How Reliable Is the Electrochemical Readout of MIP Sensors?
Abstract
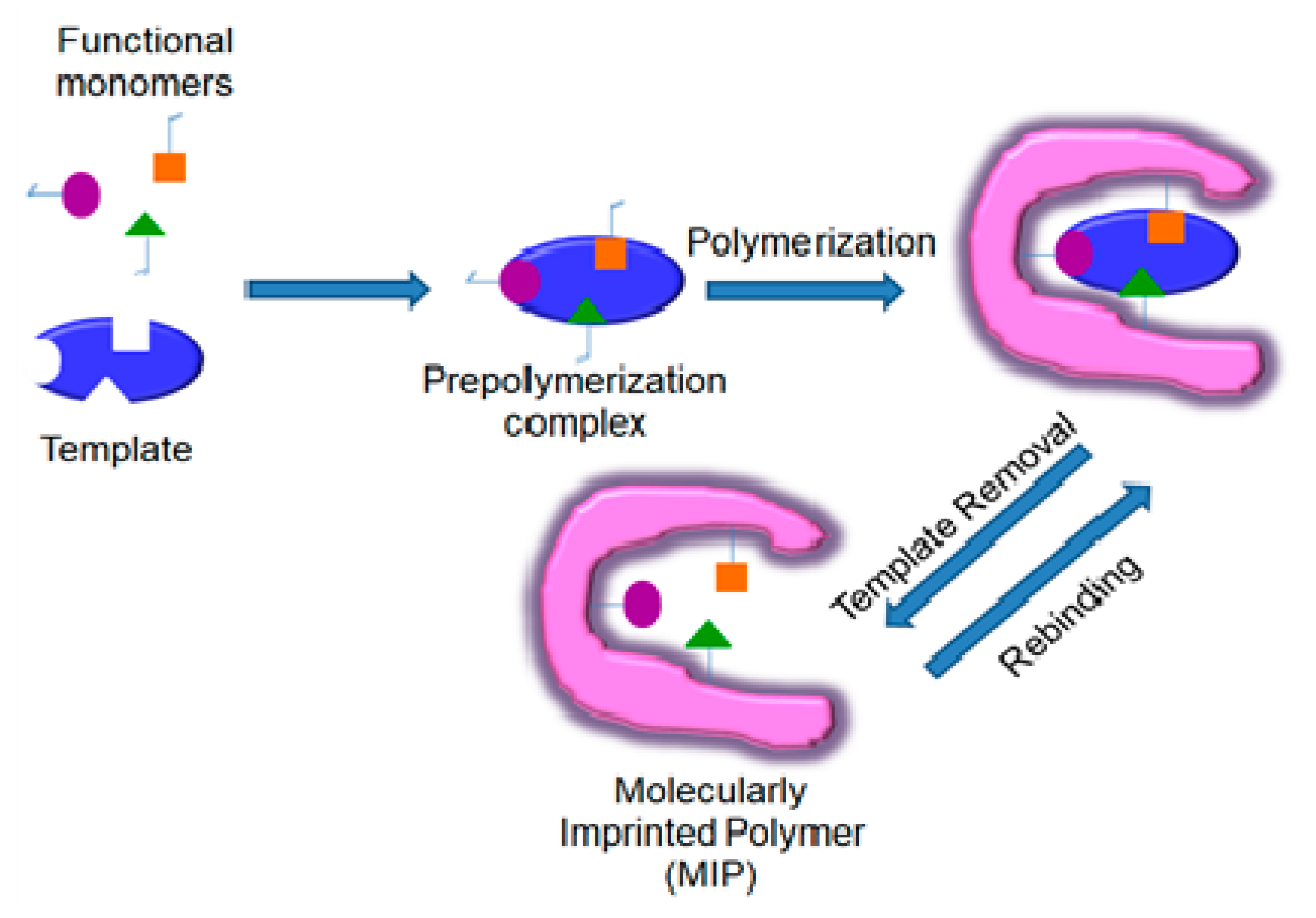
1. Introduction
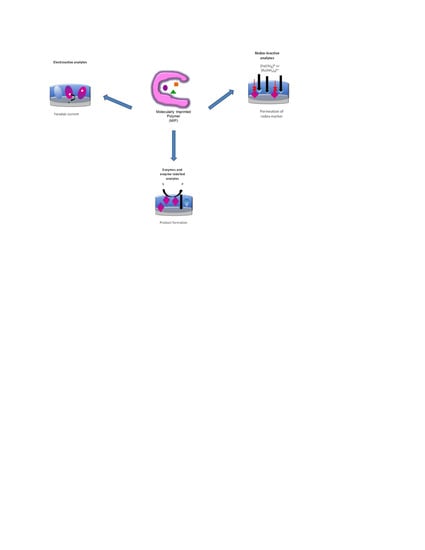
2. Electrochemical Readout
- (i)
- When the targets are redox-active, the faradaic current is measured, which is based on the direct electron transfer (DET) between the target and the underlying electrode.
- (ii)
- In the case of enzyme targets, catalytically active MIPs or enzyme-labeled targets, the enzymatic activity of the MIP layer is detected via the generation of a redox-active product at the electrode surface.
- (iii)
- Most of the research covers the flux of a redox marker. The signal modulated by the target binding is detected at the underlying electrode surface.
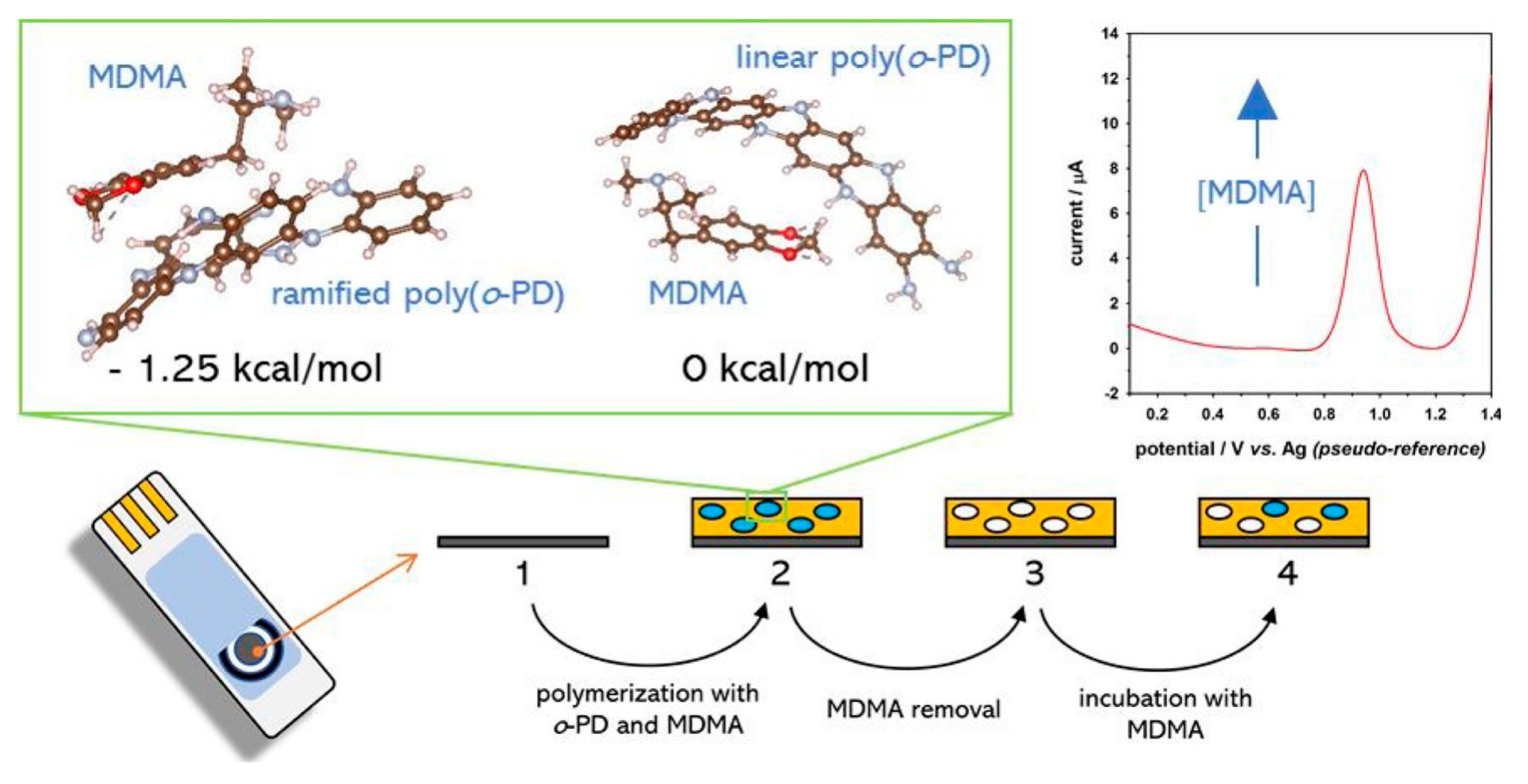
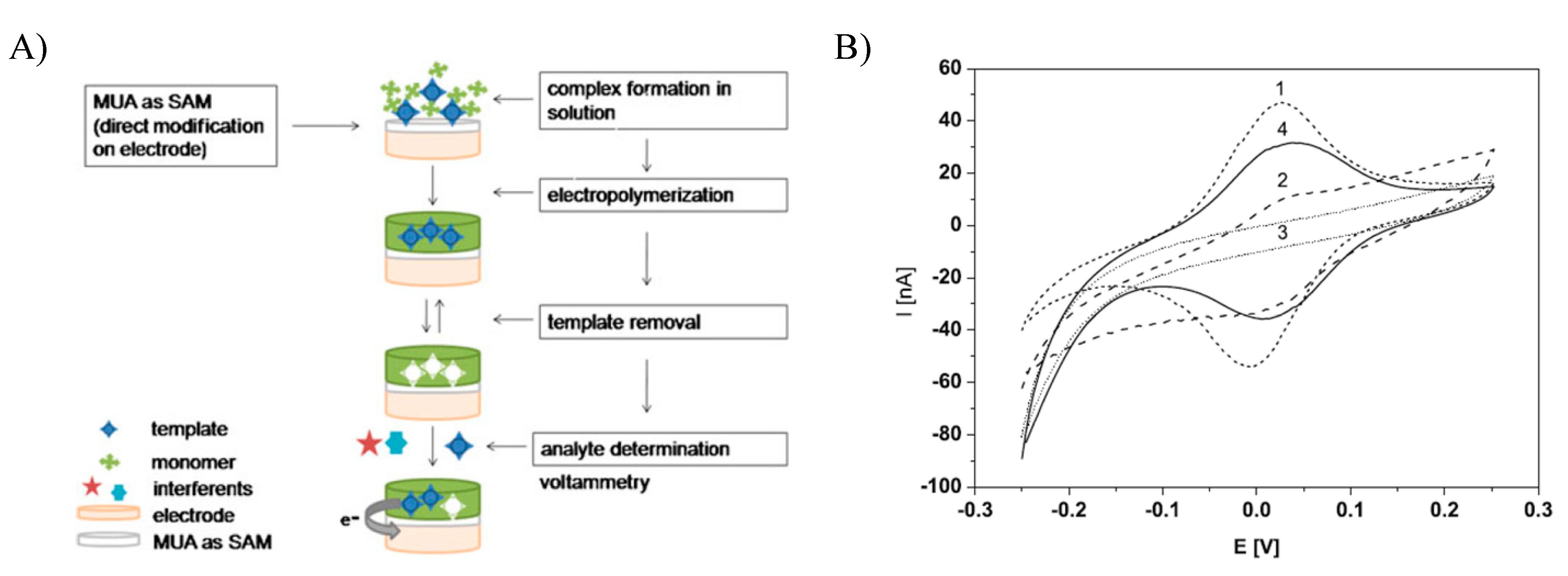
2.1. Electroactive Analytes
2.2. Catalytically Active Analytes
2.2.1. Enzymes and Enzyme-Labeled Analytes
2.2.2. Catalytically Active MIPs
2.3. Redox-Inactive Analytes
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Scheller, F.W.; Zhang, X.; Yarman, A.; Wollenberger, U.; Gyurcsányi, R.E. Molecularly imprinted polymer-based electrochemical sensors for biopolymers. Curr. Opin. Electrochem. 2019, 14, 53–59. [Google Scholar] [CrossRef]
- Wulff, G.; Sarhan, A. Macromolecular Colloquium. Angew. Chem. Int. Ed. Engl. 1972, 11, 334–342. [Google Scholar]
- Arshady, R.; Mosbach, K. Synthesis of Substrate-selective Polymers by Host-Guest Polymerizatioa. Rapid Commun. 1981, 692, 687–692. [Google Scholar]
- Hayden, O.; Lieberzeit, P.A.; Blaas, D.; Dickert, F.L. Artificial antibodies for bioanalyte detection-Sensing viruses and proteins. Adv. Funct. Mater. 2006, 16, 1269–1278. [Google Scholar] [CrossRef]
- Haupt, K.; Linares, A.V.; Bompart, M.; Bui, B.T.S. Molecularly Imprinted Polymers. In Molecular Imprinting. Topics in Current Chemistry; Haupt, K., Ed.; Springer: Berlin, Germany, 2011; pp. 1–28. [Google Scholar]
- Dickert, F.L. Molecular imprinting and functional polymers for all transducers and applications. Sensors 2018, 18, 327. [Google Scholar] [CrossRef] [PubMed]
- Whitcombe, M.J.; Kirsch, N.; Nicholls, I.A. Molecular Imprinting Science and Technology: A Survey of the Literature for the Years 2004–2011. J. Mol. Recognit. 2014, 27, 297–401. [Google Scholar] [PubMed]
- Haupt, K. Imprinted polymers-Tailor-made mimics of antibodies and receptors. Chem. Commun. 2003, 9, 171–177. [Google Scholar] [CrossRef]
- Díaz-Díaz, G.; Antuña-Jiménez, D.; Carmen Blanco-López, M.; Jesús Lobo-Castañón, M.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. New materials for analytical biomimetic assays based on affinity and catalytic receptors prepared by molecular imprinting. TrAC Trends Anal. Chem. 2012, 33, 68–80. [Google Scholar] [CrossRef]
- Resmini, M. Molecularly imprinted polymers as biomimetic catalysts. Anal. Bioanal. Chem. 2012, 402, 3021–3026. [Google Scholar] [CrossRef]
- Lahcen, A.A.; Amine, A. Recent Advances in Electrochemical Sensors Based on Molecularly Imprinted Polymers and Nanomaterials. Electroanalysis 2019, 31, 188–201. [Google Scholar] [CrossRef]
- Zhong, C.; Yang, B.; Jiang, X.; Li, J. Current Progress of Nanomaterials in Molecularly Imprinted Electrochemical Sensing. Crit. Rev. Anal. Chem. 2018, 48, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Florea, A.; Jiang, M.; Mei, Y.; Zhang, W.; Zhang, A.; Săndulescu, R.; Jaffrezic-Renault, N. Molecularly Imprinted Polymer/Metal Organic Framework Based Chemical Sensors. Coatings 2016, 6, 42. [Google Scholar] [CrossRef]
- Anik, Ü.; Timur, S.; Dursun, Z. Metal organic frameworks in electrochemical and optical sensing platforms: A review. Microchim. Acta 2019, 186, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Yarman, A.; Turner, A.P.F.; Scheller, F.W. Electropolymers for (Nano-)Imprinted Biomimetic Biosensors; Woodhead Publishing Limited: Cambridge, UK, 2014; ISBN 9780857096609. [Google Scholar]
- Branger, C.; Brisset, H. Advanced Electrochemical Molecularly Imprinted Polymer as Sensor Interfaces. Proceedings 2019, 15, 22. [Google Scholar] [CrossRef]
- Kamel, A.H.; Jiang, X.; Li, P.; Liang, R. A paper-based potentiometric sensing platform based on molecularly imprinted nanobeads for determination of bisphenol A. Anal. Methods 2018, 10, 3890–3895. [Google Scholar] [CrossRef]
- Refaat, D.; Aggour, M.G.; Farghali, A.A.; Mahajan, R.; Wiklander, J.G.; Nicholls, I.A.; Piletsky, S.A. Strategies for molecular imprinting and the evolution of MIP nanoparticles as plastic antibodies—Synthesis and applications. Int. J. Mol. Sci. 2019, 20, 304. [Google Scholar] [CrossRef] [PubMed]
- Bokeloh, F.; Ayela, C.; Haupt, K. CHAPTER 6. Micro and Nanofabrication of Molecularly Imprinted Polymers. In Molecularly Imprinted Polymers for Analytical Chemistry Applications; Wlodzimierz, K., Sharma, P.S., Eds.; The Royal Society of Chemistry: London, UK, 2018; pp. 167–196. [Google Scholar]
- Sharma, P.S.; Pietrzyk-Le, A.; D’Souza, F.; Kutner, W. Electrochemically synthesized polymers in molecular imprinting for chemical sensing. Anal. Bioanal. Chem. 2012, 402, 3177–3204. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288. [Google Scholar] [CrossRef]
