AI-Driven Framework for Recognition of Guava Plant Diseases through Machine Learning from DSLR Camera Sensor Based High Resolution Imagery
Abstract
1. Introduction
- A Guava disease classification framework based on guava plant images is proposed. The proposed framework separates the Guava images into the diseased image () and non-diseased () image. The proposed approach’s primary goal is to detect the disease present in guava plant images.
- Image-level and disease-level-based feature extraction approaches are used to obtain robust guava disease recognition.
- The corresponding disease-segmented image with a specific label is assigned a class, which gives information about the disease. Four guava diseases, such as Canker, Mummification, Dot, Rust, and one extra target class, “healthy”, are covered in the presented study.
- The proposed framework is evaluated on a high-resolution image dataset.
2. Related Work
3. Methodology
- 1.
- Pre-processing of image.
- 2.
- Segmentation of image.
- 3.
- Feature extraction.
- 4.
- Classification.
3.1. Image Pre-Processing
3.2. Image Enhancement
3.3. Color Space Transformation
3.4. Image Segmentation
3.4.1. Delta E (E)
3.4.2. Obtaining RGB Image from Binary Image
3.5. Feature Extraction
3.6. RGB and HSV Histogram Features
3.7. Local Binary Patterns
3.8. Classification
4. Results and Discussions
4.1. Dataset
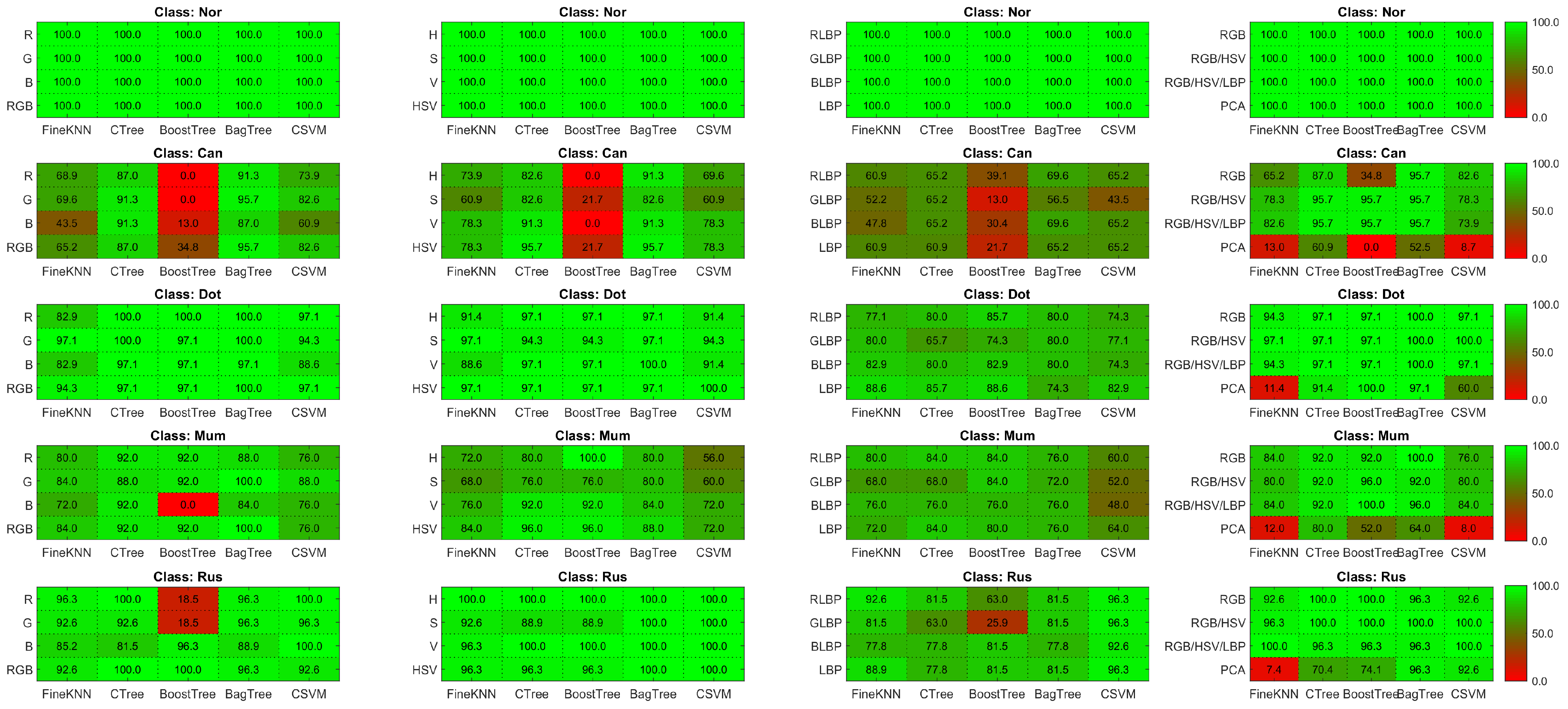
4.2. Performance Evaluation
4.3. Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohan, K.; Muralisankar, T.; Uthayakumar, V.; Chandirasekar, R.; Revathi, N.; Ganesan, A.R.; Velmurugan, K.; Sathishkumar, P.; Jayakumar, R.; Seedevi, P. Trends in the extraction, purification, characterisation and biological activities of polysaccharides from tropical and sub-tropical fruits–A comprehensive review. Carbohydr. Polym. 2020, 238, 116185. [Google Scholar] [CrossRef] [PubMed]
- Amusa, N.; Ashaye, O.; Oladapo, M.; Oni, M. Guava fruit anthracnose and the effects on its nutritional and market values in Ibadan, Nigeria. World J. Agric. Sci. 2005, 1, 169–172. [Google Scholar] [CrossRef]
- Gupta, S.D.; Rai, J. Wilt Disease of Guava (Psidium guyava L.). Curr. Sci. 1947, 16, 256–258. [Google Scholar]
- Chibber, H. A working list of diseases of vegetable pests of some of the economic plants, occurring in the Bombay Presidency. Poona Agric. Coll. Mag. 1911, 2, 180–198. [Google Scholar]
- Ruehle, G.; Brewer, C. The FDA Method. Official Method of US Food and Drug Administration, US Department of Agriculture, and National Assn. of Insecticide and Disinfectant Manufacturers for Determination of Phenol Coefficients of Disinfectants; MacNair-Dorland Co.: New York, NY, USA, 1941; pp. 189–201. [Google Scholar]
- Rana, O. Diplodia stem canker, a new disease of guava in Tarai region of Uttar Pradesh. Sci. Cult. 1981, 47, 370–371. [Google Scholar]
- Misra, A. Guava diseases—Their symptoms, causes and management. In Diseases of Fruits and Vegetables: Volume II; Springer: Berlin/Heidelberg, Germany, 2004; pp. 81–119. [Google Scholar]
- Raza, S.A.; Ali, Y.; Mehboob, F. Role of Agriculture in Economic Growth of Pakistan. Int. Res. J. Financ. Econ. 2012, 83, 180–186. [Google Scholar]
