Rapid and Sensitive Inhibitor Screening Using Magnetically Modulated Biosensors
Abstract
:1. Introduction
2. Materials and Methods
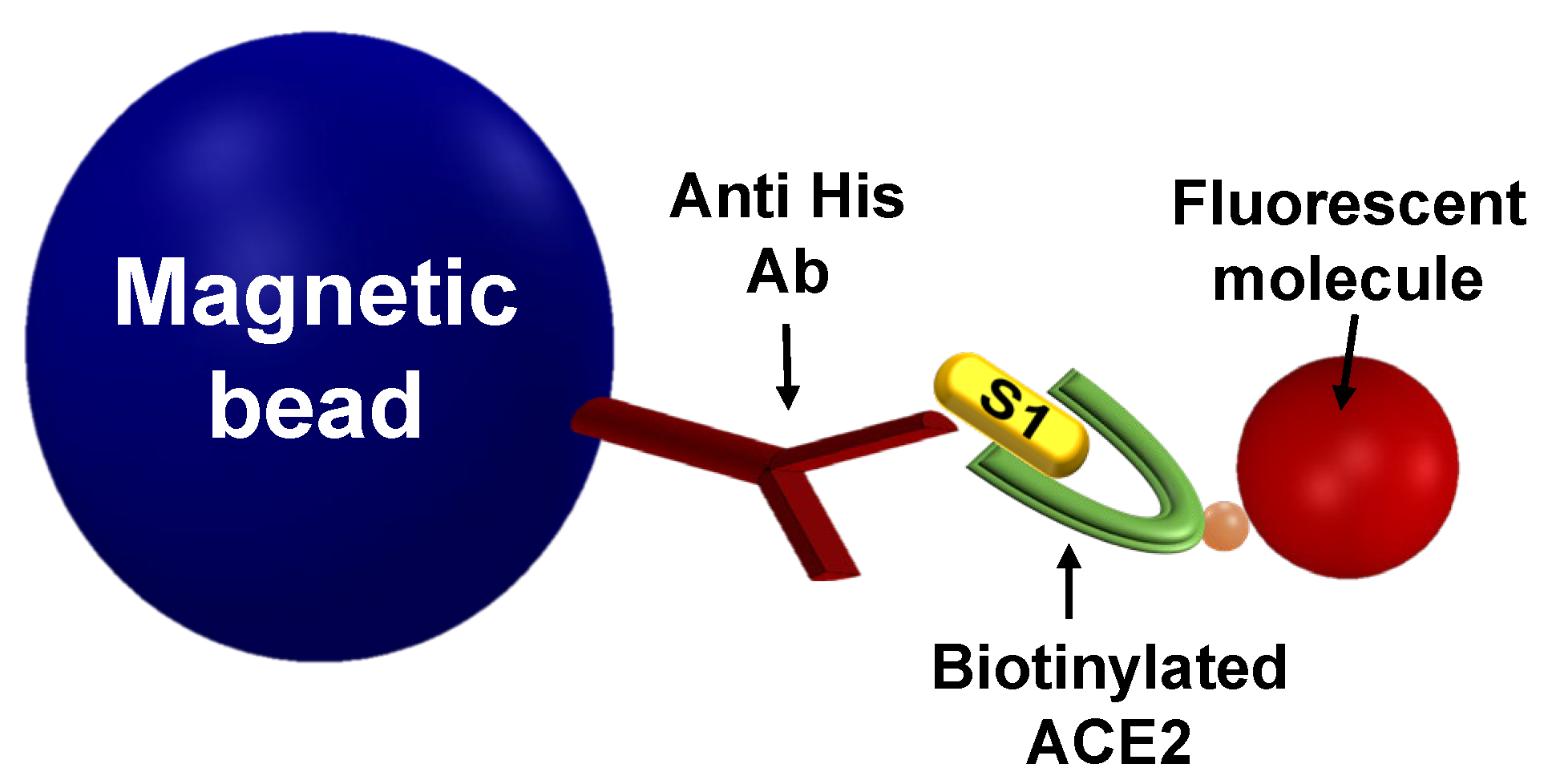
2.1. Detection of the S1-ACE2 Interaction
