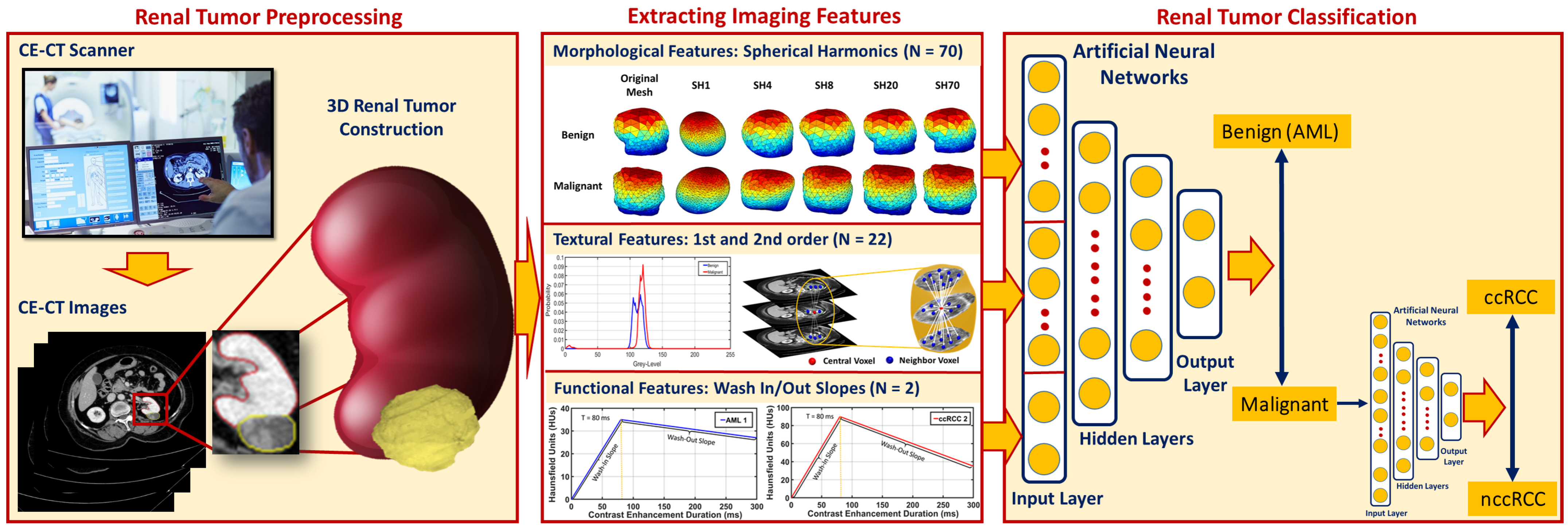
3.2. Extracting Imaging Features
For accurate identification of malignant renal tumors and the associated subtype, all 3D segmented volumes were characterized by their morphological, textural, and functional features, as described below.