- Pariona, A. Top Guava Producing Countries in the World. Worldatlas. 2017. Available online: https://www.worldatlas.com/articles/top-guava-producingcountries-in-the-world.html (accessed on 17 July 2018).
- Pujari, J.D.; Yakkundimath, R.; Byadgi, A.S. Grading and classification of anthracnose fungal disease of fruits based on statistical texture features. Int. J. Adv. Sci. Technol. 2013, 52, 121–132. [Google Scholar]
- Gilligan, C.A. Sustainable agriculture and plant diseases: An epidemiological perspective. Philos. Trans. R. Soc. B Biol. Sci. 2007, 363, 741–759. [Google Scholar] [CrossRef]
- Adenugba, F.; Misra, S.; Maskeliūnas, R.; Damaševičius, R.; Kazanavičius, E. Smart irrigation system for environmental sustainability in Africa: An Internet of Everything (IoE) approach. Math. Biosci. Eng. 2019, 16, 5490–5503. [Google Scholar] [CrossRef]
- Zhou, R.; Kaneko, S.; Tanaka, F.; Kayamori, M.; Shimizu, M. Disease detection of Cercospora Leaf Spot in sugar beet by robust template matching. Comput. Electron. Agric. 2014, 108, 58–70. [Google Scholar] [CrossRef]
- Ali, H.; Lali, M.; Nawaz, M.Z.; Sharif, M.; Saleem, B. Symptom based automated detection of citrus diseases using color histogram and textural descriptors. Comput. Electron. Agric. 2017, 138, 92–104. [Google Scholar] [CrossRef]
- Barr, M. A Novel Technique for Segmentation of High Resolution Remote Sensing Images Based on Neural Networks. Neural Process. Lett. 2020, 52, 679–692. [Google Scholar] [CrossRef]
- Horwath, J.P.; Zakharov, D.N.; Mégret, R.; Stach, E.A. Understanding important features of deep learning models for segmentation of high-resolution transmission electron microscopy images. Npj Comput. Mater. 2020, 6. [Google Scholar] [CrossRef]
- Hamuda, E.; Glavin, M.; Jones, E. A survey of image processing techniques for plant extraction and segmentation in the field. Comput. Electron. Agric. 2016, 125, 184–199. [Google Scholar] [CrossRef]
- Patrício, D.I.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Electron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef]
- Ngugi, L.C.; Abelwahab, M.; Abo-Zahhad, M. Recent advances in image processing techniques for automated leaf pest and disease recognition—A review. Inf. Process. Agric. 2021, 8, 27–51. [Google Scholar] [CrossRef]
- Gabryel, M.; Damaševičius, R. The Image Classification with Different Types of Image Features; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Zakopane, Poland, 2017; Volume 10245, pp. 497–506. [Google Scholar]
- Naranjo-Torres, J.; Mora, M.; Hernández-García, R.; Barrientos, R.J.; Fredes, C.; Valenzuela, A. A Review of Convolutional Neural Network Applied to Fruit Image Processing. Appl. Sci. 2020, 10, 3443. [Google Scholar] [CrossRef]
- Koirala, A.; Walsh, K.B.; Wang, Z.; McCarthy, C. Deep learning—Method overview and review of use for fruit detection and yield estimation. Comput. Electron. Agric. 2019, 162, 219–234. [Google Scholar] [CrossRef]
- Bhargava, A.; Bansal, A. Fruits and vegetables quality evaluation using computer vision: A review. J. King Saud Univ. Comput. Inf. Sci. 2021, 33, 243–257. [Google Scholar] [CrossRef]
