Release and Detection of microRNA by Combining Magnetic Hyperthermia and Electrochemistry Modules on a Microfluidic Chip
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Oligonucleotide Sequences
2.3. DNA Hybridization on Core–Shell Nanoparticles γ-Fe2O3 @SiO2 PEG/NH2, Release and Target Detection
2.4. Fluorometric Measurements Calibration
2.5. Electrochemical Measurements Protocol
2.5.1. Microelectrode Activation Protocol
2.5.2. Microelectrode Functionalization Protocol
2.6. Micro Nanofabrication Procedures
2.6.1. Microfluidic Chip Fabrication for On-Chip Hyperthermia
2.6.2. Microfluidic Chip Fabrication for On-Chip Hyperthermia and Electrochemistry
3. Results
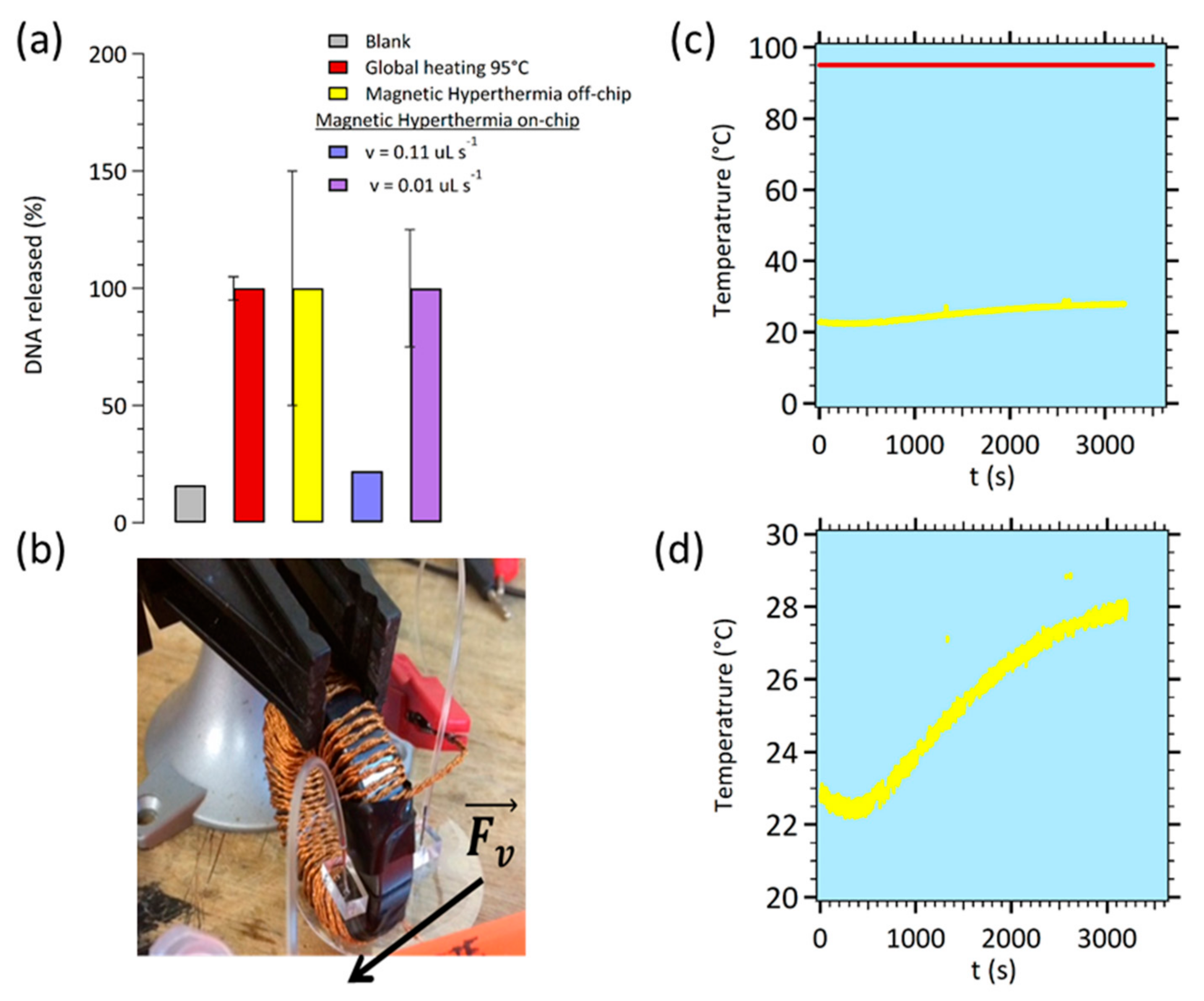
3.1. Setup for Performing Magnetic Hyperthermia On-Chip
3.1.1. Theory of Electromagnetism, an Electromagnet with a Gap
3.1.2. Magnetic Hyperthermia, Neel and Brown Contributions
3.1.3. Magnetic Hyperthermia on Chip
3.2. Coupling with Electrochemical Detection Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Synthesis of Core–Shell Nanoparticles γ-Fe2O3@SiO2 PEG/NH2

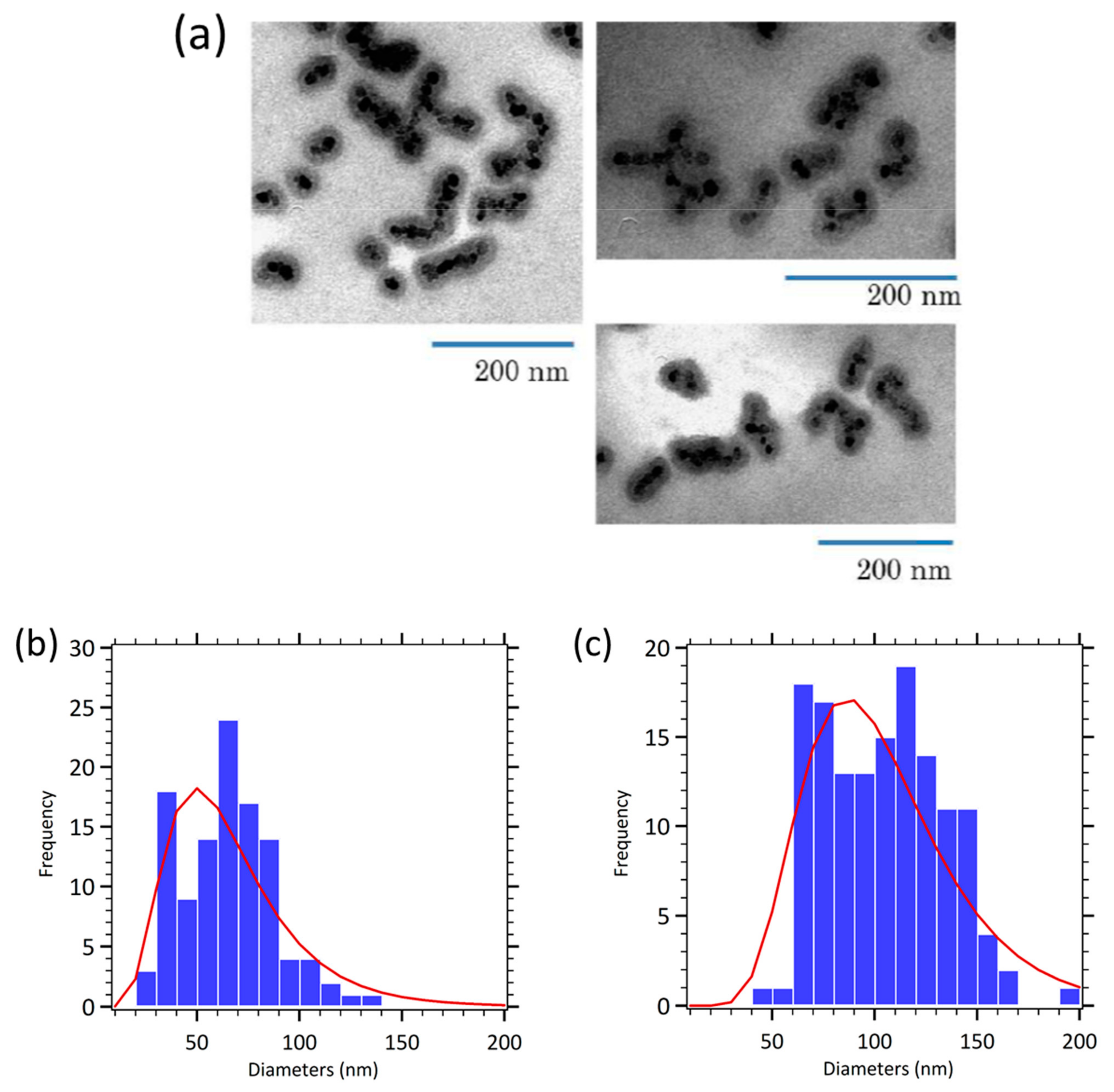
Appendix B
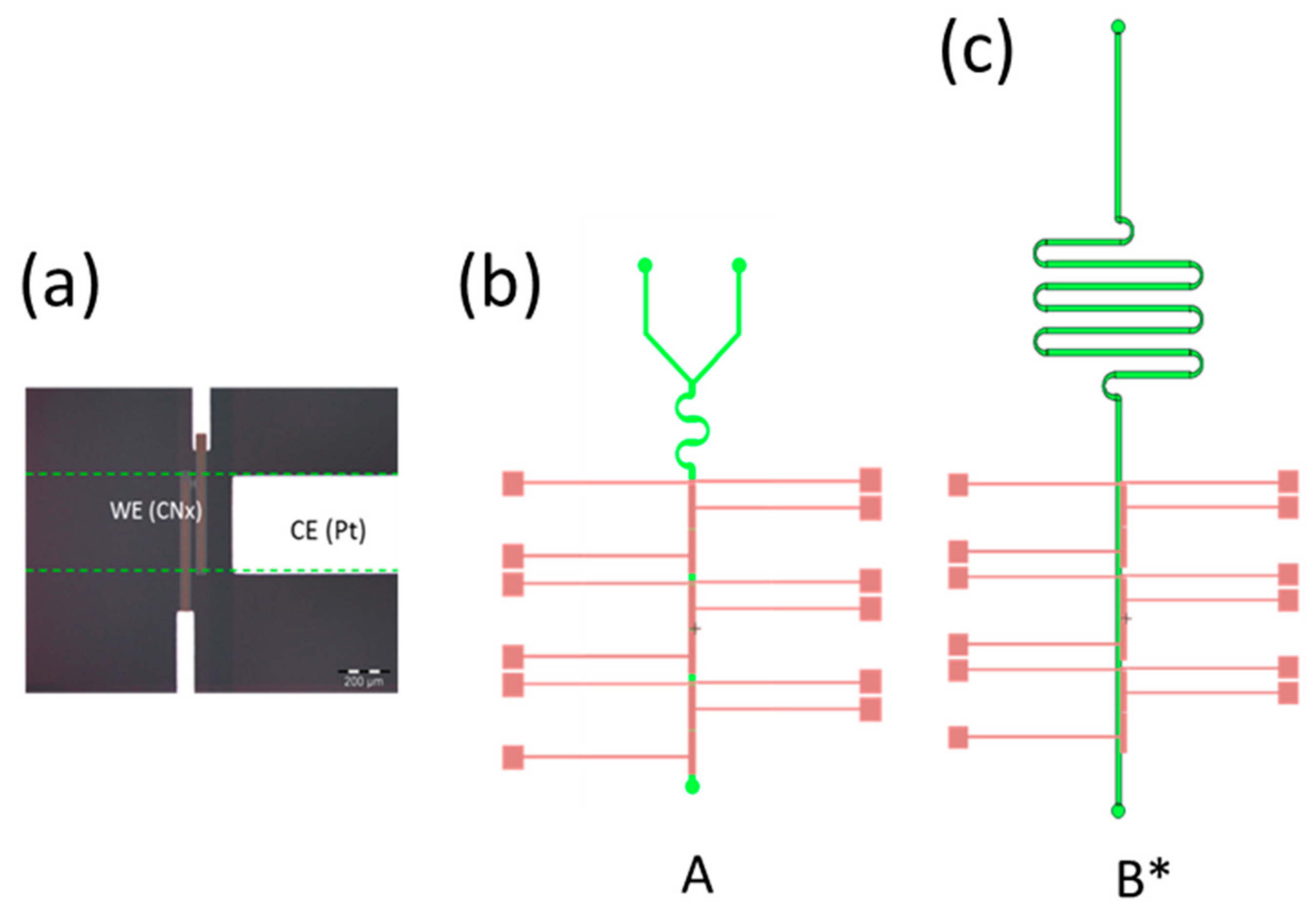
Photography and L-Edit Channel and Microelectrodes Geometries
References
- Mitchell, P. Microfluidics—Downsizing large-scale biology. Nat. Biotechnol. 2001, 19, 717–721. [Google Scholar] [CrossRef] [PubMed]
- LaVan, D.A.; McGuire, T.; Langer, R. Small-scale systems for in vivo drug delivery. Nat. Biotechnol. 2003, 21, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-B.; Chien, C.-C.; You, H.-L.; Kuo, F.-C.; Lee, M.S.; Lee, G.-B. An integrated microfluidic system for antimicrobial susceptibility testing with antibiotic combination. Lab Chip 2019, 19, 2699–2708. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.S.; Buchsbaum, S.F.; Swensen, J.S.; Hsieh, K.; Lou, X.; Soh, H.T. Integrated Microfluidic Electrochemical DNA Sensor. Anal. Chem. 2009, 81, 6503–6508. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell. Mol. Med. 2014, 18, 371–390. [Google Scholar] [CrossRef]