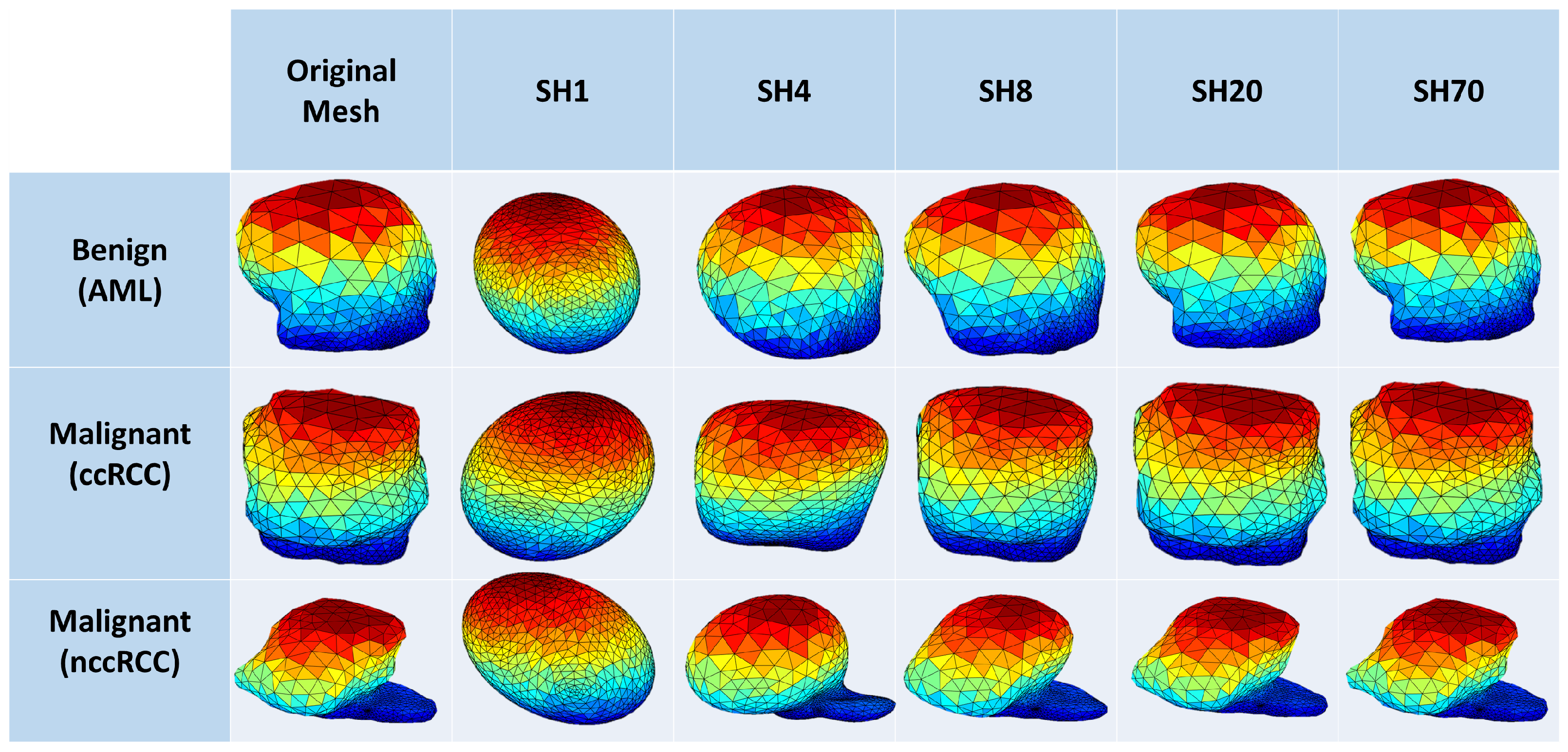
Morphological features: To enhance both the sensitivity and specificity of early renal cancer diagnosis, morphological features of the tumor are incorporated into the algorithm. These features were designed to quantify the complex shape of the tumor boundary. This was motivated by the hypothesis that rapidly growing, malignant tumors develop more irregular/complex shapes relative to more slowly growing, benign tumors. Therefore, the utilization of such shape descriptors would enhance the performance of the automatic diagnosis. Examples of this phenomenon are illustrated in
Figure 3.
Naturally, in order to measure the irregularity of the boundary, we must first construct an accurate shape model of the tumor. In this paper, we incorporated a state-of-the-art spectral decomposition in terms of spherical harmonics (SHs) [
40] to construct this shape model. An arbitrary point in the interior of the tumor, or more specifically, the interior of its convex kernel, was selected as the origin
. In this coordinate system, the tumor’s surface may be considered a function of the polar and azimuthal angle,
, which can be expressed as a linear combination of basis functions
defined on the unit sphere. Starting with a discrete approximation of the surface, i.e., a triangular mesh, the proposed algorithm uses an attraction–repulsion technique [
41] to map this mesh to the unit sphere. The mapping fixes the image of each mesh vertex at the unit distance from the origin, while preserving the mesh topology and maintaining the distance between adjacent vertices as much as possible.