- Cruz, A.C.; Luvisi, A.; Bellis, L.D.; Ampatzidis, Y. X-FIDO: An Effective Application for Detecting Olive Quick Decline Syndrome with Deep Learning and Data Fusion. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Pawar, R.; Jadhav, A. Pomogranite disease detection and classification. In Proceedings of the 2017 IEEE International Conference on Power, Control, Signals and Instrumentation Engineering (ICPCSI), Chennai, India, 21–22 September 2017; pp. 2475–2479. [Google Scholar] [CrossRef]
- Zhou, H.; Zhuang, Z.; Liu, Y.; Liu, Y.; Zhang, X. Defect Classification of Green Plums Based on Deep Learning. Sensors 2020, 20, 6993. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, R.; Xie, C.; Liu, L.; Zhang, J.; Li, R.; Wang, F.; Zhou, M.; Liu, W. A Recognition Method for Rice Plant Diseases and Pests Video Detection Based on Deep Convolutional Neural Network. Sensors 2020, 20, 578. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, A.; Yoon, S.; Kim, S.; Park, D. A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef]
- Ramcharan, A.; Baranowski, K.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep Learning for Image-Based Cassava Disease Detection. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.P.; Chouhan, S.S.; Jain, S.; Jain, S. Multilayer Convolution Neural Network for the Classification of Mango Leaves Infected by Anthracnose Disease. IEEE Access 2019, 7, 43721–43729. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Q.; Gao, L. Visual Tea Leaf Disease Recognition Using a Convolutional Neural Network Model. Symmetry 2019, 11, 343. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.; Liu, B.; He, D.; Liang, C. Real-Time Detection of Apple Leaf Diseases Using Deep Learning Approach Based on Improved Convolutional Neural Networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Iqbal, Z.; Khan, M.A.; Sharif, M.; Shah, J.H.; ur Rehman, M.H.; Javed, K. An automated detection and classification of citrus plant diseases using image processing techniques: A review. Comput. Electron. Agric. 2018, 153, 12–32. [Google Scholar] [CrossRef]
- Capizzi, G.; Lo Sciuto, G.; Napoli, C.; Tramontana, E.; Woźniak, M. A novel neural networks-based texture image processing algorithm for orange defects classification. Int. J. Comput. Sci. Appl. 2016, 13, 45–60. [Google Scholar]
- Lili, N.; Khalid, F.; Borhan, N. Classification of herbs plant diseases via hierarchical dynamic artificial neural network after image removal using kernel regression framework. Int. J. Comput. Sci. Eng. 2011, 3, 15–20. [Google Scholar]
- Aleixos, N.; Blasco, J.; Navarron, F.; Moltó, E. Multispectral inspection of citrus in real-time using machine vision and digital signal processors. Comput. Electron. Agric. 2002, 33, 121–137. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Digital image processing techniques for detecting, quantifying and classifying plant diseases. SpringerPlus 2013, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arivazhagan, S.; Shebiah, R.N.; Ananthi, S.; Varthini, S.V. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217. [Google Scholar]
- Dey, A.K.; Sharma, M.; Meshram, M. Image processing based leaf rot disease, detection of betel vine (Piper betle L.). Procedia Comput. Sci. 2016, 85, 748–754. [Google Scholar] [CrossRef]
