-Conference on the Interplay Between Natural and Artificial Computation; Springer: New York, NY, USA, 2017; Volume 10337. [Google Scholar]
- Bousleiman, H.; Zimmermann, R.; Ahmed, S.; Hardmeier, M.; Hatz, F.; Schindler, C.; Roth, V.; Gschwandtner, U.; Fuhr, P. Power spectra for screening parkinsonian patients for mild cognitive impairment. Ann. Clin. Transl. Neurol. 2014, 1, 884–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornot, J.M.S.; Wong-Lin, K.; Ahmad, A.L.; Prasad, G. Robust EEG/MEG Based Functional Connectivity with the Envelope of the Imaginary Coherence: Sensor Space Analysis. Brain Topogr. 2018, 31, 895–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Long, X.; Aarts, R.M.; van Dijk, J.P.; Arends, J.B.A.M. A broadband method of quantifying phase synchronization for discriminating seizure EEG signals. Biomed. Signal Process. Control 2019, 52, 371–383. [Google Scholar] [CrossRef]
- Bendat, J.S.; Piersol, A.G. Random Data: Analysis and Measurement Procedures; John Wiley & Sons: Hoboken, NJ, USA, 2011; Volume 729. [Google Scholar]
- Nolte, G.; Bai, O.; Wheaton, L.; Mari, Z.; Vorbach, S.; Hallett, M. Identifying true brain interaction from EEG data using the imaginary part of coherency. Clin. Neurophysiol. 2004, 115, 2292–2307. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Liu, B.; Zhang, X.; Li, J.; Qin, W.; Yu, C.; Jiang, T. The Structural Connectivity Pattern of the Default Mode Network and Its Association with Memory and Anxiety. Front. Neuroanat. 2015, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Pusil, S.; López, M.E.; Cuesta, P.; Bruña, R.; Pereda, E.; Maestú, F. Hypersynchronization in mild cognitive impairment: The ‘X’ model. Brain 2019, 142, 3936–3950. [Google Scholar] [CrossRef]
- Frutos-Lucas, J.; de Cuesta, P.; Lopez-Sanz, D.; Peral-Suarez, A.; Cuadrado-Soto, E.; Ramirez-Toraño, F.; Brown, B.M.; Ser-rano, J.M.; Laws, S.M.; Rodriguez-Rojo, I.C.; et al. The relationship between physical activity, apolipo-protein E ε4 carriage, and brain health. Alzheimers. Res. Ther. 2020, 12, 1–12. [Google Scholar]
- Koelewijn, L.; Lancaster, T.M.; Linden, D.; Dima, D.C.; Routley, B.C.; Magazzini, L.; Barawi, K.; Brindley, L.; Adams, R.; Tansey, K.E.; et al. Oscillatory hyperactivity and hyperconnectivity in young APOE-ε4 carriers and hypoconnectivity in alzheimer’s disease. eLife 2019, 8, 1–25. [Google Scholar] [CrossRef]
- López-Sanz, D.; Serrano, N.; Maestú, F. The Role of Magnetoencephalography in the Early Stages of Alzheimer’s Disease. Front. Neurosci. 2018, 12, 572. [Google Scholar] [CrossRef] [Green Version]
- Bajo, R.; Maestú, F.; Nevado, A.; Sancho, M.; Gutiérrez, R.; Campo, P.; Castellanos, N.P.; Gil, P.; Moratti, S.; Pereda, E.; et al. Functional Connectivity in Mild Cognitive Impairment During a Memory Task: Implications for the Disconnection Hypothesis. J. Alzheimer’s Dis. 2010, 22, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Rice, J.A. Mathematical Statistics and Data Analysis; China Machine Press: Beijing, China, 2003. [Google Scholar]
- Sawilowsky, S.S.; Blair, R.C. A more realistic look at the robustness and Type II error properties of the t test to departures from population normality. Psychol. Bull. 1992, 111, 352. [Google Scholar] [CrossRef]
- Ramani, S. EDF Statistics for Goodness of Fit and Some Comparisons. J. Am. Stat. Assoc. 1974, 69, 730–737. [Google Scholar]
- Stephens, M.A. Tests Based on EDF Statistics. In Goodness-of-Fit Techniques; CRC Press: Boca Raton, FL, USA, 2017; pp. 97–194. [Google Scholar]
- Daubechies, I. Ten Lectures on Wavelets. Math. Comput. 1993, 61, 941. [Google Scholar] [CrossRef]
- Yang, S.; Deravi, F. Wavelet-Based EEG Preprocessing for Biometric Applications. In Proceedings of the 2013 Fourth International Conference on Emerging Security Technologies, Washington, DC, USA, 9–11 September 2013; pp. 43–46. [Google Scholar]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification; Wiley: Hoboken, NJ, USA, 2001. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Fugal, D. Conceptual Wavelets in Digital Signal Processing: An In-Depth, Practical Approach for the Non-Mathematician; Space & Signals Technical Pub.: San Diego, CA, USA, 2009. [Google Scholar]
- Yang, S.; Deravi, F.; Hoque, S. Task sensitivity in EEG biometric recognition. Pattern Anal. Appl. 2018, 21, 105–117. [Google Scholar] [CrossRef] [Green Version]
- Differences and Approximate Derivatives—MATLAB Diff—MathWorks United Kingdom. Available online: https://uk.mathworks.com/help/matlab/ref/diff.html (accessed on 11 August 2021).