Each iteration
of the attraction-repulsion works as follows. Let
be the coordinates of the node on the unit sphere corresponding to mesh vertex
i at the beginning of iteration
. Denote the vector from node
i to node
j by
; then, the Euclidean distance between nodes
i and
j is
. Finally, let
denote the index set of neighbors of vertex
i in the triangulated mesh. Then, the attraction step updates the position of each node to keep it centered with respect to its neighbors:
The quantities
and
are implementation-defined parameters that determine the strength of the attractive force. The next step, repulsion, inflates the spherical mesh to prevent it from degenerating (the attraction step by itself would allow nodes to become arbitrarily close to one another).
Just as the attraction step, the repulsion step uses an implementation-defined parameter to set the strength of the repulsive force. Subsequently, the nodes are projected back onto the sphere by giving them the unit norm, and these are their coordinates at the beginning of the next iteration, .
At the terminal iteration
of the attraction–repulsion algorithm, the surface of the renal tumor is in a one-to-one correspondence with the unit sphere. Each node
of the original mesh is mapped to a corresponding point
with polar angle
and azimuthal angle
. Considering these points as samples of a continuous function
defining the boundary, the tumor shape may be estimated by fitting an SH series to the sample nodes, since the SHs form an orthogonal basis for functions on a sphere. The SH