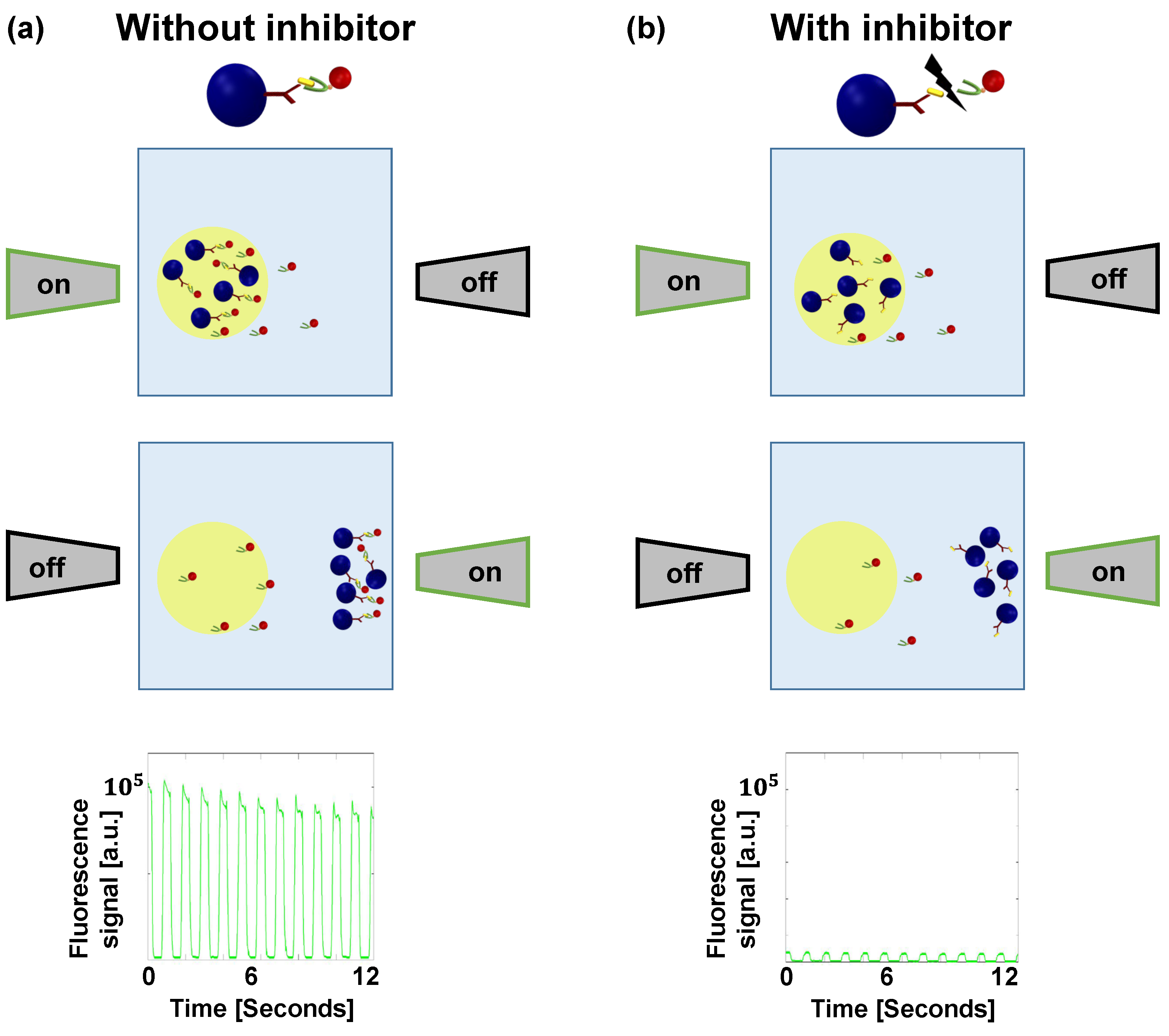
2.2. Inhibition of the S1-ACE2 Interaction
3. Results
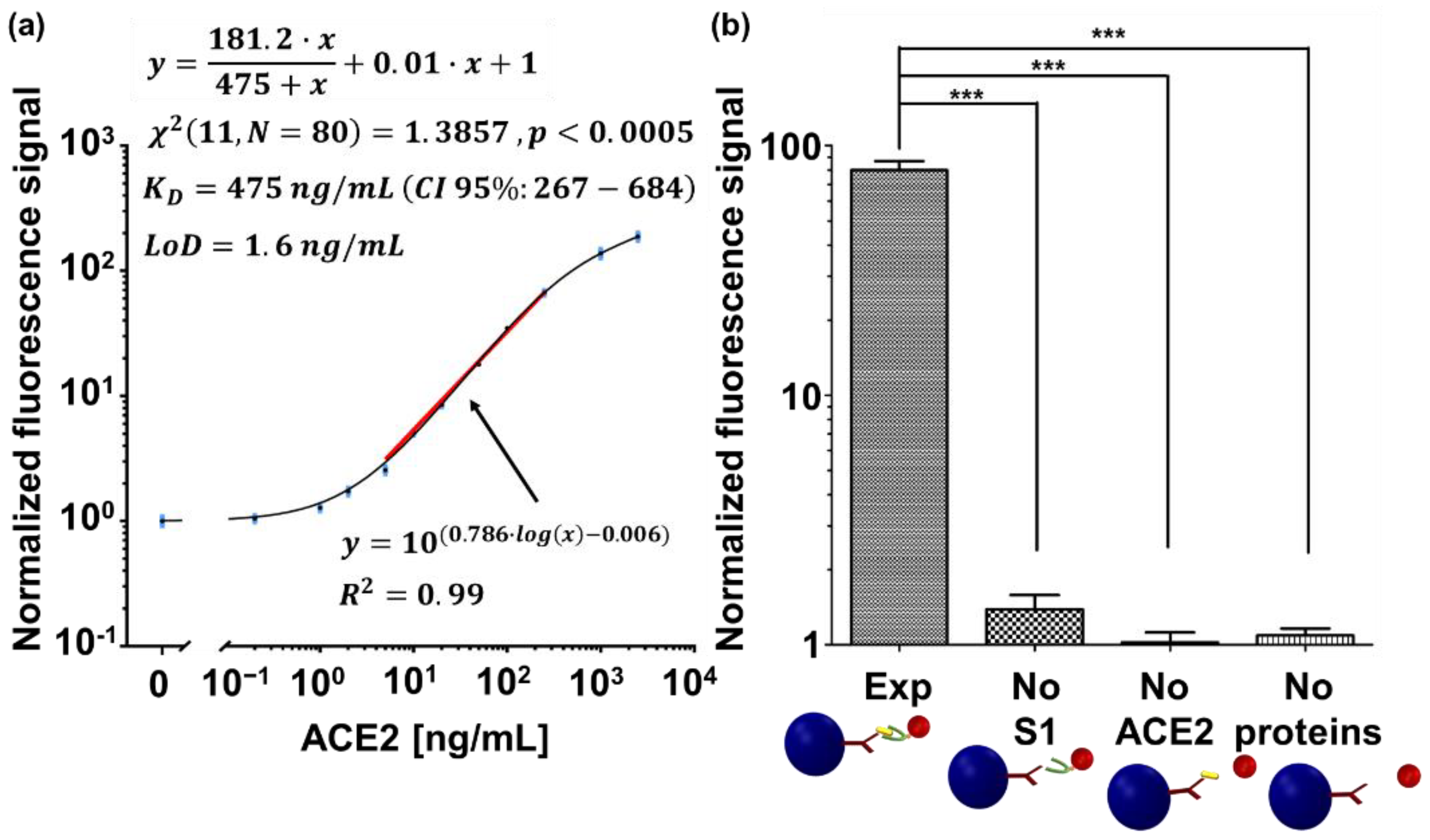
3.1. Detection of the S1-ACE2 Interaction
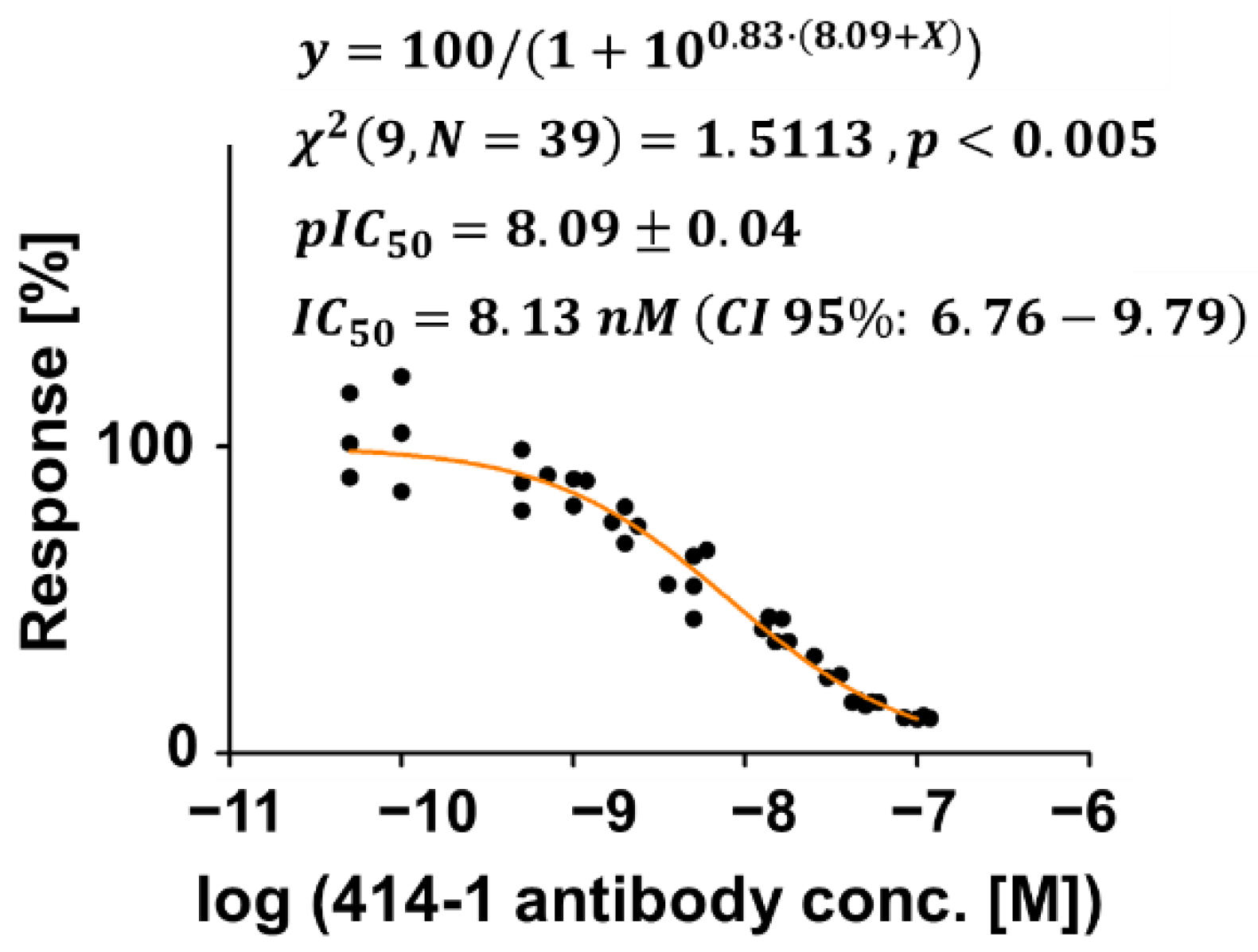