- Amatatongchai, M.; Sitanurak, J.; Sroysee, W.; Sodanat, S.; Chairam, S.; Jarujamrus, P.; Nacapricha, D.; Lieberzeit, P.A. Highly sensitive and selective electrochemical paper-based device using a graphite screen-printed electrode modified with molecularly imprinted polymers coated Fe3O4@Au@SiO2 for serotonin determination. Anal. Chim. Acta 2019, 1077, 255–265. [Google Scholar] [CrossRef]
- Karaseva, N.A.; Pluhar, B.; Beliaeva, E.A.; Ermolaeva, T.N.; Mizaikoff, B. Synthesis and application of molecularly imprinted polymers for trypsin piezoelectric sensors. Sens. Actuators B Chem. 2019, 280, 272–279. [Google Scholar] [CrossRef]
- Lowdon, J.W.; Eersels, K.; Rogosic, R.; Boonen, T.; Heidt, B.; Diliën, H.; van Grinsven, B.; Cleij, T.J. Surface grafted molecularly imprinted polymeric receptor layers for thermal detection of the New Psychoactive substance 2-methoxphenidine. Sens. Actuators A Phys. 2019, 295, 586–595. [Google Scholar] [CrossRef]
- Palladino, P.; Minunni, M.; Scarano, S. Cardiac Troponin T capture and detection in real-time via epitope-imprinted polymer and optical biosensing. Biosens. Bioelectron. 2018, 106, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Hsu, C.Y.; Thomas, J.L.; Wang, S.E.; Chen, H.C.; Chou, T.C. The microcontact imprinting of proteins: The effect of cross-linking monomers for lysozyme, ribonuclease A and myoglobin. Biosens. Bioelectron. 2006, 22, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Crapnell, R.D.; Hudson, A.; Foster, C.W.; Eersels, K.; van Grinsven, B.; Cleij, T.J.; Banks, C.E.; Peeters, M. Recent advances in electrosynthesized molecularly imprinted polymer sensing platforms for bioanalyte detection. Sensors 2019, 19, 1204. [Google Scholar] [CrossRef] [PubMed]
- Malitesta, C.; Mazzotta, E.; Picca, R.A.; Poma, A.; Chianella, I.; Piletsky, S.A. MIP sensors-The electrochemical approach. Anal. Bioanal. Chem. 2012, 402, 1827–1846. [Google Scholar] [CrossRef] [PubMed]
- Saylan, Y.; Akgönüllü, S.; Yavuz, H.; Ünal, S.; Denizli, A. Molecularly imprinted polymer based sensors for medical applications. Sensors 2019, 19, 1279. [Google Scholar] [CrossRef] [PubMed]
- Belbruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef]
- Ahmad, O.S.; Bedwell, T.S.; Esen, C.; Garcia-Cruz, A.; Piletsky, S.A. Molecularly Imprinted Polymers in Electrochemical and Optical Sensors. Trends Biotechnol. 2019, 37, 294–309. [Google Scholar] [CrossRef]
- Boysen, R.I.; Schwarz, L.J.; Nicolau, D.V.; Hearn, M.T.W. Molecularly imprinted polymer membranes and thin films for the separation and sensing of biomacromolecules. J. Sep. Sci. 2017, 40, 314–335. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Capacitive biosensors and molecularly imprinted electrodes. Sensors 2017, 17, 390. [Google Scholar] [CrossRef]
- Van Grinsven, B.; Betlem, K.; Cleij, T.J.; Banks, C.E.; Peeters, M. Evaluating the potential of thermal read-out techniques combined with molecularly imprinted polymers for the sensing of low-weight organic molecules. J. Mol. Recognit. 2017, 30, 2563. [Google Scholar] [CrossRef] [PubMed]
- Rico-Yuste, A.; Carrasco, S. Molecularly imprinted polymer-based hybrid materials for the development of optical sensors. Polymers 2019, 11, 1173. [Google Scholar] [CrossRef] [PubMed]
- Gui, R.; Jin, H. Recent advances in synthetic methods and applications of photo-luminescent molecularly imprinted polymers. J. Photochem. Photobiol. C Photochem. Rev. 2019, 41, 100315. [Google Scholar] [CrossRef]
- Erdossy, J.; Horváth, V.; Yarman, A.; Scheller, F.W.; Gyurcsányi, R.E. Electrosynthesized molecularly imprinted polymers for protein recognition. TrAC Trends Anal. Chem. 2016, 79, 179–190. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Boonsriwong, W.; Lieberzeit, P.A. Biomimetic sensors targeting oxidized-low-density lipoprotein with molecularly imprinted polymers. Anal. Chim. Acta 2020, 1116, 27–35. [Google Scholar] [CrossRef]
- Bartold, K.; Pietrzyk-Le, A.; Lisowski, W.; Golebiewska, K.; Siklitskaya, A.; Borowicz, P.; Shao, S.; D’Souza, F.; Kutner, W. Promoting bioanalytical concepts in genetics: A TATA box molecularly imprinted polymer as a small isolated fragment of the DNA damage repairing system. Mater. Sci. Eng. C 2019, 100, 1–10. [Google Scholar] [CrossRef]
- Sönmezler, M.; Özgür, E.; Yavuz, H.; Denizli, A. Quartz crystal microbalance based histidine sensor. Artif. Cells Nanomed. Biotechnol. 2019, 47, 221–227. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Canfarotta, F.; Czulak, J.; Johnson, R.; Betlem, K.; Mecozzi, F.; Down, M.P.; Eersels, K.; Van Grinsven, B.; Cleij, T.J.; et al. Thermal Detection of Cardiac Biomarkers Heart-Fatty Acid Binding Protein and ST2 Using a Molecularly Imprinted Nanoparticle-Based Multiplex Sensor Platform. ACS Sens. 2019, 4, 2838–2845. [Google Scholar] [CrossRef]
- Rajkumar, R.; Katterle, M.; Warsinke, A.; Möhwald, H.; Scheller, F.W. Thermometric MIP sensor for fructosyl valine. Biosens. Bioelectron. 2008, 23, 1195–1199. [Google Scholar] [CrossRef]
- Okan, M.; Sari, E.; Duman, M. Molecularly imprinted polymer based micromechanical cantilever sensor system for the selective determination of ciprofloxacin. Biosens. Bioelectron. 2017, 88, 258–264. [Google Scholar] [CrossRef]