of degree
and order
is defined as:
where
is the SH factor and
is the associated Legendre polynomial of degree
and order
.
In practice, of course, the SH series is truncated by discarding harmonics above degree
N, yielding an
Nth order approximation.
suffices to accurately model the surface of renal tumors. Finally, the renal tumor object is reconstructed from the SHs of Equation (
3). The first few harmonics describe the rough extent of the tumor, while higher degree harmonics provide the finer details of its surface. Therefore, benign tumors are accurately represented by a lower-order SH model, while malignant tumors, with their more complex morphology, require higher-order SH model to describe their shape.
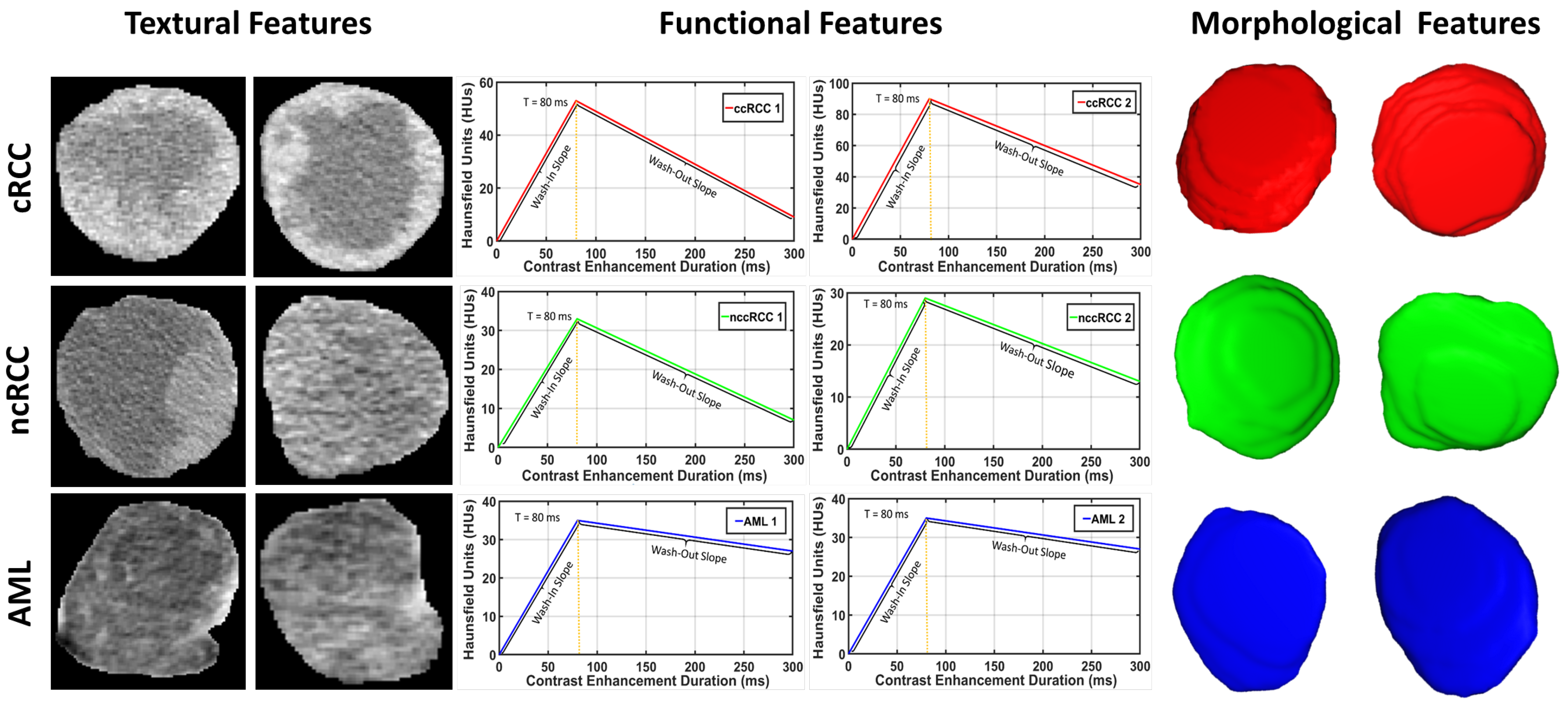
Figure 4 shows the morphology approximation for three different renal tumors: malignant ccRCC, malignant nccRCC, and benign AML tumors. A summary of the attraction–repulsion algorithm is provided below.
Initialization:
Triangulate the surface of the tumor.
Smooth the triangulated mesh with Laplacian filtering.
Initialize the spherical parameterization with an arbitrary, topology-preserving map onto the unit sphere.
Fix values of , , , and threshold T.
Attraction–repulsion:
For
- ‒
For
- *
Calculate
using Equation (
1)
- ‒
For
- *
Calculate
using Equation (
2)
- *
Let
- ‒
IfThen, let , and Stop.