- Pekin, D.; Skhiri, Y.; Baret, J.-C.; Le Corre, D.; Mazutis, L.; Ben Salem, C.; Millot, F.; El Harrak, A.; Hutchison, J.B.; Larson, J.W.; et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 2011, 11, 2156–2166. [Google Scholar] [CrossRef]
- Taly, V.; Pekin, D.; Benhaim, L.; Kotsopoulos, S.K.; Le Corre, D.; Li, X.; Atochin, I.; Link, D.R.; Griffiths, A.D.; Pallier, K.; et al. Multiplex Picodroplet Digital PCR to Detect KRAS.; Mutations in Circulating DNA from the Plasma of Colorectal Cancer Patients. Clin. Chem. 2013, 59, 1722. [Google Scholar] [CrossRef]
- Robinson, S.; Follo, M.; Haenel, D.; Mauler, M.; Stallmann, D.; Heger, L.A.; Helbing, T.; Duerschmied, D.; Peter, K.; Bode, C.; et al. Chip-based digital PCR as a novel detection method for quantifying microRNAs in acute myocardial infarction patients. Acta Pharmacol. Sin. 2018, 39, 1217–1227. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Jordi, C.; Son, M.; Van Phan, H.; Drayman, N.; Abasiyanik, M.F.; Vistain, L.; Tu, H.-L.; Tay, S. Ultra-sensitive digital quantification of proteins and mRNA in single cells. Nat. Commun. 2019, 10, 3544. [Google Scholar] [CrossRef]
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.L.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef]
- Craw, P.; Balachandran, W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: A critical review. Lab Chip 2012, 12, 2469–2486. [Google Scholar] [CrossRef] [PubMed]
- Gomez, K.F.; Lane, J.; Cunnick, G.; Grimshaw, D.; Jiang, W.G.; Mansel, R.E. From PCR to RCA: A surgical trainee’s guide to the techniques of genetic amplification. Eur. J. Surg. Oncol. 2002, 28, 554–556. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Chang, W.-H.; Wang, C.-H.; Wang, J.-H.; Mai, J.D.; Lee, G.-B. Nucleic acid amplification using microfluidic systems. Lab Chip 2013, 13, 1225–1242. [Google Scholar] [CrossRef] [PubMed]