3.2. Assessing an Anti-S1 Antibody as an Inhibitor of the S1-ACE2 Interaction
3.3. Assessing a Small Molecule (SSAA09E2) as an Inhibitor of the S1-ACE2 Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Magnetically Modulated Biosensors
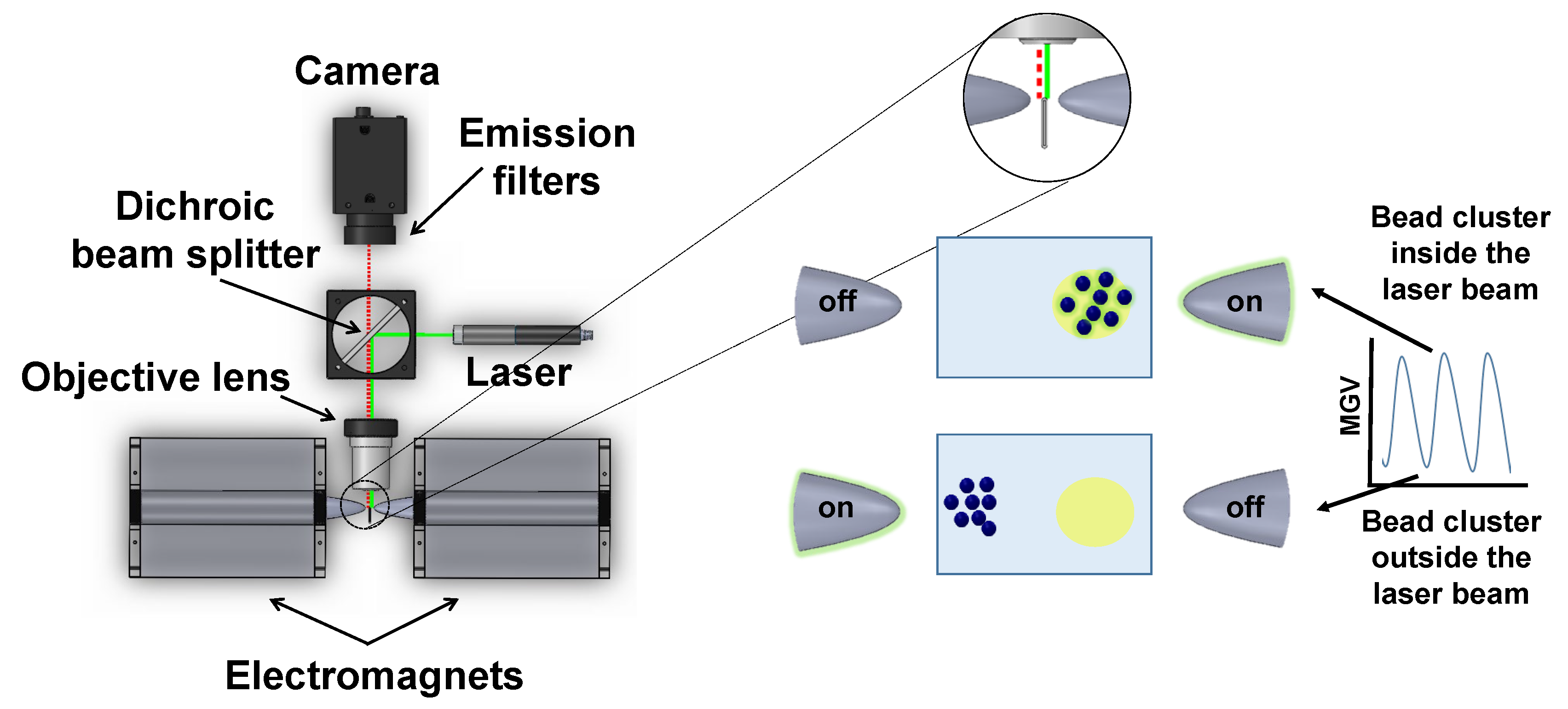
Appendix A.2. Z-Factor Calculation
Appendix A.3. Curve Fitting
- is the maximum specific binding [normalized fluorescent signal],
- is the equilibrium dissociation constant [ng/mL],
- is the slope of nonspecific binding [normalized fluorescent signal/(ng/mL)],
- is the average normalized fluorescent signal measured without a fluorescent marker (i.e., 0 ng/mL of biotinylated ACE2 protein).