- Kawabata, N. A Nonstationary Analysis of the Electroencephalogram. IEEE Trans. Biomed. Eng. 1973, 20, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Hoque, S.; Deravi, F. Improved Time-Frequency Features and Electrode Placement for EEG-Based Biometric Person Recognition. IEEE Access 2019, 7, 49604–49613. [Google Scholar] [CrossRef]
- Bucholc, M.; Ding, X.; Wang, H.; Glass, D.H.; Wang, H.; Prasad, G.; Maguire, L.P.; Bjourson, A.J.; McClean, P.L.; Todd, S.; et al. A practical computerized decision support system for predicting the severity of Alzheimer’s disease of an individual. Expert Syst. Appl. 2019, 130, 157–171. [Google Scholar] [CrossRef] [Green Version]
- Richardson, A. Nonparametric Statistics for Non-Statisticians: A Step-by-Step Approach by Gregory W. Corder, Dale I. Foreman. Int. Stat. Rev. 2010, 78, 451–452. [Google Scholar] [CrossRef]
- Miller, R.G. Nonparametric Techniques. In Simultaneous Statistical Inference; Springer: New York, NY, USA, 1981; pp. 129–188. [Google Scholar]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Brain Anatomy, Anatomy of the Human Brain. Available online: https://mayfieldclinic.com/pe-anatbrain.htm (accessed on 16 August 2021).
- Hillary, F.G.; Roman, C.A.; Venkatesan, U.; Rajtmajer, S.M.; Bajo, R.; Castellanos, N.D. Hyperconnectivity is a Fundamental Response to Neurological Disruption Introduction: Disconnecting the Brain. Neuropsychology 2015, 29, 59–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.; Bucholc, M.; Wang, H.; Glass, D.H.; Wang, H.; Clarke, D.H.; Bjourson, A.J.; Dowey, L.R.C.; O’Kane, M.; Prasad, G.; et al. A hybrid computational approach for efficient Alzheimer’s disease classification based on heterogeneous data. Sci. Rep. 2018, 8, 9774. [Google Scholar] [CrossRef] [PubMed]
- Youssofzadeh, V.; McGuinness, B.; Maguire, L.P.; Wong-Lin, K. Corrigendum: Multi-Kernel Learning with Dartel Improves Combined MRI-PET Classification of Alzheimer’s Disease in AIBL Data: Group and Individual Analyses. Front. Hum. Neurosci. 2017, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadel, F.; Bock, E.; Niso, G.; Mosher, J.C.; Cousineau, M.; Pantazis, D.; Leahy, R.M.; Baillet, S. Brainstorm. 2011. Available online: https://neuroimage.usc.edu/brainstorm/CiteBrainstorm (accessed on 6 June 2020).