- Houssin, T.; Cramer, J.; Grojsman, R.; Bellahsene, L. Ultrafast, sensitive and large-volume on-chip real-time PCR for the molecular diagnosis of bacterial and viral infections. Lab Chip 2016, 16, 1401–1411. [Google Scholar] [CrossRef]
- Frisbie, C.D.; Fritsch-Faules, I.; Wollman, E.W.; Wrighton, M.S. Preparation and characterization of redox active molecular assemblies on microelectrode arrays. Thin Solid Films 1992, 210–211, 341–347. [Google Scholar] [CrossRef]
- Compton, R.G.; Fisher, A.C.; Wellington, R.G.; Dobson, P.J.; Leigh, P.A. Hydrodynamic voltammetry with microelectrodes: Channel microband electrodes; theory and experiment. J. Phys. Chem. 1993, 97, 10410–10415. [Google Scholar] [CrossRef]
- Rossier, J.S.; Schwarz, A.; Reymond, F.; Ferrigno, R.; Bianchi, F.; Girault, H.H. Microchannel networks for electrophoretic separations. Electrophoresis 1999, 20, 727–731. [Google Scholar] [CrossRef]
- Gamby, J.; Abid, J.-P.; Girault, H.H. Supercapacitive Admittance Tomoscopy. J. Am. Chem. Soc. 2005, 127, 13300–13304. [Google Scholar] [CrossRef] [Green Version]
- Streeter, I.; Fietkau, N.; Del Campo, J.; Mas, R.; Munoz, F.X.; Compton, R.G. Voltammetry at Regular Microband Electrode Arrays: Theory and Experiment. J. Phys. Chem. C 2007, 111, 12058–12066. [Google Scholar] [CrossRef]
- Amatore, C.; Da Mota, N.; Sella, C.; Thouin, L. General Concept of High-Performance Amperometric Detector for Microfluidic (Bio)Analytical Chips. Anal. Chem. 2008, 80, 4976–4985. [Google Scholar] [CrossRef]
- Faure, M.; Pallandre, A.; Chebil, S.; Le Potier, I.; Taverna, M.; Tribollet, B.; Deslouis, C.; Haghiri-Gosnet, A.-M.; Gamby, J. Improved electrochemical detection of a transthyretin synthetic peptide in the nanomolar range with a two-electrode system integrated in a glass/PDMS microchip. Lab Chip 2014, 14, 2800–2805. [Google Scholar] [CrossRef] [PubMed]