- VijayaLakshmi, B.; Mohan, V. Kernel-based PSO and FRVM: An automatic plant leaf type detection using texture, shape, and color features. Comput. Electron. Agric. 2016, 125, 99–112. [Google Scholar] [CrossRef]
- Bhange, M.; Hingoliwala, H. Smart farming: Pomegranate disease detection using image processing. Procedia Comput. Sci. 2015, 58, 280–288. [Google Scholar] [CrossRef]
- Wu, L.; Wen, Y.; Deng, X.; Peng, H. Identification of weed/corn using BP network based on wavelet features and fractal dimension. Sci. Res. Essays 2009, 4, 1194–1200. [Google Scholar]
- Pydipati, R.; Burks, T.; Lee, W. Identification of citrus disease using color texture features and discriminant analysis. Comput. Electron. Agric. 2006, 52, 49–59. [Google Scholar] [CrossRef]
- Napoli, C.; Pappalardo, G.; Tramontana, E.; Marszalek, Z.; Polap, D.; Wozniak, M. Simplified firefly algorithm for 2D image key-points search. In Proceedings of the 2014 IEEE Symposium on Computational Intelligence for Human-like Intelligence (CIHLI), Orlando, FL, USA, 9–12 December 2014. [Google Scholar]
- Zhang, H.; Wu, Q.M.J.; Zheng, Y.; Nguyen, T.M.; Wang, D. Effective fuzzy clustering algorithm with Bayesian model and mean template for image segmentation. IET Image Process. 2014, 8, 571–581. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Guyer, D.; Miles, G.; Gaultney, L.; Schreiber, M. Application of machine vision to shape analysis in leaf and plant identification. Trans. ASAE 1993, 36, 163–172. [Google Scholar] [CrossRef]
- Crowe, T.; Delwiche, M. Real-time defect detection in fruit—Part II: An algorithm and performance of a prototype system. Trans. ASAE 1996, 39, 2309–2317. [Google Scholar] [CrossRef]
- Sweet, H.C.; Edwards, G.J. Citrus blight assessment using a microcomputer; quantifying damage using an apple computer to solve reflectance spectra of entire trees. Fla Sci. 1986, 49, 48–54. [Google Scholar]
- Guru, D.; Mallikarjuna, P.; Manjunath, S. Segmentation and classification of tobacco seedling diseases. In Proceedings of the Fourth Annual ACM Bangalore Conference, Bangalore, India, 25–26 March 2011; pp. 1–5. [Google Scholar]
- Moshou, D.; Bravo, C.; Oberti, R.; West, J.; Ramon, H.; Vougioukas, S.; Bochtis, D. Intelligent multi-sensor system for the detection and treatment of fungal diseases in arable crops. Biosyst. Eng. 2011, 108, 311–321. [Google Scholar] [CrossRef]
- Khamparia, A.; Saini, G.; Gupta, D.; Khanna, A.; Tiwari, S.; de Albuquerque, V.H.C. Seasonal Crops Disease Prediction and Classification Using Deep Convolutional Encoder Network. Circuits Syst. Signal Process. 2020, 39, 818–836. [Google Scholar] [CrossRef]
- Qin, J.; Burks, T.F.; Ritenour, M.A.; Bonn, W.G. Detection of citrus canker using hyperspectral reflectance imaging with spectral information divergence. J. Food Eng. 2009, 93, 183–191. [Google Scholar] [CrossRef]
- Garcia-Ruiz, F.; Sankaran, S.; Maja, J.M.; Lee, W.S.; Rasmussen, J.; Ehsani, R. Comparison of two aerial imaging platforms for identification of Huanglongbing-infected citrus trees. Comput. Electron. Agric. 2013, 91, 106–115. [Google Scholar] [CrossRef]