- is the concentration of inhibitor that gives a response halfway between bottom and top,
- describes the steepness of the family of curves.
References
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Wan, J.; Xing, S.; Ding, L.; Wang, Y.; Gu, C.; Wu, Y.; Rong, B.; Li, C.; Wang, S.; Chen, K.; et al. Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep. 2020, 32, 107918. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Beer, J.C.; Sankaranarayanan, N.V.; Swanson-Mungerson, M.; Desai, U.R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today 2020, 25, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Meenakshisundaram, S.; Manickam, M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today 2020, 25, 668–688. [Google Scholar] [CrossRef] [PubMed]
- Olaleye, O.A.; Kaur, M.; Onyenaka, C.; Adebusuyi, T. Discovery of Clioquinol and Analogues as Novel Inhibitors of Severe Acute Respiratory Syndrome Coronavirus 2 Infection, ACE2 and ACE2—Spike Protein Interaction In Vitro. BioRxiv 2020. [Google Scholar] [CrossRef]
- Magro, G. COVID-19: Review on latest available drugs and therapies against SARS-CoV-2. Coagulation and inflammation cross-talking. Virus Res. 2020, 286, 198070. [Google Scholar] [CrossRef]
- Jawaid Akhtar, M. COVID19 inhibitors: A prospective therapeutics. Bioorg. Chem. 2020, 101, 104027. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Honko, A.; Zhou, J.; Gong, H.; Downs, S.N.; Vasquez, J.H.; Fang, R.H.; Gao, W.; Griffiths, A.; Zhang, L. Cellular Nanosponges Inhibit SARS-CoV-2 Infectivity. Nano Lett. 2020, 20, 5570–5574. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, L.; Chen, K.; Shen, X.; Jiang, H. Techniques used for the discovery of therapeutic compounds: The case of SARS. Drug Discov. Today Technol. 2006, 3, 277–283. [Google Scholar] [CrossRef]
- Hosseini, S.; Martinez-Chapa, S.O.; Rito-Palomares, M.; Vázquez-Villegas, P. Advantages, Disadvantages and Modifications of Conventional ELISA in Enzyme-linked Immunosorbent Assay (ELISA): From A to Z. In SpringerBriefs in Forensic and Medical Bioinformatics, Online Resource (XI, 115 Pages 38 Illustrations, 34 Illustrations in Color), 1st ed.; Springer: Singapore; pp. 67–115.
- Roman, D.L.; Talbot, J.N.; Roof, R.A.; Sunahara, R.K.; Traynor, J.R.; Neubig, R.R. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol. Pharmacol. 2007, 71, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Biosensors and Bioanalytical Devices based on Magnetic Particles: A Review. Curr. Med. Chem. 2021, 28, 2828–2841. [Google Scholar] [CrossRef]
- Huergo, L.F.; Selim, K.A.; Conzentino, M.S.; Gerhardt, E.C.M.; Santos, A.R.S.; Wagner, B.; Alford, J.T.; Deobald, N.; Pedrosa, F.O.; de Souza, E.M.; et al. Magnetic Bead-Based Immunoassay Allows Rapid, Inexpensive, and Quantitative Detection of Human SARS-CoV-2 Antibodies. ACS Sens. 2021, 6, 703–708. [Google Scholar] [CrossRef]
- Abedini-Nassab, R.; Pouryosef Miandoab, M.; Sasmaz, M. Microfluidic Synthesis, Control, and Sensing of Magnetic Nanoparticles: A Review. Micromachines 2021, 12, 768. [Google Scholar] [CrossRef]
- Chen, Y.T.; Kolhatkar, A.G.; Zenasni, O.; Xu, S.; Lee, T.R. Biosensing Using Magnetic Particle Detection Techniques. Sensors 2017, 17, 2300. [Google Scholar] [CrossRef]