- Veselinovic, J.; Li, Z.; Daggumati, P.; Seker, E. Electrically Guided DNA Immobilization and Multiplexed DNA Detection with Nanoporous Gold Electrodes. Nanomaterials 2018, 8, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rant, U.; Arinaga, K.; Fujita, S.; Yokoyama, N.; Abstreiter, G.; Tornow, M. Electrical manipulation of oligonucleotides grafted to charged surfaces. Org. Biomol. Chem. 2006, 4, 3448–3455. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Khandurina, J.; McKnight, T.E.; Jacobson, S.C.; Waters, L.C.; Foote, R.S.; Ramsey, J.M. Integrated System for Rapid PCR-Based DNA Analysis in Microfluidic Devices. Anal. Chem. 2000, 72, 2995–3000. [Google Scholar] [CrossRef]
- Ruano-Lopez, J.M.; Agirregabiria, M.; Olabarria, G.; Verdoy, D.; Bang, D.D.; Bu, M.; Wolff, A.; Voigt, A.; Dziuban, J.A.; Walczak, R.; et al. The SmartBioPhone™, a point of care vision under development through two European projects: OPTOLABCARD and LABONFOIL. Lab Chip 2009, 9, 1495–1499. [Google Scholar] [CrossRef]
- Jordan, A.; Scholz, R.; Wust, P.; Fähling, H.; Roland, F. Magnetic fluid hyperthermia (MFH): Cancer treatment with AC magnetic field induced excitation of biocompatible superparamagnetic nanoparticles. J. Magn. Magn. Mater. 1999, 201, 413–419. [Google Scholar] [CrossRef]
- Hirsch, L.R.; Stafford, R.J.; Bankson, J.A.; Sershen, S.R.; Rivera, B.; Price, R.E.; Hazle, J.D.; Halas, N.J.; West, J.L. Nanoshell-mediated near-infrared thermal therapy of tumors under magnetic resonance guidance. Proc. Natl. Acad. Sci. USA 2003, 100, 13549–13554. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Jang, J.-T.; Choi, J.-S.; Moon, S.H.; Noh, S.-H.; Kim, J.-W.; Kim, J.-G.; Kim, I.-S.; Park, K.I.; Cheon, J. Exchange-coupled magnetic nanoparticles for efficient heat induction. Nat. Nanotechnol. 2011, 6, 418–422. [Google Scholar] [CrossRef]