- Hossain, E.; Hossain, M.F.; Rahaman, M.A. A color and texture based approach for the detection and classification of plant leaf disease using KNN classifier. In Proceedings of the International Conference on Electrical, Computer and Communication Engineering (ECCE), Bazar, Bangladesh, 7–9 February 2019; pp. 1–6. [Google Scholar]
- Rauf, H.T.; Saleem, B.A.; Lali, M.I.U.; Khan, M.A.; Sharif, M.; Bukhari, S.A.C. A citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning. Data Brief 2019, 26, 104340. [Google Scholar] [CrossRef] [PubMed]
- Leitzke Betemps, D.; Vahl de Paula, B.; Parent, S.É.; Galarça, S.P.; Mayer, N.A.; Marodin, G.A.; Rozane, D.E.; Natale, W.; Melo, G.W.B.; Parent, L.E.; et al. Humboldtian diagnosis of peach tree (prunus persica) nutrition using machine-learning and compositional methods. Agronomy 2020, 10, 900. [Google Scholar] [CrossRef]
- Mary, N.A.B.; Singh, A.R.; Athisayamani, S. Classification of Banana Leaf Diseases Using Enhanced Gabor Feature Descriptor. In Inventive Communication and Computational Technologies; Springer: Berlin/Heidelberg, Germany, 2020; pp. 229–242. [Google Scholar]
- Pham, T.N.; Van Tran, L.; Dao, S.V.T. Early Disease Classification of Mango Leaves Using Feed-Forward Neural Network and Hybrid Metaheuristic Feature Selection. IEEE Access 2020, 8, 189960–189973. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, S.; Tanta, A.; Gupta, R. Extraction of Multiple Diseases in Apple Leaf Using Machine Learning. Int. J. Image Graph. 2021, 2140009. [Google Scholar] [CrossRef]
- Kianat, J.; Khan, M.A.; Sharif, M.; Akram, T.; Rehman, A.; Saba, T. A Joint Framework of Feature Reduction and Robust Feature Selection for Cucumber Leaf Diseases Recognition. Optik 2021, 166566. [Google Scholar] [CrossRef]
- Oyewola, D.O.; Dada, E.G.; Misra, S.; Damaševičius, R. Detecting cassava mosaic disease using a deep residual convolutional neural network with distinct block processing. PeerJ Comput. Sci. 2021, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rehman, Z.U.; Khan, M.A.; Ahmed, F.; Damaševičius, R.; Naqvi, S.R.; Nisar, W.; Javed, K. Recognizing apple leaf diseases using a novel parallel real-time processing framework based on MASK RCNN and transfer learning: An application for smart agriculture. IET Image Process. 2021. [Google Scholar] [CrossRef]
- Lee, K.; Seong, J.; Han, Y.; Lee, W.H. Evaluation of Applicability of Various Color Space Techniques of UAV Images for Evaluating Cool Roof Performance. Energies 2020, 13, 4213. [Google Scholar] [CrossRef]
- Shaik, K.B.; Ganesan, P.; Kalist, V.; Sathish, B.; Jenitha, J.M.M. Comparative study of skin color detection and segmentation in HSV and YCbCr color space. Procedia Comput. Sci. 2015, 57, 41–48. [Google Scholar] [CrossRef]
- Cugmas, B.; Štruc, E. Accuracy of an Affordable Smartphone-Based Teledermoscopy System for Color Measurements in Canine Skin. Sensors 2020, 20, 6234. [Google Scholar] [CrossRef]
- Pizer, S.M.; Amburn, E.P.; Austin, J.D.; Cromartie, R.; Geselowitz, A.; Greer, T.; ter Haar Romeny, B.; Zimmerman, J.B.; Zuiderveld, K. Adaptive histogram equalization and its variations. Comput. Vision Graph. Image Process. 1987, 39, 355–368. [Google Scholar] [CrossRef]