- El Kirat, K.; Bartkowski, M.; Haupt, K. Probing the recognition specificity of a protein molecularly imprinted polymer using force spectroscopy. Biosens. Bioelectron. 2009, 24, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Iskierko, Z.; Sharma, P.S.; Bartold, K.; Pietrzyk-Le, A.; Noworyta, K.; Kutner, W. Molecularly imprinted polymers for separating and sensing of macromolecular compounds and microorganisms. Biotechnol. Adv. 2016, 34, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Bosserdt, M.; Gajovic-Eichelman, N.; Scheller, F.W. Modulation of direct electron transfer of cytochrome c by use of a molecularly imprinted thin film. Anal. Bioanal. Chem. 2013, 405, 6437–6444. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, J.; Wang, X.; Peng, H.; Xiong, H.; Chen, L. Strategies of molecular imprinting-based fluorescence sensors for chemical and biological analysis. Biosens. Bioelectron. 2018, 112, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.T.C.; Sharma, S.; Dutra, R.A.F.; Noronha, J.P.C.; Cass, A.E.G.; Sales, M.G.F. Protein-responsive polymers for point-of-care detection of cardiac biomarker. Sens. Actuators B Chem. 2014, 196, 123–132. [Google Scholar] [CrossRef]
- Zhang, X.; Yarman, A.; Erdossy, J.; Katz, S.; Zebger, I.; Jetzschmann, K.J.; Altintas, Z.; Wollenberger, U.; Gyurcsányi, R.E.; Scheller, F.W. Electrosynthesized MIPs for transferrin: Plastibodies or nano-filters? Biosens. Bioelectron. 2018, 105, 29–35. [Google Scholar] [CrossRef]
- Saylan, Y.; Denizli, A. Molecular fingerprints of hemoglobin on a nanofilm chip. Sensors 2018, 18, 3016. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Singsanan, S.; Lieberzeit, P.A. Sensing array based on molecularly imprinted polymers for simultaneous assessment of lipoproteins. Sens. Actuators B Chem. 2019, 298, 126828. [Google Scholar] [CrossRef]
- Bosserdt, M.; Erdossy, J.; Lautner, G.; Witt, J.; Köhler, K.; Gajovic-Eichelmann, N.; Yarman, A.; Wittstock, G.; Scheller, F.W.; Gyurcsányi, R.E. Microelectrospotting as a new method for electrosynthesis of surface-imprinted polymer microarrays for protein recognition. Biosens. Bioelectron. 2015, 73, 123–129. [Google Scholar] [CrossRef]
- Clark, L.C.; Lyons, C. Electrode Systems for Continuous Monitoring in Cardiovascular Surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef]
- Warsinke, A.; Benkert, A.; Scheller, F.W. Electrochemical immunoassays. Fresenius. J. Anal. Chem. 2000, 366, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Ghindilis, A.L.; Smith, M.W.; Schwarzkopf, K.R.; Roth, K.M.; Peyvan, K.; Munro, S.B.; Lodes, M.J.; Stöver, A.G.; Bernards, K.; Dill, K.; et al. CombiMatrix oligonucleotide arrays: Genotyping and gene expression assays employing electrochemical detection. Biosens. Bioelectron. 2007, 22, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Menger, M.; Yarman, A.; Erdossy, J.; Yildiz, H.B.; Gyurcsányi, R.E.; Scheller, F.W. MIPs and aptamers for recognition of proteins in biomimetic sensing. Biosensors 2016, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Hedborg, E.; Winquist, F.; Lundström, I.; Andersson, L.I.; Mosbach, K. Some studies of molecularly-imprinted polymer membranes in combination with field-effect devices. Sens. Actuators A Phys. 1993, 37–38, 796–799. [Google Scholar] [CrossRef]
- Iskierko, Z.; Sharma, P.S.; Noworyta, K.R.; Borowicz, P.; Cieplak, M.; Kutner, W.; Bossi, A.M. Selective PQQPFPQQ Gluten Epitope Chemical Sensor with a Molecularly Imprinted Polymer Recognition Unit and an Extended-Gate Field-Effect Transistor Transduction Unit. Anal. Chem. 2019, 91, 4537–4543. [Google Scholar] [CrossRef] [PubMed]
- Frasco, M.F.; Truta, L.A.A.N.A.; Sales, M.G.F.; Moreira, F.T.C. Imprinting technology in electrochemical biomimetic sensors. Sensors 2017, 17, 523. [Google Scholar] [CrossRef]
- Yarman, A.; Scheller, F.W. The first electrochemical MIP sensor for tamoxifen. Sensors 2014, 14, 7647–7654. [Google Scholar] [CrossRef]
- Özcan, L.; Şahin, Y. Determination of paracetamol based on electropolymerized-molecularly imprinted polypyrrole modified pencil graphite electrode. Sens. Actuators B Chem. 2007, 127, 362–369. [Google Scholar] [CrossRef]
- Lin, L.; Lian, H.-T.; Sun, X.-Y.; Yu, Y.-M.; Liu, B. An L-dopa electrochemical sensor based on a graphene doped molecularly imprinted chitosan film. Anal. Methods 2015, 7, 1387–1394. [Google Scholar] [CrossRef]
- Moro, G.; Bottari, F.; Sleegers, N.; Florea, A.; Cowen, T.; Moretto, L.M.; Piletsky, S.; De Wael, K. Conductive imprinted polymers for the direct electrochemical detection of β-lactam antibiotics: The case of cefquinome. Sens. Actuators B Chem. 2019, 297, 126786. [Google Scholar] [CrossRef]
- Zembrzuska, D.; Kalecki, J.; Cieplak, M.; Lisowski, W.; Borowicz, P.; Noworyta, K.; Sharma, P.S. Electrochemically initiated co-polymerization of monomers of different oxidation potentials for molecular imprinting of electroactive analyte. Sens. Actuators B Chem. 2019, 298, 126884. [Google Scholar] [CrossRef]