Textural features: Recently, TA has become a popular research topic, particularly in the field of medical imaging. New techniques of TA provide different quantitative patterns/descriptors by combining the grey values of each pixel/voxel in a tumor image/volume. As a result of these abilities, TA has been used in the diagnosis of several tumors and their related subtypes with encouraging classification abilities [
24,
25,
42,
43,
44,
45,
46,
47,
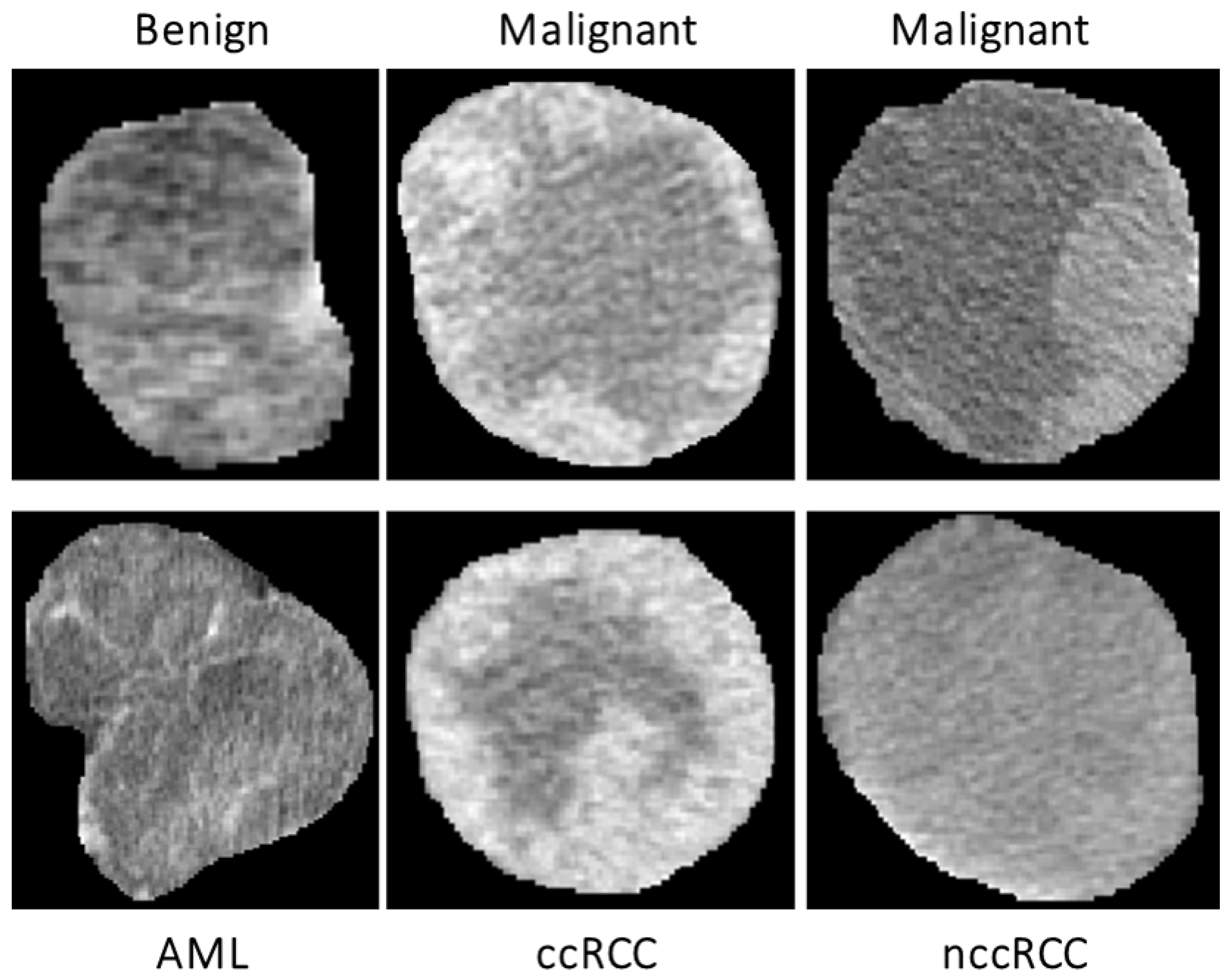
48]. Therefore, in this manuscript, TA techniques were applied on the segmented 3D renal tumor volumes to precisely extract first- and second-order textural features that best describe the homogeneity/heterogeneity between renal tumors with different diagnoses. The use of such comprehensive textural features relies on the fact that malignant tumors mostly show high textural heterogeneity when compared to benign ones. The success of these findings would enhance the sensitivity and the specificity towards an early identification of renal cancer tumors.
Figure 5 demonstrates the lesion texture differences of two malignant ccRCC subjects, two malignant nccRCC subjects, and two benign (AML) subjects.
First-order textural features: These textural features include any quantity that can be derived from the gray-level histogram of the tumor volume. In particular, mean, variance, standard deviation, entropy, skewness, kurtosis, cumulative distribution functions, and the grey-level percentiles [
49] were extracted.