- Dawei, Z.; Hongqiu, H.; Mengmeng, L.; Zhixia, M.; Shunxing, G. A Novel Assay for Screening Inhibitors Targeting HIV Integrase LEDGF/p75 Interaction Based on Ni2+ Coated Magnetic Agarose Beads. Sci. Rep. 2016, 6, 33477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelson, Y.; Lustig, Y.; Avivi, S.; Schwartz, E.; Danielli, A. Highly Sensitive and Specific Zika Virus Serological Assays Using a Magnetic Modulation Biosensing System. J. Infect. Dis. 2019, 219, 1035–1043. [Google Scholar] [CrossRef]
- Verbarg, J.; Hadass, O.; Olivo, P.D.; Danielli, A. High sensitivity detection of a protein biomarker interleukin-8 utilizing a magnetic modulation biosensing system. Sens. Actuators B Chem. 2017, 241, 614–618. [Google Scholar] [CrossRef]
- Margulis, M.; Danielli, A. Rapid and Sensitive Detection of Repetitive Nucleic Acid Sequences Using Magnetically Modulated Biosensors. ACS Omega 2019, 4, 11749–11755. [Google Scholar] [CrossRef]
- Margulis, M.; Ashri, S.; Cohen, M.; Danielli, A. Detecting nucleic acid fragments in serum using a magnetically modulated sandwich assay. J. Biophotonics 2019, 12, e201900104. [Google Scholar] [CrossRef]
- Danielli, A.; Porat, N.; Arie, A.; Ehrlich, M. Rapid homogenous detection of the Ibaraki virus NS3 cDNA at picomolar concentrations by magnetic modulation. Biosens. Bioelectron. 2009, 25, 858–863. [Google Scholar] [CrossRef]
- Roth, S.; Zander, I.; Michelson, Y.; Ben-David, Y.; Banin, E.; Danielli, A. Identification of protein-protein interactions using a magnetic modulation biosensing system. Sens. Actuators B Chem. 2020, 303, 127228. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, S.; Hadass, O.; Cohen, M.; Verbarg, J.; Wilsey, J.; Danielli, A. Improving the Sensitivity of Fluorescence-Based Immunoassays by Photobleaching the Autofluorescence of Magnetic Beads. Small 2019, 15, 1803751. [Google Scholar] [CrossRef] [PubMed]
- Borysiak, M.D.; Thompson, M.J.; Posner, J.D. Translating diagnostic assays from the laboratory to the clinic: Analytical and clinical metrics for device development and evaluation. Lab Chip 2016, 16, 1293–1313. [Google Scholar] [CrossRef]
- SARS-CoV-2 Spike Antibody, 414–1 (AM001414). Available online: https://www.activemotif.com/catalog/details/91361/sars-cov-2-spike-antibody-am001414 (accessed on 11 April 2021).
- Adedeji, A.O.; Severson, W.; Jonsson, C.; Singh, K.; Weiss, S.R.; Sarafianos, S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 2013, 87, 8017–8028. [Google Scholar] [CrossRef] [Green Version]
- Prajapat, M.; Sarma, P.; Shekhar, N.; Avti, P.; Sinha, S.; Kaur, H.; Kumar, S.; Bhattacharyya, A.; Kumar, H.; Bansal, S.; et al. Drug targets for corona virus: A systematic review. Indian J. Pharmacol. 2020, 52, 56–65. [Google Scholar]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- SARS-CoV-2 (COVID-19) S1 Protein, His Tag. Available online: https://www.acrobiosystems.com/P3103-SARS-CoV-2-%28COVID-19%29-S1-protein-His-Tag.html (accessed on 11 April 2021).
- Hulme, E.C.; Trevethick, M.A. Ligand binding assays at equilibrium: Validation and interpretation. Br. J. Pharmacol. 2010, 161, 1219–1237. [Google Scholar] [CrossRef] [Green Version]
- Sauer Robert, T. Review of Chemical Thermodynamics 7.51. Available online: https://ocw.mit.edu/courses/biology/7-51-graduate-biochemistry-fall-2001/lecture-notes/fa01lec07.pdf (accessed on 13 July 2021).