- Ghodsi, J.; Rafati, A.A. A novel molecularly imprinted sensor for imidacloprid pesticide based on poly(levodopa) electro-polymerized/TiO2 nanoparticles composite. Anal. Bioanal. Chem. 2018, 410, 7621–7633. [Google Scholar] [CrossRef] [PubMed]
- Khadem, M.; Faridbod, F.; Norouzi, P.; Foroushani, A.R.; Ganjali, M.R.; Shahtaheri, S.J. Biomimetic electrochemical sensor based on molecularly imprinted polymer for dicloran pesticide determination in biological and environmental samples. J. Iran. Chem. Soc. 2016, 13, 2077–2084. [Google Scholar] [CrossRef]
- Pacheco, J.G.; Castro, M.; Machado, S.; Barroso, M.F.; Nouws, H.P.A.; Delerue-Matos, C. Molecularly imprinted electrochemical sensor for ochratoxin A detection in food samples. Sens. Actuators B Chem. 2015, 215, 107–112. [Google Scholar] [CrossRef]
- Couto, R.A.S.; Costa, S.S.; Mounssef, B.; Pacheco, J.G.; Fernandes, E.; Carvalho, F.; Rodrigues, C.M.P.; Delerue-Matos, C.; Braga, A.A.C.; Moreira Gonçalves, L.; et al. Electrochemical sensing of ecstasy with electropolymerized molecularly imprinted poly(o-phenylenediamine) polymer on the surface of disposable screen-printed carbon electrodes. Sens. Actuators B Chem. 2019, 290, 378–386. [Google Scholar] [CrossRef]
- Kriz, D.; Mosbach, K. Competitive amperometric morphine sensor based on an agarose immobilised molecularly imprinted polymer. Anal. Chim. Acta 1995, 300, 71–75. [Google Scholar] [CrossRef]
- Li, J.; Xu, Z.; Liu, M.; Deng, P.; Tang, S.; Jiang, J.; Feng, H.; Qian, D.; He, L. Ag/N-doped reduced graphene oxide incorporated with molecularly imprinted polymer: An advanced electrochemical sensing platform for salbutamol determination. Biosens. Bioelectron. 2017, 90, 210–216. [Google Scholar] [CrossRef]
- Rohani Moghadam, M.; Salehi, L.; Jafari, S.; Nasirizadeh, N.; Ghasemi, J. Voltammetric sensing of oxacillin by using a screen-printed electrode modified with molecularly imprinted polyaniline, gold nanourchins and graphene oxide. Microchim. Acta 2019, 186, 798. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, M.; Liu, W.; Yu, S.; Niu, L.; Li, G.; Li, H.; Liu, W. Electrochemical sensor based on molecularly imprinted polymer/reduced graphene oxide composite for simultaneous determination of uric acid and tyrosine. J. Electroanal. Chem. 2018, 813, 75–82. [Google Scholar] [CrossRef]
- Lu, Z.; Li, Y.; Liu, T.; Wang, G.; Sun, M.; Jiang, Y.; He, H.; Wang, Y.; Zou, P.; Wang, X.; et al. A dual-template imprinted polymer electrochemical sensor based on AuNPs and nitrogen-doped graphene oxide quantum dots coated on NiS2/biomass carbon for simultaneous determination of dopamine and chlorpromazine. Chem. Eng. J. 2020, 389, 124417. [Google Scholar] [CrossRef]
- Rawool, C.R.; Srivastava, A.K. A dual template imprinted polymer modified electrochemical sensor based on Cu metal organic framework/mesoporous carbon for highly sensitive and selective recognition of rifampicin and isoniazid. Sens. Actuators B Chem. 2019, 288, 493–506. [Google Scholar] [CrossRef]
- Yarman, A.; Scheller, F.W. Coupling biocatalysis with molecular imprinting in a biomimetic sensor. Angew. Chem. Int. Ed. 2013, 52, 11521–11525. [Google Scholar] [CrossRef] [PubMed]
- Yarman, A.; Scheller, F.W. MIP-esterase/Tyrosinase Combinations for Paracetamol and Phenacetin. Electroanalysis 2016, 28, 2222–2227. [Google Scholar] [CrossRef]
- Mazurenko, I.; Hitaishi, V.P.; Lojou, E. Recent advances in surface chemistry of electrodes to promote direct enzymatic bioelectrocatalysis. Curr. Opin. Electrochem. 2020, 19, 113–121. [Google Scholar] [CrossRef]
- Peng, L.; Yarman, A.; Jetzschmann, K.J.; Jeoung, J.H.; Schad, D.; Dobbek, H.; Wollenberger, U.; Scheller, F.W. Molecularly imprinted electropolymer for a hexameric heme protein with direct electron transfer and peroxide electrocatalysis. Sensors 2016, 16, 272. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Ni, X.; Cao, Y.; Cao, G. Electrochemical sensor based on magnetic molecularly imprinted nanoparticles modified magnetic electrode for determination of Hb. Biosens. Bioelectron. 2017, 91, 354–358. [Google Scholar] [CrossRef]
- Pandey, I.; Tiwari, J.D. A novel dual imprinted conducting nanocubes based flexible sensor for simultaneous detection of hemoglobin and glycated haemoglobin in gestational diabetes mellitus patients. Sens. Actuators B Chem. 2019, 285, 470–478. [Google Scholar] [CrossRef]
- Ertürk, G.; Hedström, M.; Mattiasson, B. A sensitive and real-time assay of trypsin by using molecular imprinting-based capacitive biosensor. Biosens. Bioelectron. 2016, 86, 557–565. [Google Scholar] [CrossRef]
- Bossi, A.; Piletsky, S.A.; Piletska, E.V.; Righetti, P.G.; Turner, A.P.F. Surface-grafted molecularly imprinted polymers for protein recognition. Anal. Chem. 2001, 73, 5281–5286. [Google Scholar] [CrossRef]
- Jetzschmann, K.J.; Yarman, A.; Rustam, L.; Kielb, P.; Urlacher, V.B.; Fischer, A.; Weidinger, I.M.; Wollenberger, U.; Scheller, F.W. Molecular LEGO by domain-imprinting of cytochrome P450 BM3. Colloids Surf. B Biointerfaces 2018, 164, 240–246. [Google Scholar] [CrossRef]
- Jetzschmann, K.J.; Jágerszki, G.; Dechtrirat, D.; Yarman, A.; Gajovic-Eichelmann, N.; Gilsing, H.D.; Schulz, B.; Gyurcsányi, R.E.; Scheller, F.W. Vectorially Imprinted Hybrid Nanofilm for Acetylcholinesterase Recognition. Adv. Funct. Mater. 2015, 25, 5178–5183. [Google Scholar] [CrossRef]