- Rao, W.; Zhang, W.; Poventud-Fuentes, I.; Wang, Y.; Lei, Y.; Agarwal, P.; Weekes, B.; Li, C.; Lu, X.; Yu, J.; et al. Thermally responsive nanoparticle-encapsulated curcumin and its combination with mild hyperthermia for enhanced cancer cell destruction. Acta Biomater. 2014, 10, 831–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, J.T.; Moros, M.A.A. DNA as a molecular local thermal probe for the analysis of magnetic hyperthermia. Angew. Chem. Int. Ed. 2013, 52, 11526–11529. [Google Scholar] [CrossRef] [PubMed]
- Jangpatarapongsa, K.; Polpanich, D.; Yamkamon, V.; Dittharot, Y.; Peng-On, J.; Thiramanas, R.; Hongeng, S.; Jootar, S.; Charoenmak, L.; Tangboriboonrat, P. DNA detection of chronic myelogenous leukemia by magnetic nanoparticles. Analyst 2011, 136, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zink, J.I. Taking the temperature of the interiors of magnetically heated nanoparticles. ACS Nano 2014, 8, 5199–5207. [Google Scholar] [CrossRef] [PubMed]
- Faure, M.; Billon, F.; Le Potier, I.; Haghiri-Gosnet, A.M.; Tribollet, B.; Pailleret, A.; Deslouis, C.; Gamby, J. Improvement of electrochemical detection of transthyretin synthetic peptide and its amino acids on carbon electrodes: Glassy carbon versus amorphous carbon nitride a-CNx. Electrochim. Acta 2019, 296, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Horny, M.C.; Billon, F.; Deslouis, C.; Lazerges, M.; Dupuis, V.; Siaugue, J.M.; Pailleret, A.; Gamby, J. microRNA electrochemical biosensor based on amorphous carbon nitride microband integrated in a microfluidic device. Electrochim. Acta 2020. In revision (under review). [Google Scholar]
- Iliescu, C.; Jing, J.; Tay, F.E.H.; Miao, J.; Sun, T. Characterization of masking layers for deep wet etching of glass in an improved HF/HCl solution. Surf. Coat. Technol. 2005, 198, 314–318. [Google Scholar] [CrossRef]