- Rauf, H.T. A Guava Fruits and Leaves Dataset for Detection and Classification of Guava Diseases through Machine Learning. Mendeley Data 2021. [Google Scholar] [CrossRef]


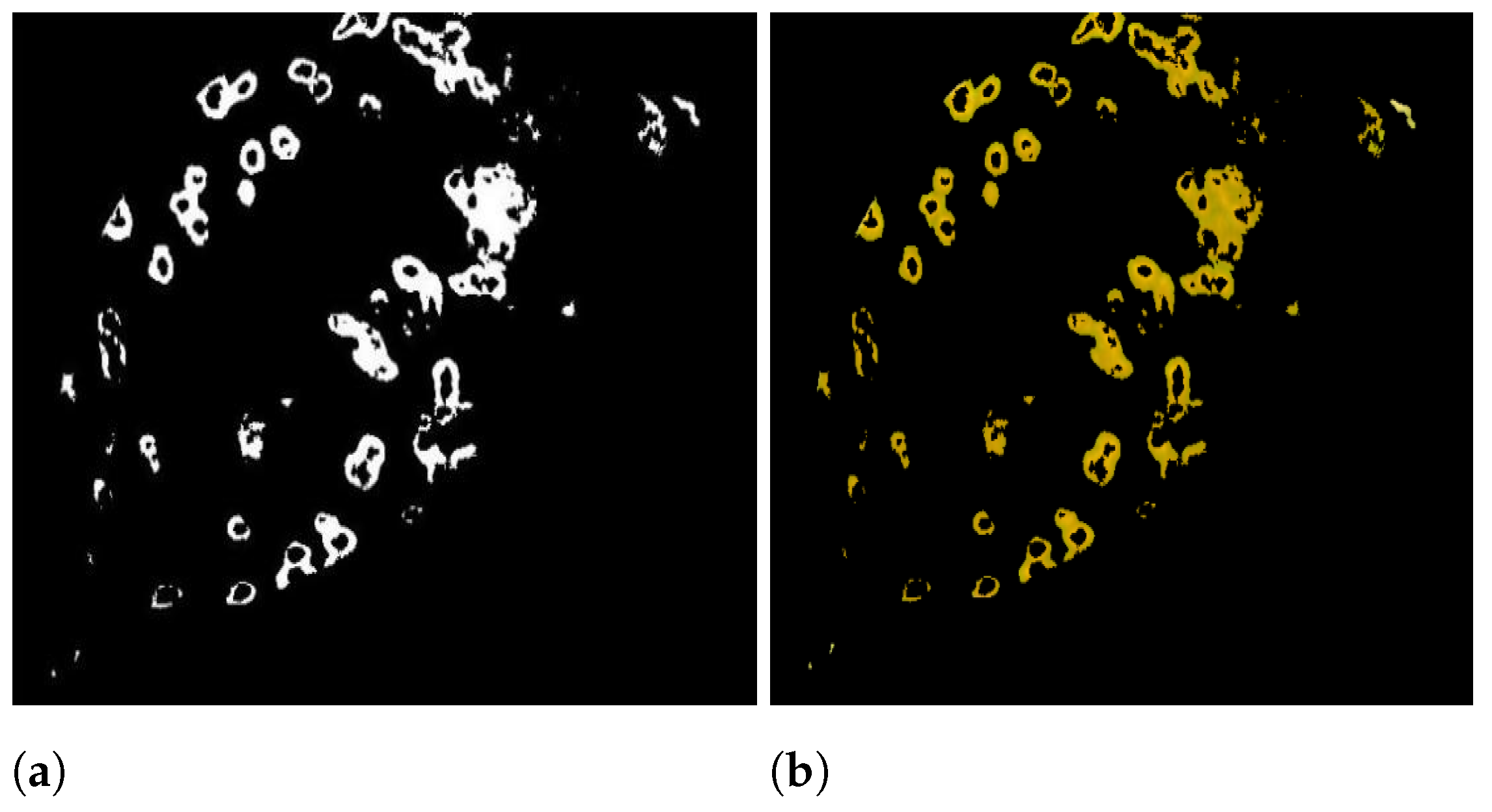
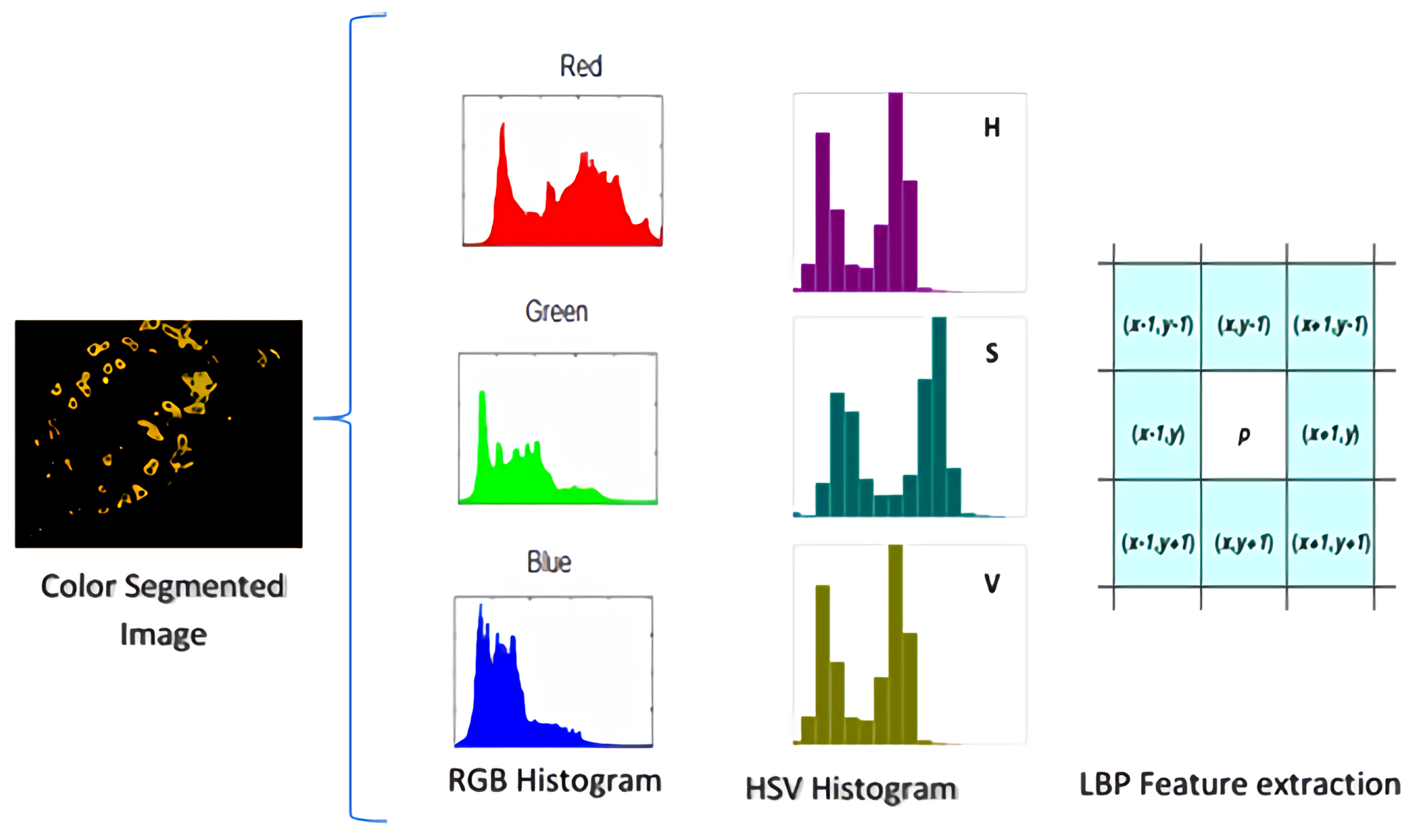


| Ref | Plants | Method | Segmentation | Feature Extraction | Classification Technique |
|---|---|---|---|---|---|
| [42] | Weeds/corn | Using color features, determine whether the apples are ripe enough to harvest. | color-based background subtraction | Energy features | BPNN |
| [53] | Redgrape fruit | Suppurating and usual images, as well as disease symptoms | - | Textural features | Spectral information |
| [54] | Citrus | Data are obtained using a citrus UAV, and images of plants are analyzed using sensors | - | Regression analysis | Stepwise SVM, LDA and QDA |
| [13] | Beats | Leaf blight spot, leaf rust, and powdery mildew are classified. | - | Nine spectral vegetation | SVM |
| [40] | Multiple plants | PSO feature selection which is kernel-based, is used for optimal feature selection and leaf classification. | Region of Interest | GLCM + LBP | (FRVM) |
| [34] | Orange fruit | By calculating gray level co-occurrence matrix (GLCM), texture and gray level features of defect area are extracted, and Probabilistic Neural Networks (RBPNN) is used for classification | Hue and Saturation histograms | GLCM | RBPNN |
| [14] | Citrus | For classification of disease textural features and color histogram were used. | Delta E | RGB, HSV histogram features + LBP | KNN and SVM |
| [55] | Multiple plants | Plant leaf disease using KNN classifier | color Segmentation | Textural features | KNsVMN |
| [56] | Citrus plants | Citrus diseases detection using machine-learning feature selection, extraction and classification. | Weighted Segmentation | Textural + color + Geometric features | M-SVM |
| [57] | Peach tree | Humboldtian diagnosis of peach tree using random forest | - | Meteorological indices and soil and tissue tests | Random Forest |
| [58] | Banana plants | Banana leaf diseases using enhanced Gabor feature descriptor | - | Gabor filter and 2D log Gabor filter descriptor | KNN |
| [59] | Mango plants | Disease of mango leaves detection through ANN and Hybrid Metaheuristic descriptor | Binary Segmentation | Textural+ Statistical | ANN |
| [60] | Apple tree | Used Brightness-preserving dynamic fuzzy histogram equalization | Histogram Equalization | Automatic feature extraction | KNN |
| [61] | Cucumber plant | Feature fusion and selection techniques for cucumber diseases detection | - | Probability distribution-based entropy | Multiple Classifiers |
| [62] | Cassava leaves | Cassava mosaic disease recognition using a deep residual convolution neural network (DRNN) with distinct block processing | Distinct block processing | - | DRNN |
| [63] | Apple leaves | MASK RCNN to detect infected regions, CNN for feature extraction and Kapur’s entropy along multiclass SVM for feature selection | Mask RNN | Kapur’s entropy with multiclass SVM | Ensemble subspace discriminant analysis |
| Ref | Advantages | Drawbacks |
|---|---|---|
| [42] | Wavelet decomposition using different color textures to obtain color bands | Ignored image foregrounds |