- Yarman, A. Electrosynthesized Molecularly Imprinted Polymer for Laccase Using the Inactivated Enzyme as the Target. Bull. Korean Chem. Soc. 2018, 39, 483–488. [Google Scholar] [CrossRef]
- Yarman, A. Development of a molecularly imprinted polymer-based electrochemical sensor for tyrosinase. Turkish J. Chem. 2018, 42, 346–354. [Google Scholar] [CrossRef]
- Ozcelikay, G.; Kurbanoglu, S.; Zhang, X.; Soz, C.K.; Wollenberger, U.; Ozkan, S.A.; Yarman, A.; Scheller, F.W. Electrochemical MIP sensor for butyrylcholinesterase. Polymers 2019, 11, 1970. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Bai, C.; Teng, Y.; Zhang, J.; Peng, J.; Fang, Z.; Xu, W. Study of horseradish peroxidase and hydrogen peroxide bi-analyte sensor with boronate affinity-based molecularly imprinted film. Can. J. Chem. 2019, 97, 833–839. [Google Scholar] [CrossRef]
- Reddy, S.M.; Sette, G.; Phan, Q. Electrochemical probing of selective haemoglobin binding in hydrogel-based molecularly imprinted polymers. Electrochim. Acta 2011, 56, 9203–9208. [Google Scholar] [CrossRef]
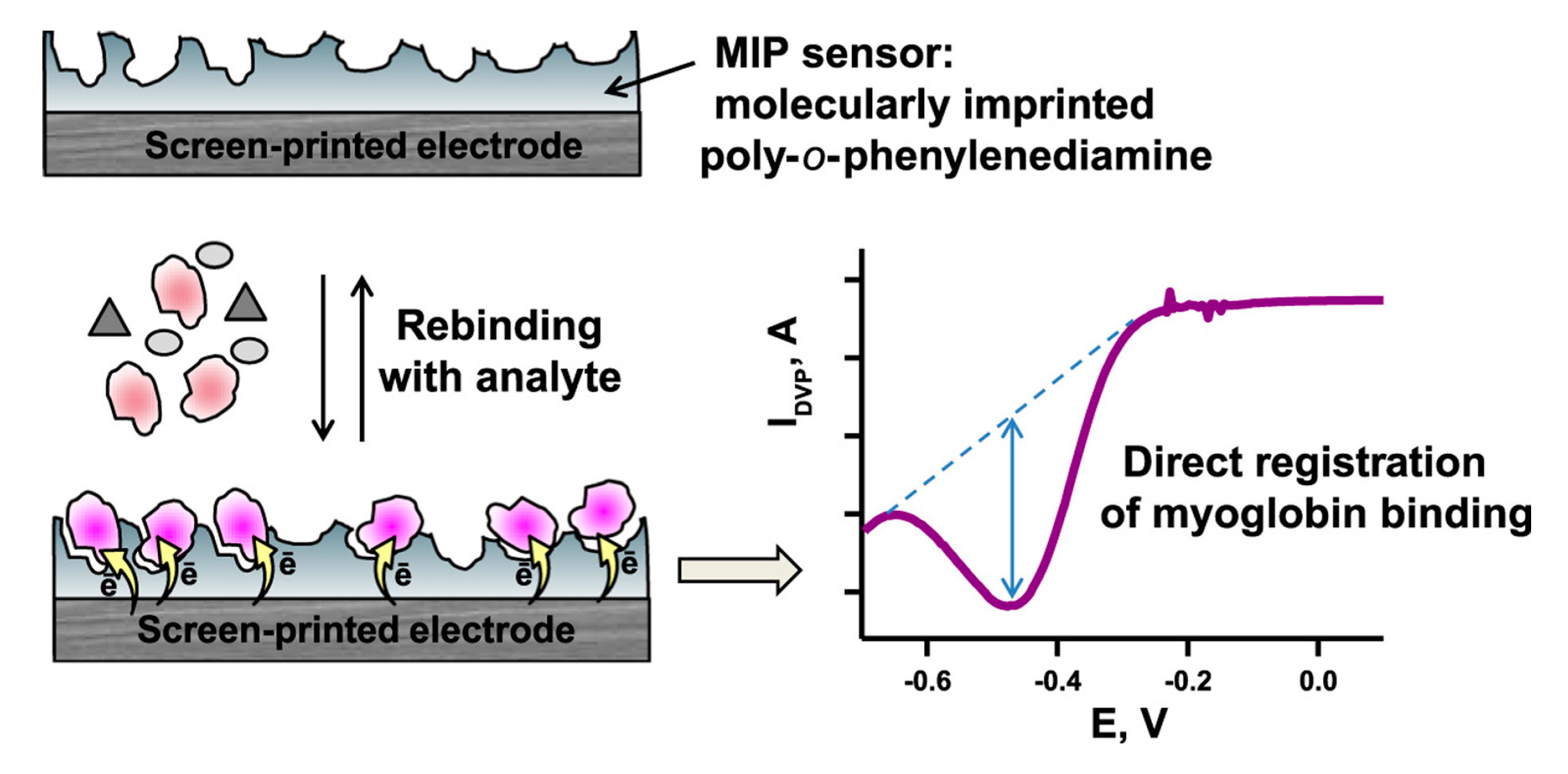
- Shumyantseva, V.V.; Bulko, T.V.; Sigolaeva, L.V.; Kuzikov, A.V.; Archakov, A.I. Electrosynthesis and binding properties of molecularly imprinted poly-o-phenylenediamine for selective recognition and direct electrochemical detection of myoglobin. Biosens. Bioelectron. 2016, 86, 330–336. [Google Scholar] [CrossRef]
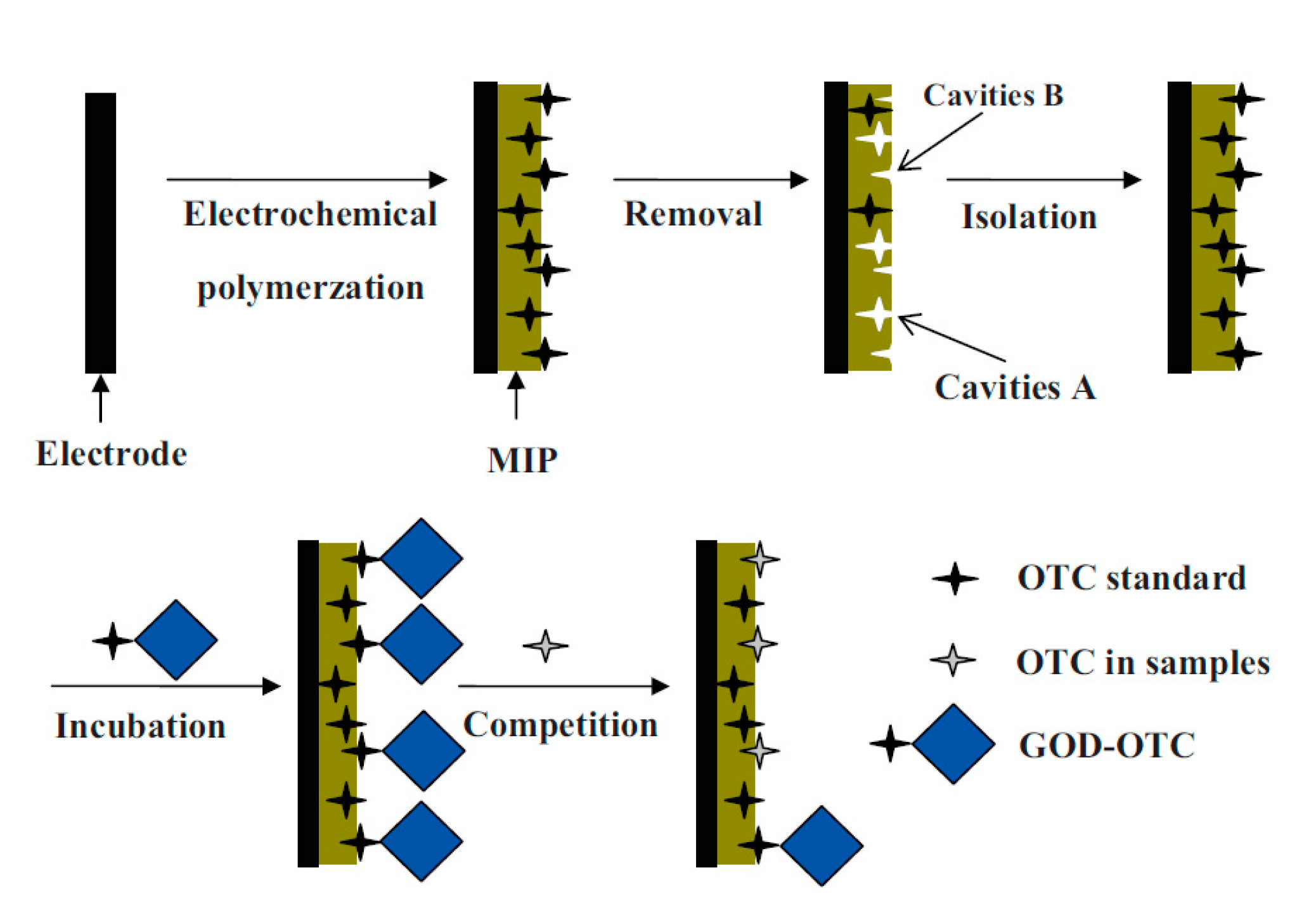
- Li, J.; Jiang, F.; Wei, X. Molecularly imprinted sensor based on an enzyme amplifier for ultratrace oxytetracycline determination. Anal. Chem. 2010, 82, 6074–6078. [Google Scholar] [CrossRef]
- Li, J.; Jiang, F.; Li, Y.; Chen, Z. Fabrication of an oxytetracycline molecular-imprinted sensor based on the competition reaction via a GOD-enzymatic amplifier. Biosens. Bioelectron. 2011, 26, 2097–2101. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Zhang, Y.; Wei, G. Highly sensitive molecularly imprinted electrochemical sensor based on the double amplification by an inorganic prussian blue catalytic polymer and the enzymatic effect of glucose oxidase. Anal. Chem. 2012, 84, 1888–1893. [Google Scholar] [CrossRef]
- Karyakin, A.A.; Karyakina, E.E.; Gorton, L. On the mechanism of H2O2 reduction at Prussian Blue modified electrodes. Electrochem. Commun. 1999, 1, 78–82. [Google Scholar] [CrossRef]
- Que, X.; Liu, B.; Fu, L.; Zhuang, J.; Chen, G.; Tang, D. Molecular Imprint for Electrochemical Detection of Streptomycin Residues Using Enzyme Signal Amplification. Electroanalysis 2013, 25, 531–537. [Google Scholar] [CrossRef]
- Wulff, G. Enzyme-like catalysis by molecularly imprinted polymers. Chem. Rev. 2002, 102, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Garcia Lopez, J.; Piletska, E.V.; Whitcombe, M.J.; Czulak, J.; Piletsky, S.A. Application of molecularly imprinted polymer nanoparticles for degradation of the bacterial autoinducer: N-hexanoyl homoserine lactone. Chem. Commun. 2019, 55, 2664–2667. [Google Scholar] [CrossRef] [PubMed]
- Robinsona, D.K.; Mosbachb, K. Molecular Imprinting of a Transition State Analogue Leads to a Polymer Exhibiting Esterolytic Activity. J. Chem. Soc. Chem. Commun. 1989, 969–970. [Google Scholar] [CrossRef]
- Mathew, D.; Thomas, B.; Devaky, K.S. Phosphonate TSA-Built Macromatric Polymer Catalysts as Chymotrypsin Mimics for the Amidolysis of Amino Acid P-Nitroanilides: Effect of the Nature and Extent of Crosslinker on Amidase Activities; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; Volume 111, ISBN 9148127310. [Google Scholar]
- Mirata, F.; Resmini, M. Metal Complexes and Imprinted Polymers for Shape-Selective Catalysis. In Effects of Nanoconfinement on Catalysis; Springer: Berlin, Germany, 2017; pp. 83–104. [Google Scholar]
- Lakshmi, D.; Bossi, A.; Whitcombe, M.J.; Chianella, I.; Fowler, S.A.; Subrahmanyam, S.; Piletska, E.V.; Piletsky, S.A. Electrochemical sensor for catechol and dopamine based on a catalytic molecularly imprinted polymer-conducting polymer hybrid recognition element. Anal. Chem. 2009, 81, 3576–3584. [Google Scholar] [CrossRef]
- Huang, X.; Yin, Y.; Liu, Y.; Bai, X.; Zhang, Z.; Xu, J.; Shen, J.; Liu, J. Incorporation of glutathione peroxidase active site into polymer based on imprinting strategy. Biosens. Bioelectron. 2009, 25, 657–660. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, L.; Li, Y. Synthesis of an enzyme-like imprinted polymer with the substrate as the template, and its catalytic properties under aqueous conditions. Chem. A Eur. J. 2004, 10, 3555–3561. [Google Scholar] [CrossRef]
- De Jesus Rodrigues Santos, W.; Lima, P.R.; Tarley, C.R.T.; Kubota, L.T. A catalytically active molecularly imprinted polymer that mimics peroxidase based on hemin: Application to the determination of p-aminophenol. Anal. Bioanal. Chem. 2007, 389, 1919–1929. [Google Scholar] [CrossRef]
- De Santos, W.J.R.; Lima, P.R.; Tarley, C.R.T.; Höehr, N.F.; Kubota, L.T. Synthesis and application of a peroxidase-like molecularly imprinted polymer based on hemin for selective determination of serotonin in blood serum. Anal. Chim. Acta 2009, 631, 170–176. [Google Scholar] [CrossRef]