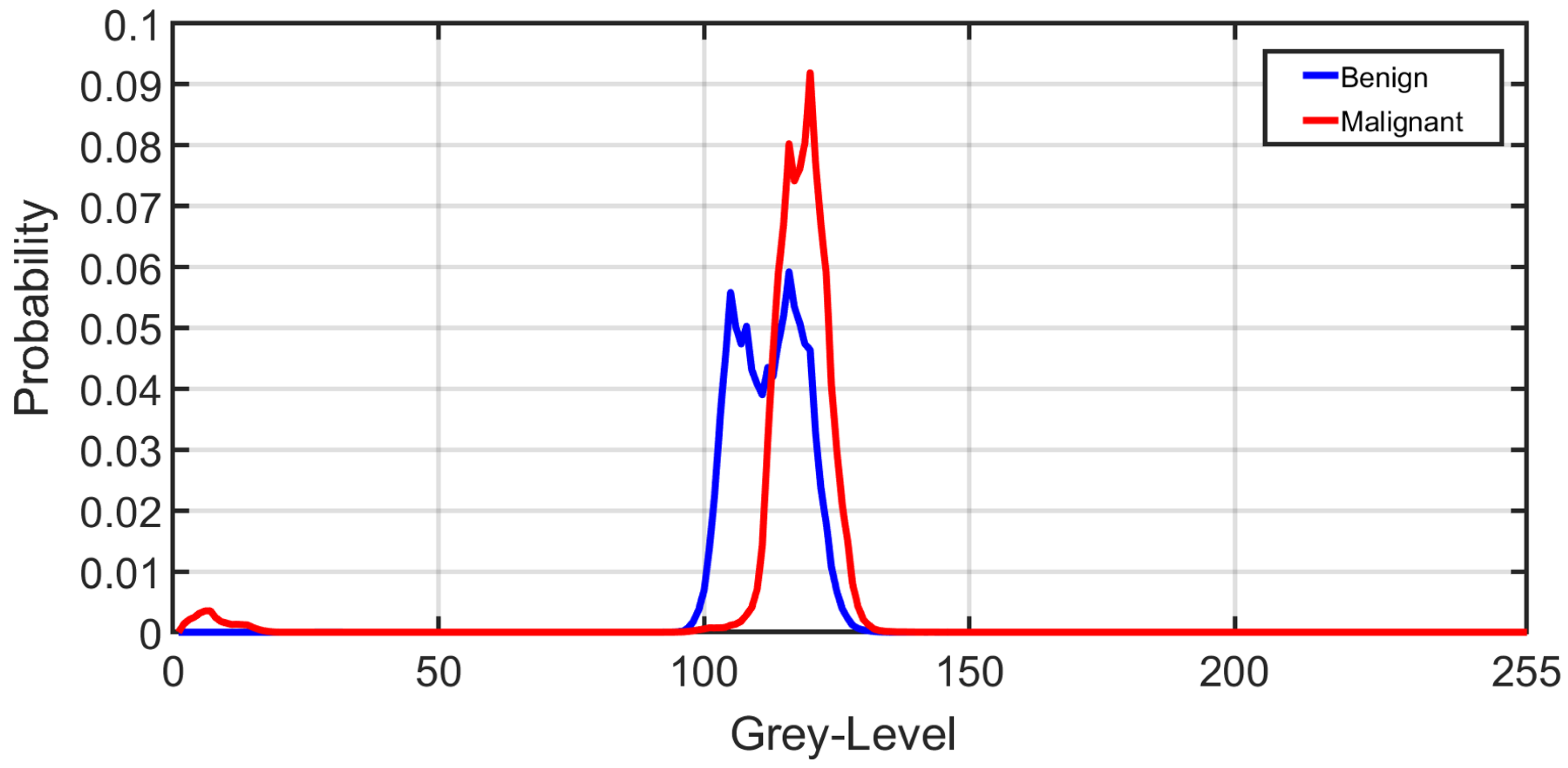
Figure 6 shows the average normalized histogram curves for all benign subjects (blue) vs. malignant (red). To construct these curves, the grey-level range was normalized first by dividing by the maximum grey-level value obtained from all subjects. Then, all histograms were constructed for all subjects within the new normalized grey-level range from 0 to 255. For each subject, the individual grey-level probability was obtained by dividing the histogram values by the corresponding number of voxels. Then, all normalized histograms from a particular group (malignant or benign) were averaged pointwise to obtain the final curve.