- Walker, S.N.; Chokkalingam, N.; Reuschel, E.L.; Purwar, M.; Xu, Z.; Gary, E.N.; Kim, K.Y.; Helble, M.; Schultheis, K.; Walters, J.; et al. SARS-CoV-2 assays to detect functional antibody responses that block ACE2 recognition in vaccinated animals and infected patients. J. Clin. Microbiol. 2020, 58, e01533-20. [Google Scholar] [CrossRef]
- SARS-CoV-2 (COVID-19) Inhibitor screening Kit. Available online: https://www.acrobiosystems.com/P3144-SARS-CoV-2-%28COVID-19%29-Inhibitor-screening-Kit.html (accessed on 11 April 2021).
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [Green Version]
- Fiedler, S.; Piziorska, M.A.; Denninger, V.; Morgunov, A.S.; Ilsley, A.; Malik, A.Y.; Schneider, M.M.; Devenish, S.R.A.; Meisl, G.; Aguzzi, A.; et al. In vitro measurements of protein–protein interactions show that antibody affinity governs the inhibition of SARS-CoV-2 spike/ACE2 binding in convalescent serum. BioRxiv 2020. [Google Scholar] [CrossRef]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef]
- Molek, P. Estimation of dissociation constant by modified ELISA assay. Available online: https://www.youtube.com/watch?v=in0ykyFRLCw (accessed on 7 July 2021).
- Friguet, B.; Chaffotte, A.F.; Djavadi-Ohaniance, L.; Goldberg, M.E. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods 1985, 77, 305–319. [Google Scholar] [CrossRef]
- Rammler, D.H. The effect of DMSO on several enzyme systems. Ann. N. Y. Acad. Sci. 1967, 141, 291–299. [Google Scholar] [CrossRef]
- Kumar, A.; Darreh-Shori, T. DMSO: A Mixed-Competitive Inhibitor of Human Acetylcholinesterase. ACS Chem. Neurosci. 2017, 8, 2618–2625. [Google Scholar] [CrossRef] [PubMed]
- Gil-Moles, M.; Basu, U.; Bussing, R.; Hoffmeister, H.; Turck, S.; Varchmin, A.; Ott, I. Gold Metallodrugs to Target Coronavirus Proteins: Inhibitory Effects on the Spike-ACE2 Interaction and on PLpro Protease Activity by Auranofin and Gold Organometallics. Chemistry 2020, 26, 15140–15144. [Google Scholar] [CrossRef]
- Hanson, Q.M.; Wilson, K.M.; Shen, M.; Itkin, Z.; Eastman, R.T.; Shinn, P.; Hall, M.D. Targeting ACE2–RBD Interaction as a Platform for COVID-19 Therapeutics: Development and Drug-Repurposing Screen of an AlphaLISA Proximity Assay. ACS Pharmacol. Transl. Sci. 2020, 3, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H.J. GraphPad Curve Fitting Guide-Equation: One Site—Total Binding. Available online: https://www.graphpad.com/guides/prism/8/curve-fitting/REG_One_site_Total.htm (accessed on 7 July 2021).
- Motulsky, H.J. GraphPad Curve Fitting Guide-Equation: Log (Inhibitor) vs. Normalized Response—Variable Slope. Available online: https://www.graphpad.com/guides/prism/8/curve-fitting/REG_DR_inhibit_normalized_variable.htm (accessed on 8 July 2021).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roth, S.; Danielli, A. Rapid and Sensitive Inhibitor Screening Using Magnetically Modulated Biosensors. Sensors 2021, 21, 4814. https://doi.org/10.3390/s21144814
Roth S, Danielli A. Rapid and Sensitive Inhibitor Screening Using Magnetically Modulated Biosensors. Sensors. 2021; 21(14):4814. https://doi.org/10.3390/s21144814
Chicago/Turabian StyleRoth, Shira, and Amos Danielli. 2021. "Rapid and Sensitive Inhibitor Screening Using Magnetically Modulated Biosensors" Sensors 21, no. 14: 4814. https://doi.org/10.3390/s21144814
APA StyleRoth, S., & Danielli, A. (2021). Rapid and Sensitive Inhibitor Screening Using Magnetically Modulated Biosensors. Sensors, 21(14), 4814. https://doi.org/10.3390/s21144814