- Desikan, R.S.; Ségonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T.; et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 2006, 31, 968–980. [Google Scholar] [CrossRef] [PubMed]






| Stats. | Mean | Max | Min | SD | ||||
|---|---|---|---|---|---|---|---|---|
| ROI | HC | MCI | HC | MCI | HC | MCI | HC | MCI |
| LF↑ | 3.36 | 3.53 | 5.55 | 5.81 | 2.26 | 2.29 | 0.51 | 0.56 |
| RF↓ | 3.54 | 3.53 | 9.49 | 7.87 | 1.91 | 2.17 | 0.80 | 0.66 |
| LT↓ | 3.20 | 3.19 | 5.76 | 5.53 | 2.00 | 2.15 | 0.47 | 0.44 |
| RT↓ | 3.65 | 3.53 | 7.40 | 6.22 | 2.13 | 2.25 | 0.67 | 0.58 |
| LC↓ | 3.21 | 3.19 | 5.09 | 4.89 | 2.01 | 2.29 | 0.42 | 0.39 |
| RC↑ | 3.25 | 3.26 | 5.34 | 5.30 | 2.23 | 2.05 | 0.46 | 0.44 |
| LP↓ | 3.42 | 3.40 | 8.40 | 6.86 | 2.04 | 2.07 | 0.66 | 0.54 |
| RP↓ | 3.29 | 3.26 | 8.76 | 5.55 | 2.16 | 2.20 | 0.62 | 0.49 |
| LO↓ | 3.65 | 3.62 | 8.92 | 6.35 | 2.26 | 2.10 | 0.74 | 0.61 |
| RO↓ | 3.32 | 3.31 | 5.65 | 5.24 | 2.00 | 2.12 | 0.51 | 0.47 |
| ROI Group and Scenario | Mean Sensitivity | Mean Specificity | Balanced Accuracy |
|---|---|---|---|
| Hyper-COH ROI with MCR | 92.72% | 94.94% | 93.83% |
| Hypo-COH ROI with MCR | 91.44% | 91.33% | 91.39% |
| Hyper-COH ROI with LOO | 86.61% | 76.67% | 81.64% |
| Hypo-COH ROI with LOO | 32.06% | 90.61% | 61.33% |
| Stats. | Two-Sample t-Test | Kruskal–Wallis Test | ||
|---|---|---|---|---|
| Band (Hz) | p-Value | Corrected p | p-Value | Corrected p |
| 25–50 | ||||
| 37.5–50 | 0.0306 | 0.0306 | 0.0203 | 0.0203 |
| 50–75 | 0.0014 | 0.0062 | 0.0043 | 0.0214 |
| 50–62.5 | 0.0012 | 0.0062 | 0.0066 | 0.0266 |
| 62.5–75 | 0.0029 | 0.0088 | 0.0068 | 0.0266 |
| 75–100 | 0.0045 | 0.0089 | 0.0080 | 0.0204 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Bornot, J.M.S.; Fernandez, R.B.; Deravi, F.; Hoque, S.; Wong-Lin, K.; Prasad, G. Detection of Mild Cognitive Impairment with MEG Functional Connectivity Using Wavelet-Based Neuromarkers. Sensors 2021, 21, 6210. https://doi.org/10.3390/s21186210
Yang S, Bornot JMS, Fernandez RB, Deravi F, Hoque S, Wong-Lin K, Prasad G. Detection of Mild Cognitive Impairment with MEG Functional Connectivity Using Wavelet-Based Neuromarkers. Sensors. 2021; 21(18):6210. https://doi.org/10.3390/s21186210
Chicago/Turabian StyleYang, Su, Jose Miguel Sanchez Bornot, Ricardo Bruña Fernandez, Farzin Deravi, Sanaul Hoque, KongFatt Wong-Lin, and Girijesh Prasad. 2021. "Detection of Mild Cognitive Impairment with MEG Functional Connectivity Using Wavelet-Based Neuromarkers" Sensors 21, no. 18: 6210. https://doi.org/10.3390/s21186210
APA StyleYang, S., Bornot, J. M. S., Fernandez, R. B., Deravi, F., Hoque, S., Wong-Lin, K., & Prasad, G. (2021). Detection of Mild Cognitive Impairment with MEG Functional Connectivity Using Wavelet-Based Neuromarkers. Sensors, 21(18), 6210. https://doi.org/10.3390/s21186210