- Nanteuil, C.; Gosnet-Haghiri, A.M. Method for Manufacturing a Microfluidic Chip, and Related Chip and Plate. U.S. Patent 8,747,604, 10 June 2014. [Google Scholar]
- Lacroix, L.M.; Malaki, R.B.; Carrey, J.; Lachaize, S.; Respaud, M.; Goya, G.F.; Chaudret, B. Magnetic hyperthermia in single-domain monodisperse FeCo nanoparticles: Evidences for Stoner-Wohlfarth behavior and large losses. J. Appl. Phys. 2009, 105. [Google Scholar] [CrossRef]
- De Lacheisserie, E.D.T. Magnétisme, Tome 1: Fondements (Coll. Grenoble Sciences); UGA: Saint-Martin-d’Hères, France, 2000. [Google Scholar]
- Shliomis, M.I. Magnetic fluids. Soviet Phys. Uspekhi 1974, 17, 153. [Google Scholar] [CrossRef]
- Fortin, J.P.; Wilhelm, C.; Servais, J.; Mnager, C.; Bacri, J.C.; Gazeau, F. Size-Sorted Anionic Iron Oxide Nanomagnets as Colloidal Mediators for Magnetic Hyperthermia. J. Am. Chem. Soc. 2007, 129, 2628–2635. [Google Scholar] [CrossRef]
- Horny, M.C.; Lazerges, M.; Siaugue, J.M.; Pallandre, A.; Rose, D.; Bedioui, F.; Deslouis, C.; Haghiri-Gosnet, A.M.; Gamby, J. Electrochemical DNA biosensors based on long-range electron transfer: Investigating the efficiency of a fluidic channel microelectrode compared to an ultramicroelectrode in a two-electrode setup. Lab Chip 2016, 16, 4373–4381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Liu, J.; Wu, K.; Klein, T.; Jiang, Y.; Wang, J.-P. Evaluation of Hyperthermia of Magnetic Nanoparticles by Dehydrating DNA. Sci. Rep. 2014, 4, 7216. [Google Scholar] [CrossRef] [PubMed]
- Meunier-Prest, R.; Raveau, S.; Finot, E.; Legay, G.; Cherkaoui-Malki, M.; Latruffe, N. Direct measurement of the melting temperature of supported DNA by electrochemical method. Nucleic Acids Res. 2003, 31, e150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, D.; Wang, X.; Williams, K.; Levicky, R. Thermostable DNA Immobilization and Temperature Effects on Surface Hybridization. Langmuir 2012, 28, 8446–8455. [Google Scholar] [CrossRef] [PubMed]