- Sartori, L.R.; Santos, W.D.J.R.; Kubota, L.T.; Segatelli, M.G.; Tarley, C.R.T. Flow-based method for epinephrine determination using a solid reactor based on molecularly imprinted poly(FePP-MAA-EGDMA). Mater. Sci. Eng. C 2011, 31, 114–119. [Google Scholar] [CrossRef]
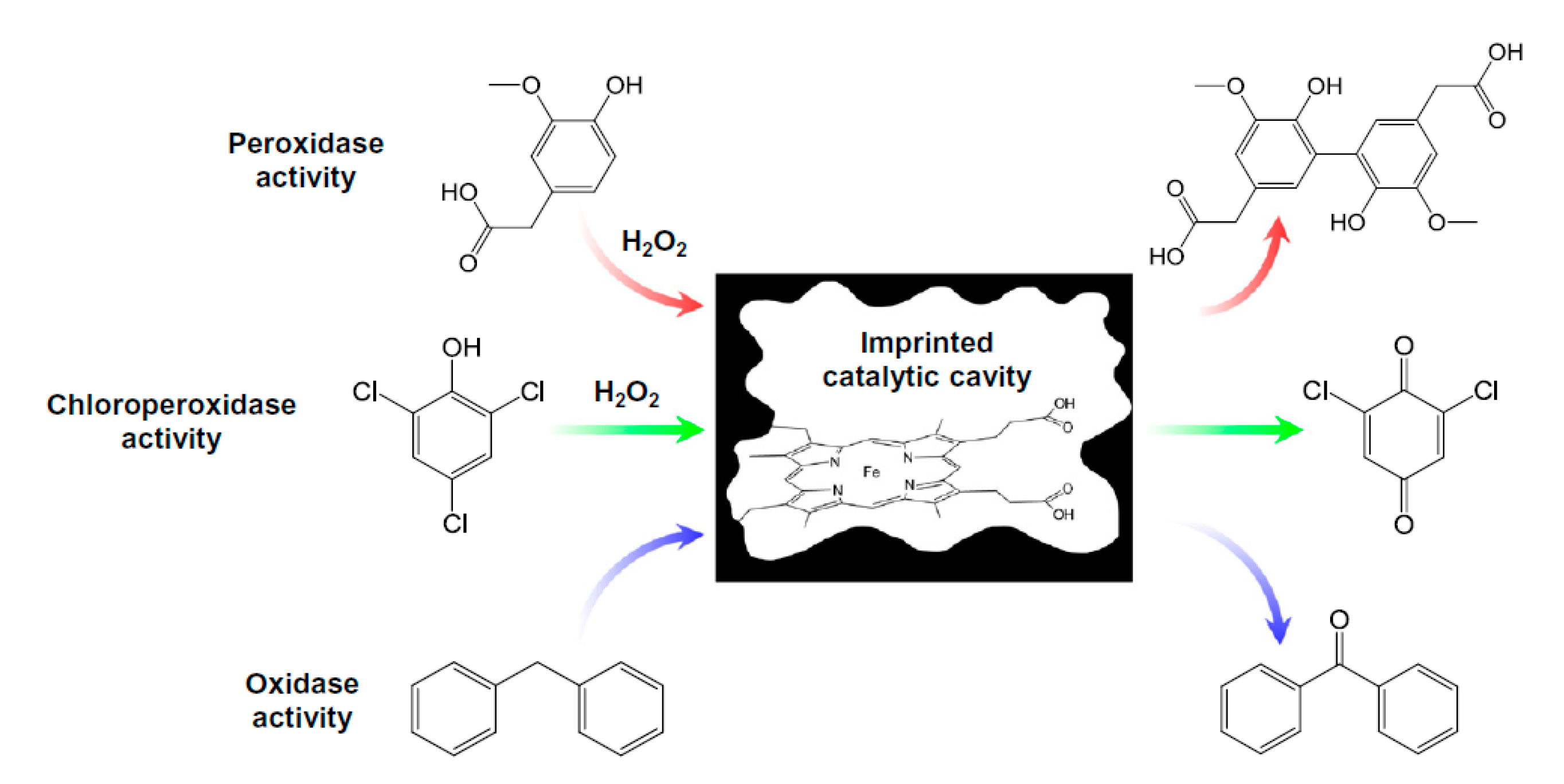
- Díaz-Díaz, G.; Diñeiro, Y.; Menéndez, M.I.; Blanco-López, M.C.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Molecularly imprinted catalytic polymers with biomimetic chloroperoxidase activity. Polymer (Guildf) 2011, 52, 2468–2473. [Google Scholar] [CrossRef]
- Zhang, J.; Lei, J.; Ju, H.; Wang, C. Electrochemical sensor based on chlorohemin modified molecularly imprinted microgel for determination of 2,4-dichlorophenol. Anal. Chim. Acta 2013, 786, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Antuña-Jiménez, D.; Blanco-López, M.C.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Artificial enzyme with magnetic properties and peroxidase activity on indoleamine metabolite tumor marker. Polymer (Guildf) 2014, 55, 1113–1119. [Google Scholar] [CrossRef]
- Sode, K.; Ohta, S.; Yanai, Y.; Yamazaki, T. Construction of a molecular imprinting catalyst using target analogue template and its application for an amperometric fructosylamine sensor. Biosens. Bioelectron. 2003, 18, 1485–1490. [Google Scholar] [CrossRef]
- Guo, L.; Zheng, H.; Zhang, C.; Qu, L.; Yu, L. A novel molecularly imprinted sensor based on PtCu bimetallic nanoparticle deposited on PSS functionalized graphene with peroxidase-like activity for selective determination of puerarin. Talanta 2020, 210, 120621. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Wei, X.; Li, J. A Sensitive and renewable chlortoluron molecularly imprinted polymer sensor based on the gate-controlled catalytic electrooxidation of H2O2 on Magnetic Nano-NiO. Electroanalysis 2013, 25, 1286–1293. [Google Scholar] [CrossRef]
- Zeng, L.; Cui, H.; Chao, J.; Huang, K.; Wang, X.; Zhou, Y.; Jing, T. Colorimetric determination of tetrabromobisphenol A based on enzyme-mimicking activity and molecular recognition of metal-organic framework-based molecularly imprinted polymers. Microchim. Acta 2020, 187, 142. [Google Scholar] [CrossRef]
- Yuan, Y.; Yang, Y.; Faheem, M.; Zou, X.; Ma, X.; Wang, Z.; Meng, Q.; Wang, L.; Zhao, S.; Zhu, G. Molecularly Imprinted Porous Aromatic Frameworks Serving as Porous Artificial Enzymes. Adv. Mater. 2018, 30, 1–9. [Google Scholar] [CrossRef]
- Lach, P.; Cieplak, M.; Majewska, M.; Noworyta, K.R.; Sharma, P.S.; Kutner, W. “gate Effect” in p-Synephrine Electrochemical Sensing with a Molecularly Imprinted Polymer and Redox Probes. Anal. Chem. 2019, 91, 7546–7553. [Google Scholar] [CrossRef]
- Cai, D.; Ren, L.; Zhao, H.; Xu, C.; Zhang, L.; Yu, Y.; Wang, H.; Lan, Y.; Roberts, M.F.; Chuang, J.H.; et al. A molecular-imprint nanosensor for ultrasensitive detection of proteins. Nat. Nanotechnol. 2010, 5, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Bozal-Palabiyik, B.; Lettieri, M.; Uslu, B.; Marrazza, G. Electrochemical Detection of Vascular Endothelial Growth Factor by Molecularly Imprinted Polymer. Electroanalysis 2019, 31, 1475–1481. [Google Scholar] [CrossRef]
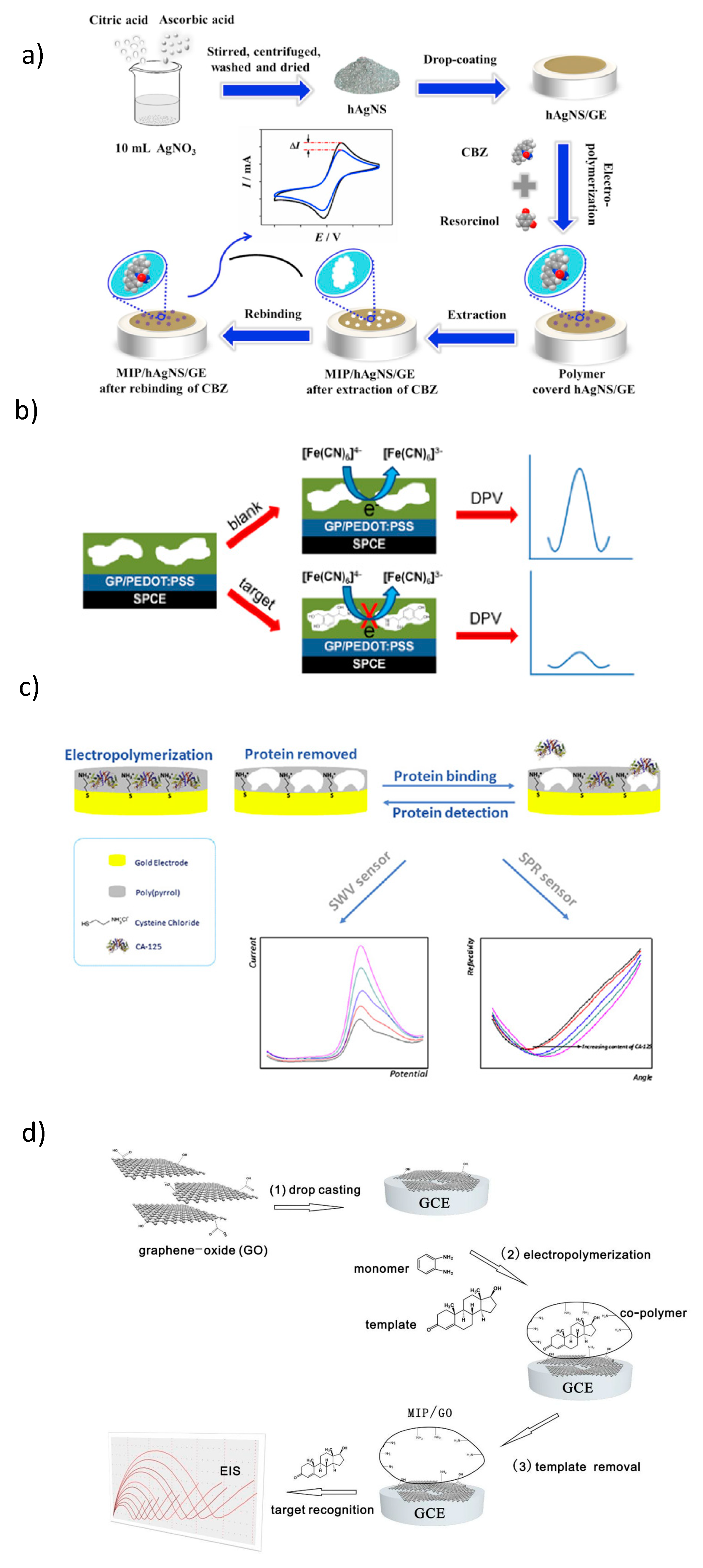
- Lee, M.H.; Thomas, J.L.; Liu, W.C.; Zhang, Z.X.; Liu, B.D.; Yang, C.H.; Lin, H.Y. A multichannel system integrating molecularly imprinted conductive polymers for ultrasensitive voltammetric determination of four steroid hormones in urine. Microchim. Acta 2019, 186, 695. [Google Scholar] [CrossRef] [PubMed]
- Dechtrirat, D.; Sookcharoenpinyo, B.; Prajongtat, P.; Sriprachuabwong, C.; Sanguankiat, A.; Tuantranont, A.; Hannongbua, S. An electrochemical MIP sensor for selective detection of salbutamol based on a graphene/PEDOT:PSS modified screen printed carbon electrode. RSC Adv. 2018, 8, 206–212. [Google Scholar] [CrossRef]
- Rebelo, T.S.C.R.; Costa, R.; Brandão, A.T.S.C.; Silva, A.F.; Sales, M.G.F.; Pereira, C.M. Molecularly imprinted polymer SPE sensor for analysis of CA-125 on serum. Anal. Chim. Acta 2019, 1082, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ma, Y.; Sun, G.; Wang, S.; Deng, J.; Wei, H. Molecularly imprinted polymers on graphene oxide surface for EIS sensing of testosterone. Biosens. Bioelectron. 2017, 92, 305–312. [Google Scholar] [CrossRef]
- Yoshimi, Y.; Ohdaira, R.; Iiyama, C.; Sakai, K. ‘Gate effect’ of thin layer of molecularly-imprinted poly(methacrylic acid-co-ethyleneglycol dimethacrylate). Sens. Actuators B Chem. 2001, 73, 49–53. [Google Scholar] [CrossRef]
- Sharma, P.S.; Garcia-Cruz, A.; Cieplak, M.; Noworyta, K.R.; Kutner, W. ‘Gate effect’ in molecularly imprinted polymers: The current state of understanding. Curr. Opin. Electrochem. 2019, 16, 50–56. [Google Scholar] [CrossRef]
- Jiang, M.; Braiek, M.; Florea, A.; Chrouda, A.; Farre, C.; Bonhomme, A.; Bessueille, F.; Vocanson, F.; Zhang, A.; Jaffrezic-Renault, N. Aflatoxin B1 detection using a highly-sensitive molecularly-imprinted electrochemical sensor based on an electropolymerized metal organic framework. Toxins 2015, 7, 3540–3553. [Google Scholar] [CrossRef]