| [53] | Considered Spectral information | Missing statistical features |
| [54] | Different RGB ranges (R = 900 nm, G = 690 nm and B = 560 nm) used to extraction different color intensities. | Lack k-fold cross validaiton |
| [13] | Used adaptive template matching for disease development observation | The under-classification problem happened mainly in limited lighting conditions. |
| [40] | Optimal feature selection using PSO | Only considered ROI |
| [34] | Used Radial Basis Probabilistic Neural Networks | Lack k-fold cross validaiton |
| [14] | Combination of ML and Computer-Vision-based approaches | Lack of deep learning-based approaches |
| [55] | Clustered the corresponding diseases based on color and texture | Lack of deep learning-based approaches |
| [56] | Combination of ML and Computer-Vision-based approaches | Lack of deep learning-based approaches |
| [57] | Analyzed Soil conditions | Experiments performed on a small dataset |
| [58] | Used Gabor filter and 2D log Gabor filter | Lack of deep learning-based approaches |
| [59] | Extract both Textural + Statistical feature vectors | Adopted Binary Segmentation |
| [60] | Used Brightness-preserving dynamic fuzzy histogram equalization | Lack of deep learning-based approaches |
| [61] | Used data augmentation with different angles rotations. | Lack of deep learning-based approaches |
| [63] | Adopted Kapur’s entropy with multiclass SVM | Lack of handcrafted feature vectors |
| Sr # | Feature Sets | Dimensions |
|---|---|---|
| 1 | {H} | 255 |
| 2 | {S} | 255 |
| 3 | {V} | 255 |
| 4 | {HSV} | 768 |
| 5 | {R} | 255 |
| 6 | {G} | 255 |
| 7 | {B} | 255 |
| 8 | {RGB} | 768 |
| 9 | {LBP(R)} | 255 |
| 10 | {LBP(G)} | 255 |
| 11 | {LBP(B)} | 255 |
| 12 | {RGB HSV} | 1536 |
| 13 | PCA{RGB HSV LBP} | 195 |
| 14 | {RGB HSV LBP} | 2304 |
| 15 | {LBP(R) LBP(G) LBP(B)} | 768 |
| Description | No of Images |
|---|---|
| Normal | 87 |
| Rust | 70 |
| Canker | 77 |
| Mummification | 83 |
| Dot | 76 |
| Total | 393 |
| Channel | R | G | B | RGB | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measure | TPR | TNR | ACC | TPR | TNR | ACC | TPR | TNR | ACC | TPR | TNR | ACC |
| Class | P | N | % | P | N | % | P | N | % | P | N | % |
| Fine KNN | 99.1% | 100% | 99.5% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Cubic SVM | 94.5% | 100% | 97% | 97.3% | 100% | 98.5% | 99.1% | 100% | 99.5% | 99.1% | 100% | 99.5% |
| Boosted Tree | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 0% | 55.8% | 100% | 0% | 55.8% |
| Bagged Tree | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Complex Tree | 99.1% | 100% | 99.5% | 100% | 100% | 100% | 99.1% | 100% | 99.5% | 99.1% | 100% | 99.5% |
| Channel | H | S | V | HSV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measure | TPR | TNR | ACC | TPR | TNR | ACC | TPR | TNR | ACC | TPR | TNR | ACC |
| Class | P | N | P | N | P | N | P | N | ||||
| Fine KNN | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Cubic SVM | 97.3% | 100% | 98.5% | 96.4% | 100% | 98% | 97.3% | 100% | 98.5% | 94.5% | 100% | 97% |
| Boosted Tree | 100% | 0% | 55.8% | 98.2% | 100% | 99% | 99.1% | 100% | 99.5% | 100% | 0% | 55.8% |