Second-order textural features: Since the first-order textural features might not be sufficient, with their range of values exhibiting significant overlap across classes, especially between subtypes of malignant tumor, second-order textural features were incorporated into the system. These features describe the joint distribution of gray values in multiple voxels that are considered to be neighbors of each other. In particular, the grey-level co-occurrence matrix (GLCM) [
50] was used to capture the heterogeneous appearance of renal tumors.
To construct the GLCM, we must count the number of times an ordered pair of two grey values occurs in two neighboring voxels within the renal tumor object. This technique is continued until all conceivable occurrence frequencies within the grey-level range of the renal tumor item are found, which covers all possible pairs of neighbors. For this, we first contrast stretched the renal tumor object’s original grey-level range to fit the desired span 0–255, yielding a GLCM matrix with a size of 256 × 256. Then, all feasible pair combinations were identified to construct the GLCM matrix (i.e., neighbors with gray levels
i and
j contribute to row
i, column
j of the GLCM). To define our neighborhoods, we used a distance criterion that voxels must be separated by ≤
mm, making the calculations rotation invariant (see
Figure 7). The resultant GLCM was then normalized and used to extracting the following second-order texture features [
49,
50]: contrast, dissimilarity, homogeneity, angular second moment (ASM), energy, and correlation.
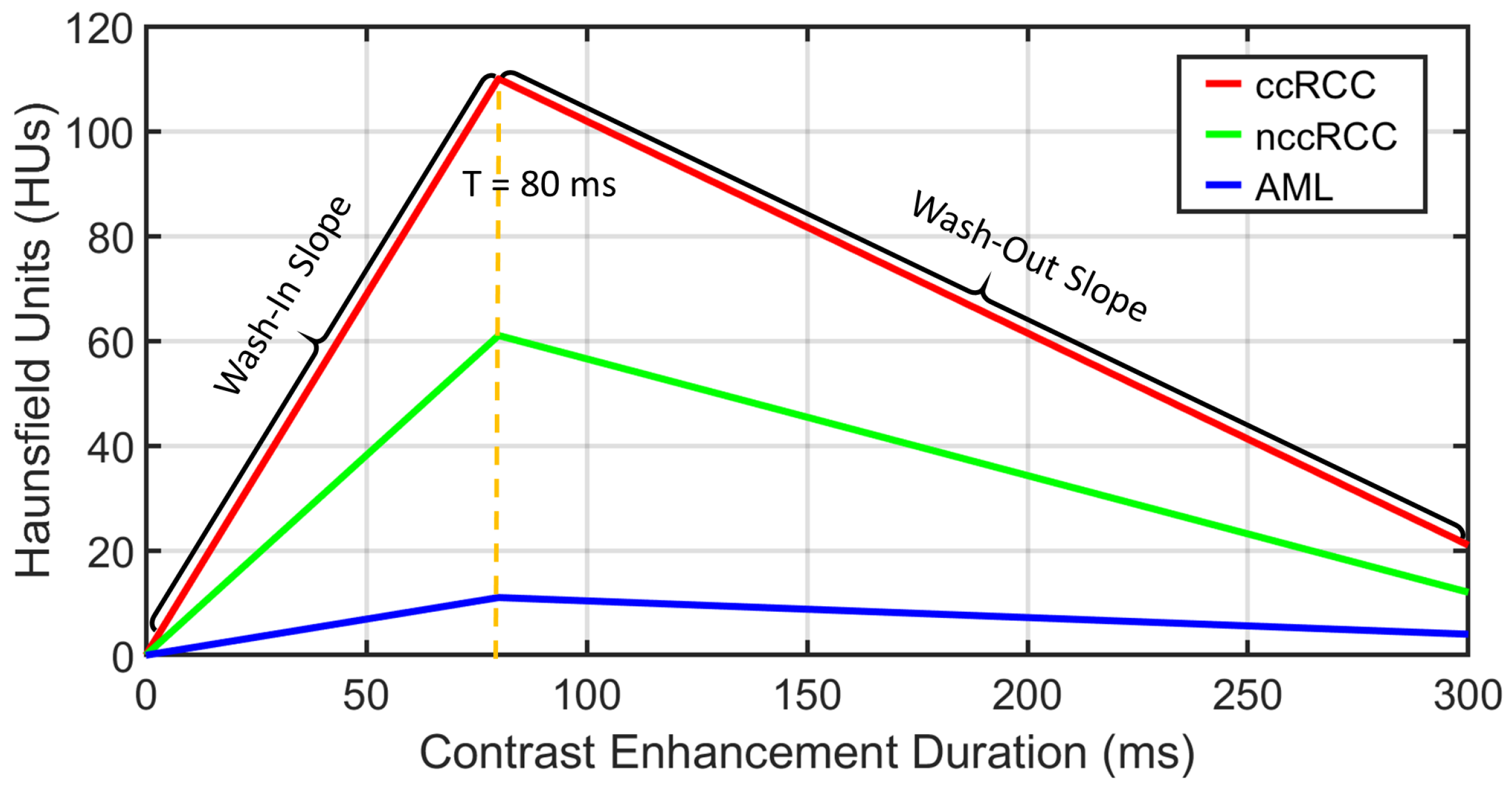
Functional features: Discriminating RCC from AML, as well as ccRCC from nccRCC might be achieved using time-dependent characteristics of CE-CT imaging. The most relevant CE-CT findings for this purpose are generally homogenous and prolonged enhancement patterns [
51]. The time dependency can be expressed by the slopes of wash-in and wash-out. Wash-in is described as the rate of increasing attenuation (in HU) from the precontrast to portal-venous phase. Similarly, wash-out is the rate of decrease in attenuation between the portal-venous and delayed-contrast phase [
52]. Higher slopes of wash-in and wash-out are typically associated with malignancy. Moreover, nccRCC demonstrates wash-in and wash-out slopes intermediate between those of AML and those of ccRCC [