- Duan, D.; Si, X.; Ding, Y.; Li, L.; Ma, G.; Zhang, L.; Jian, B. A novel molecularly imprinted electrochemical sensor based on double sensitization by MOF/CNTs and Prussian blue for detection of 17β-estradiol. Bioelectrochemistry 2019, 129, 211–217. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Ding, S.; Hong, C.; Wang, Z. A novel sensitive and selective electrochemical sensor based on integration of molecularly imprinted with hollow silver nanospheres for determination of carbamazepine. Microchem. J. 2019, 147, 191–197. [Google Scholar] [CrossRef]
- Lian, W.; Liu, S.; Wang, L.; Liu, H. A novel strategy to improve the sensitivity of antibiotics determination based on bioelectrocatalysis at molecularly imprinted polymer film electrodes. Biosens. Bioelectron. 2015, 73, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, B.; Boroujeni, M.K.; Ensafi, A.A. A novel electrochemical nanocomposite imprinted sensor for the determination of lorazepam based on modified polypyrrole@sol-gel@gold nanoparticles/pencil graphite electrode. Electrochim. Acta 2014, 123, 332–339. [Google Scholar] [CrossRef]
- Han, S.; Li, B.; Song, Z.; Pan, S.; Zhang, Z.; Yao, H.; Zhu, S.; Xu, G. A kanamycin sensor based on an electrosynthesized molecularly imprinted poly-o-phenylenediamine film on a single-walled carbon nanohorn modified glassy carbon electrode. Analyst 2017, 142, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, Y.; Yang, R.; Li, J.; Qu, L. A highly sensitive and selective electrochemical sensor based on polydopamine functionalized graphene and molecularly imprinted polymer for the 2,4-dichlorophenol recognition and detection. Talanta 2019, 195, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Waffo, A.F.T.; Yesildag, C.; Caserta, G.; Katz, S.; Zebger, I.; Lensen, M.C.; Wollenberger, U.; Scheller, F.W.; Altintas, Z. Fully electrochemical MIP sensor for artemisinin. Sens. Actuators B Chem. 2018, 275, 163–173. [Google Scholar] [CrossRef]
- Florea, A.; Guo, Z.; Cristea, C.; Bessueille, F.; Vocanson, F.; Goutaland, F.; Dzyadevych, S.; Săndulescu, R.; Jaffrezic-Renault, N. Anticancer drug detection using a highly sensitive molecularly imprinted electrochemical sensor based on an electropolymerized microporous metal organic framework. Talanta 2015, 138, 71–76. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; El-Wekil, M.M.; Mahnashi, M.H.; Ali, M.F.B.; Alkahtani, S.A. Modification of N,S co-doped graphene quantum dots with p-aminothiophenol-functionalized gold nanoparticles for molecular imprint-based voltammetric determination of the antiviral drug sofosbuvir. Microchim. Acta 2019, 186, 617. [Google Scholar] [CrossRef]
- Zhang, H.; Gui, Y.; Wang, M.L.B. Molecularly Imprinted Sensor based on o-phenylenediamine for Electrochemical Detection of Sulfamethoxazole. Int. J. Electrochem. Sci. 2019, 14, 11630–11640. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Zhang, Y.; Yu, F.; Wang, F.; Peng, Z.; Li, Y. Voltammetric lidocaine sensor by using a glassy carbon electrode modified with porous carbon prepared from a MOF, and with a molecularly imprinted polymer. Microchim. Acta 2018, 185, 3–10. [Google Scholar] [CrossRef]
- Ji, J.; Zhou, Z.; Zhao, X.; Sun, J.; Sun, X. Electrochemical sensor based on molecularly imprinted film at Au nanoparticles-carbon nanotubes modified electrode for determination of cholesterol. Biosens. Bioelectron. 2015, 66, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, J.G.; Rebelo, P.; Cagide, F.; Gonçalves, L.M.; Borges, F.; Rodrigues, J.A.; Delerue-Matos, C. Electrochemical sensing of the thyroid hormone thyronamine (T0AM) via molecular imprinted polymers (MIPs). Talanta 2019, 194, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, H.; Yu, S.; Zhang, J.; Zheng, W.; Niu, L.; Li, G. Poly(3,6-diamino-9-ethylcarbazole) based molecularly imprinted polymer sensor for ultra-sensitive and selective detection of 17-β-estradiol in biological fluids. Biosens. Bioelectron. 2018, 104, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Buffon, E.; Stradiotto, N.R. Electrochemical sensor based on molecularly imprinted poly(ortho-phenylenediamine) for determination of hexahydrofarnesol in aviation biokerosene. Sens. Actuators B Chem. 2019, 287, 371–379. [Google Scholar] [CrossRef]
- Roushani, M.; Rahmati, Z. Development of electrochemical sensor based on molecularly imprinted copolymer for detection of nitrofurantoin. J. Iran. Chem. Soc. 2019, 16, 999–1006. [Google Scholar] [CrossRef]
- Bolat, G.; Yaman, Y.T.; Abaci, S. Molecularly imprinted electrochemical impedance sensor for sensitive dibutyl phthalate (DBP) determination. Sens. Actuators B Chem. 2019, 299, 127000. [Google Scholar] [CrossRef]
- Uygun, Z.O.; Dilgin, Y. A novel impedimetric sensor based on molecularly imprinted polypyrrole modified pencil graphite electrode for trace level determination of chlorpyrifos. Sens. Actuators B Chem. 2013, 188, 78–84. [Google Scholar] [CrossRef]
- Stojanovic, Z.; Erdőssy, J.; Keltai, K.; Scheller, F.W.; Gyurcsányi, R.E. Electrosynthesized molecularly imprinted polyscopoletin nanofilms for human serum albumin detection. Anal. Chim. Acta 2017, 977, 1–9. [Google Scholar] [CrossRef]