- Tamiasso-Martinhon, P.; Cachet, H.; Deslouis, C.; Vivier, V. Amorphous carbon nitride a-CNx microelectrode: Fabrication and characterization. Electrochem. Commun. 2010, 12, 1074–1076. [Google Scholar] [CrossRef]
- Kelley, S.O.; Holmlin, R.E.; Stemp, E.D.A.; Barton, J.K. Photoinduced Electron Transfer in Ethidium-Modified DNA Duplexes: Dependence on Distance and Base Stacking. J. Am. Chem. Soc. 1997, 119, 9861–9870. [Google Scholar] [CrossRef]
- Kaplan, J.-C.; Delpech, M. Biologie Moléculaire et Médecine; Flammarion Médecine-Sciences: Paris, France, 1989. [Google Scholar]
- Kelley, S.O.; Barton, J.K.; Jackson, N.M.; Hill, M.G. Electrochemistry of Methylene Blue Bound to a DNA-Modified Electrode. Bioconj. Chem. 1997, 8, 31–37. [Google Scholar] [CrossRef]
- Kelley, S.O.; Jackson, N.M.; Hill, M.G.; Barton, J.K. Long-range electron transfer through DNA films. Angew. Chem. Int. Ed. 1999, 38, 941–945. [Google Scholar] [CrossRef]
- Massart, R.; Dubois, E.; Cabuil, V.; Hasmonay, E. Preparation and properties of monodisperse magnetic fluids. J. Magn. Magn. Mater. 1995, 149, 1–5. [Google Scholar] [CrossRef]
- Lefebure, S.; Dubois, E.; Cabuil, V.; Neveu, S.; Massart, R. Monodisperse magnetic nanoparticles: Preparation and dispersion in water and oils. J. Mater. Res. 1998, 13. [Google Scholar] [CrossRef]
- Schöneborn, H.; Raudzus, F.; Secret, E.; Otten, N.; Michel, A.; Fresnais, J.; Ménager, C.; Siaugue, J.-M.; Zaehres, H.; Dietzel, I.D.; et al. Novel Tools towards Magnetic Guidance of Neurite Growth: (I) Guidance of Magnetic Nanoparticles into Neurite Extensions of Induced Human Neurons and In Vitro Functionalization with RAS Regulating Proteins. J. Funct. Biomater. 2019, 10, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurice, V.; Georgelin, T.; Siaugue, J.-M.; Cabuil, V. Synthesis and characterization of functionalized core–shell γFe2O3–SiO2 nanoparticles. J. Magn. Magn. Mater. 2009, 321, 1408–1413. [Google Scholar] [CrossRef]