53]. Therefore, we constructed both wash-in and wash-out slopes for all renal tumor subjects for the classification of the renal tumor status. Examples of wash-in/-out slopes showing the differences across ccRCC, nccRCC, and AML are shown in
Figure 8.
3.3. Feature Integration and Renal Tumor Classification
Following the extraction of morphological, textural, and functional features from all given renal tumors, RC-CAD proceeds with two-stage diagnostic classification. The first stage aims to differentiate malignant (RCC) from benign (AML) tumors. In the case of malignancy, the second stage provides the classification of RCC tumors as ccRCC or nccRCC.
The multilayer perceptron (MLP) artificial neural network (ANN) consists of at least three layers: an input layer, one or more hidden layers, and an output layer, each with arbitrarily many activation/processing units, known as nodes/neurons. Each layer is fully connected to the next layer in sequence. Neurons use nonlinear activation functions to give the MLP-ANN the capability to divide the feature space into arbitrarily complex regions. The MLP-ANN mainly utilizes supervised backpropagation learning technique in the training phase, in which gradient descent methods are utilized to update the connection weights and additive biases in order to minimize the loss function. To achieve our goal, we utilized the MLP-ANN in both classification stages to obtain the final diagnosis. Classifier performance was assessed using five different feature sets (
Table 2) as the ANN input in both stages. Feature Set 1 includes first-order histogram textural features (
N = 6; mean, variance, standard deviation, skewness, kurtosis, and entropy); Feature Set 2 includes first-order percentile textural features (
N = 10; from the 10th to the 100th percentile in 10% point steps); Feature Set 3 includes second-order GLCM textural features (
N = 6; contrast, dissimilarity, homogeneity, ASM, energy, and correlation); Feature Set 4 includes SH reconstruction error (SHRE) morphological features (
N = 70); and Feature Set 5 includes functional features (
N = 2; wash-in slope and wash-out slope). At each classification stage, the individual feature sets were concatenated to obtain the combined features (
N = 94) and were fed to a MLP-ANN to obtain the final diagnosis.