| Bagged Tree | 100% | 100% | 100% | 98.2% | 100% | 99% | 100% | 100% | 100% | 100% | 100% | 100% |
| Complex Tree | 98.2% | 100% | 99% | 98.2% | 100% | 99% | 100% | 100% | 100% | 98.2% | 100% | 99% |
| Features | RLBP | GLBP | BLBP | LBP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measure | TPR | TNR | ACC | TPR | TNR | ACC | TPR | TNR | ACC | TPR | TNR | ACC |
| Class | P | N | P | N | P | N | P | N | ||||
| Fine KNN | 100% | 100% | 100% | 100% | 100% | 100% | 98.2% | 100% | 99% | 100% | 100% | 100% |
| Cubic SVM | 96.4% | 100% | 98% | 91.8% | 100% | 95.4% | 95.5% | 19.5% | 61.9% | 100% | 100% | 100% |
| Boosted Tree | 100% | 0% | 55.8% | 100% | 0% | 55.8% | 100% | 0% | 55.8% | 100% | 0% | 55.8% |
| Bagged Tree | 99.1% | 100% | 99.5% | 98.2% | 100% | 99% | 99.1% | 100% | 99.5% | 99.1% | 100% | 99.5% |
| Complex Tree | 99.1% | 100% | 99.5% | 98.2% | 100% | 99% | 98.2% | 100% | 99% | 99.1% | 100% | 99.5% |
| Features | {RGB, HSV} | {RGB, HSV, LBP} | PCA{RGB, HSV, LBP} | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Measure | TPR | TNR | ACC | TPR | TNR | ACC | TPR | TNR | ACC |
| Class | P | N | % | P | N | % | P | N | % |
| Fine KNN | 100% | 100% | 100% | 100% | 100% | 100% | 15.5% | 100% | 52.8% |
| Cubic SVM | 99.1% | 100% | 99.5% | 100% | 100% | 100% | 59.1% | 100% | 72.5% |
| Boosted Tree | 100% | 0% | 55.8% | 100% | 0% | 55.8% | 100% | 100% | 100% |
| Bagged Tree | 100% | 100% | 100% | 99.1% | 100% | 99.5% | 100% | 100% | 100% |
| Complex Tree | 99.1% | 100% | 99.5% | 99.1% | 100% | 99.5% | 97.3% | 100% | 98.5% |
| {RGB, HSV} | {RGB, HSV, LBP} | CA{RGB, HSV, LBP} | ||||
|---|---|---|---|---|---|---|
| Classifier | Rank | Z | Rank | Z | Rank | Z |
| Bagged Tree | 4.5 | 1.06 | 2.5 | −0.35 | 4.5 | 1.06 |
| Boosted Tree | 1.0 | −1.41 | 1.0 | −1.41 | 4.5 | 1.06 |
| Complex Tree | 2.5 | −0.35 | 2.5 | −0.35 | 3.0 | 0.00 |
| Cubic SVM | 2.5 | −0.35 | 4.5 | 1.06 | 2.0 | −0.71 |
| Fine KNN | 4.5 | 1.06 | 4.5 | 1.06 | 1.0 | −1.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almadhor, A.; Rauf, H.T.; Lali, M.I.U.; Damaševičius, R.; Alouffi, B.; Alharbi, A. AI-Driven Framework for Recognition of Guava Plant Diseases through Machine Learning from DSLR Camera Sensor Based High Resolution Imagery. Sensors 2021, 21, 3830. https://doi.org/10.3390/s21113830
Almadhor A, Rauf HT, Lali MIU, Damaševičius R, Alouffi B, Alharbi A. AI-Driven Framework for Recognition of Guava Plant Diseases through Machine Learning from DSLR Camera Sensor Based High Resolution Imagery. Sensors. 2021; 21(11):3830. https://doi.org/10.3390/s21113830
Chicago/Turabian StyleAlmadhor, Ahmad, Hafiz Tayyab Rauf, Muhammad Ikram Ullah Lali, Robertas Damaševičius, Bader Alouffi, and Abdullah Alharbi. 2021. "AI-Driven Framework for Recognition of Guava Plant Diseases through Machine Learning from DSLR Camera Sensor Based High Resolution Imagery" Sensors 21, no. 11: 3830. https://doi.org/10.3390/s21113830
APA StyleAlmadhor, A., Rauf, H. T., Lali, M. I. U., Damaševičius, R., Alouffi, B., & Alharbi, A. (2021). AI-Driven Framework for Recognition of Guava Plant Diseases through Machine Learning from DSLR Camera Sensor Based High Resolution Imagery. Sensors, 21(11), 3830. https://doi.org/10.3390/s21113830