- Georgelin, T.; Bombard, S.; Siaugue, J.-M.; Cabuil, V. Nanoparticle-Mediated Delivery of Bleomycin. Angew. Chem. Int. Ed. 2010, 49, 8897–8901. [Google Scholar] [CrossRef] [PubMed]
- Etoc, F.; Vicario, C.; Lisse, D.; Siaugue, J.-M.; Piehler, J.; Coppey, M.; Dahan, M. Magnetogenetic Control of Protein Gradients Inside Living Cells with High Spatial and Temporal Resolution. Nano Lett. 2015, 15, 3487–3494. [Google Scholar] [CrossRef]



| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| DNA Probe (P) for MNPs | 5′-Carboxy C6-CAA ACA CCA TTG TCA CAC TGC-3′ |
| DNA Probe (P’) for microelectrodes | 5′-Amino C6-CAA ACA CCA TTG TCA CAC TGC-3′ |
| Target (T) | 5′-GC AGT GTG ACA ATG GTG TTT G-3′ |
| Fluidic Geometry | (µL s−1) | Residence Time (t) | Volume of Detection (µL) | Δt § |
|---|---|---|---|---|
| A | 0.11 | 3 s | 400 | 2 h 48 min |
| A | 0.01 | 12 s | 100 | 2 h 48 min |
| Fluidic Geometry | V# (µL s−1) | Δt § | Sensitivity (M) |
|---|---|---|---|
| Geometry A | 0.01 | 2 h 48 min | 10−8 |
| Geometry B * | 0.056 | 30 min | 10−8 |
| Geometry B * | 0.056 | 30 min | <10−16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horny, M.-C.; Dupuis, V.; Siaugue, J.-M.; Gamby, J. Release and Detection of microRNA by Combining Magnetic Hyperthermia and Electrochemistry Modules on a Microfluidic Chip. Sensors 2021, 21, 185. https://doi.org/10.3390/s21010185
Horny M-C, Dupuis V, Siaugue J-M, Gamby J. Release and Detection of microRNA by Combining Magnetic Hyperthermia and Electrochemistry Modules on a Microfluidic Chip. Sensors. 2021; 21(1):185. https://doi.org/10.3390/s21010185
Chicago/Turabian StyleHorny, Marie-Charlotte, Vincent Dupuis, Jean-Michel Siaugue, and Jean Gamby. 2021. "Release and Detection of microRNA by Combining Magnetic Hyperthermia and Electrochemistry Modules on a Microfluidic Chip" Sensors 21, no. 1: 185. https://doi.org/10.3390/s21010185