- Cieplak, M.; Szwabinska, K.; Sosnowska, M.; Chandra, B.K.C.; Borowicz, P.; Noworyta, K.; D’Souza, F.; Kutner, W. Selective electrochemical sensing of human serum albumin by semi-covalent molecular imprinting. Biosens. Bioelectron. 2015, 74, 960–966. [Google Scholar] [CrossRef]
- Rebelo, T.S.C.R.; Pereira, C.M.; Sales, M.G.F.; Noronha, J.P.; Silva, F. Protein Imprinted Material electrochemical sensor for determination of Annexin A3 in biological samples. Electrochim. Acta 2016, 190, 887–893. [Google Scholar] [CrossRef]
- Yazdani, Z.; Yadegari, H.; Heli, H. A molecularly imprinted electrochemical nanobiosensor for prostate specific antigen determination. Anal. Biochem. 2019, 566, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.R.; de Sá, M.H.; Sales, M.G.F. An impedimetric molecularly-imprinted biosensor for Interleukin-1β determination, prepared by in-situ electropolymerization on carbon screen-printed electrodes. Bioelectrochemistry 2019, 130, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.R.; Moreira, F.T.C.; Riu, J.; Sales, M.G.F. Plastic antibody for the electrochemical detection of bacterial surface proteins. Sens. Actuators B Chem. 2016, 233, 697–704. [Google Scholar] [CrossRef]
- Pacheco, J.G.; Rebelo, P.; Freitas, M.; Nouws, H.P.A.; Delerue-Matos, C. Breast cancer biomarker (HER2-ECD) detection using a molecularly imprinted electrochemical sensor. Sens. Actuators B Chem. 2018, 273, 1008–1014. [Google Scholar] [CrossRef]
- Karimi, M.; Rabiee, M.; Tahriri, M.; Salarian, R.; Tayebi, L. A graphene based–biomimetic molecularly imprinted polyaniline sensor for ultrasensitive detection of human cardiac troponin T (cTnT). Synth. Met. 2019, 256, 116136. [Google Scholar] [CrossRef]


| Template | Monomer | Electrode | Detection Method | Measuring Range and LOD | Reference |
|---|---|---|---|---|---|
| Tamoxifen | o-PD/Resorcinol | GCE | CV | 1–100 nM | [60] |
| Lorazepam | Pyrrole | Sol-gel-AuNP-Graphite | SWV | 0.2–2 nM and 2–20 nM; LOD: 0.09 nM | [128] |
| Kanamycin | o-PD | SWCNH-GCE | LSV | 0.1–50 µM; LOD: 0.1 µM | [129] |
| 2,4-dichlorophenol | o-PD | PDA-rGO-GCE | DPV | 2–10 nM and 10–100 nM; LOD: 0.8 nM | [130] |
| p-Synephrine | Functionalized thiophene | Pt electrode | EIS | 0.1–0.99 μM; LOD: 5.69 nM | [115] |
| Artemisinin | o-PD | Au electrode | CV, SWV | 0.01–1.36 µM; LOD: 0.01 µM | [131] |
| Gemcitabine | PATP | AuNP-Au electrode | LSV | 3.8 fM–38 nM; LOD: 3 fM | [132] |
| Sofosbuvir | PATP | N,S@GQDs-AuNP-PGE | DPV | 0.1–40.0 × 10−8 M; LOD: 0.036 × 10−8 M | [133] |
| Sulfamethoxazole | o-PD | GCE | SWV | 0.2–1.4 µM; LOD: 0.05 µM | [134] |
| Lidocaine | Resorcinol | Porous C-GCE | CV | 0.2 pM–8 nM; LOD: 67 fM | [135] |
| Carbamazepine | Resorcinol | hAgNS-Au electrode | CV | 8.0 × 10−11–6.0 × 10−8 M; LOD: 3.2 × 10−11 M | [126] |
| Cholesterol | PATP | AuNP-MWCNT-GCE | DPV | 1 × 10−13–1 × 10−9 M; LOD: 3.3 × 10−14 M | [136] |
| Thyronamine | 4-ABA | Carbon-SPE | SWV | up to 10 µM; LOD: 0.081 µM | [137] |
| 17-β-estradiol | 3,6-diamino-9-ethylcarbazole | GCE | EIS | 1 aM–10 µM; LOD: 0.36 aM | [138] |
| Hexahydrofarnesol | o-PD | GCE | DPV | 4.0 × 10−8–1.5 × 10−7 M and 1.5 × 10−7–1.5 × 10−6 M; LOD: 1.2 × 10−8 M | [139] |
| Nitrofurantoin | m-Dihydroxy-benzene/o-Aminophenol | GCE | EIS | 0.001–0.05 µM and 0.1–1 µM; LOD: 0.3 nM | [140] |
| Dibutyl phthalate | Pyrrole | PGE | EIS | 0.01–1.0 μM; LOD: 4.5 nM | [141] |
| Chlorpyrifos | Pyrrole | PGE | EIS | 20–300 µg/L; LOD: 4.5 µg/L | [142] |
| Transferrin | Scopoletin | Au electrode | SWV | 0.1–1 µM | [49] |
| HSA | Scopoletin | Au electrode | CV | 20–100 mg/dm3; LOD: 3.7 mg/dm3 | [143] |
| HSA | Bithiophene derivatives | Au electrode | DPV | 12–300 pM; LOD: 0.25 pM | [144] |
| Ferritin | Phenol | Nanotube arrays | EIS | 1 × 10−12 × 10−7 g/L | [116] |
| Annexin A3 | Caffeic acid | Carbon-SPE | SWV | 0.1–200 ng/mL; LOD: 0.095 ng/mL | [145] |
| Tyrosinase | o-PD | GCE | DPV | up to 50 nM; LOD: 3.97 nM | [86] |
| PSA | Pyrrole | Au-SPE | DPV | 0.01–4 ng/mL LOD: 2 pg/mL | [146] |
| CA-125 | Pyrrole | Au-SPE | SWV | 0.01–500 U/mL; LOD: 0.01 U/mL | [120] |
| IL-1β | Eriochrome black T | EDOT-PATP-Carbon-SPE | EIS | 60 pM–600 nM LOD: 1.5 pM | [147] |
| Protein A | 3-aminophenol | SWCNT-SPE | EIS | LOD: 0.60 nM | [148] |
| HER2-ECD | Phenol | Au-SPE | DPV | 10–70 ng/mL; LOD: 1.6 ng/L | [149] |
| cTnT | Aniline/Carboxylated aniline | rGO/C-SPE | DPV | 0.02–0.09 ng/mL; LOD: 0.008 ng/mL | [150] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yarman, A.; Scheller, F.W. How Reliable Is the Electrochemical Readout of MIP Sensors? Sensors 2020, 20, 2677. https://doi.org/10.3390/s20092677
Yarman A, Scheller FW. How Reliable Is the Electrochemical Readout of MIP Sensors? Sensors. 2020; 20(9):2677. https://doi.org/10.3390/s20092677
Chicago/Turabian StyleYarman, Aysu, and Frieder W. Scheller. 2020. "How Reliable Is the Electrochemical Readout of MIP Sensors?" Sensors 20, no. 9: 2677. https://doi.org/10.3390/s20092677
APA StyleYarman, A., & Scheller, F. W. (2020). How Reliable Is the Electrochemical Readout of MIP Sensors? Sensors, 20(9), 2677. https://doi.org/10.3390/s20092677