Recognition of Patient Groups with Sleep Related Disorders using Bio-signal Processing and Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. EMG Analysis
2.1.1. EMG Features from Moments
2.1.2. EMG Features from Entropy Measurement
2.2. ECG Analysis
2.2.1. Synchrosqueezed Wavelet Transform
2.2.2. Inverse Synchrosqueezed Wavelet Transform
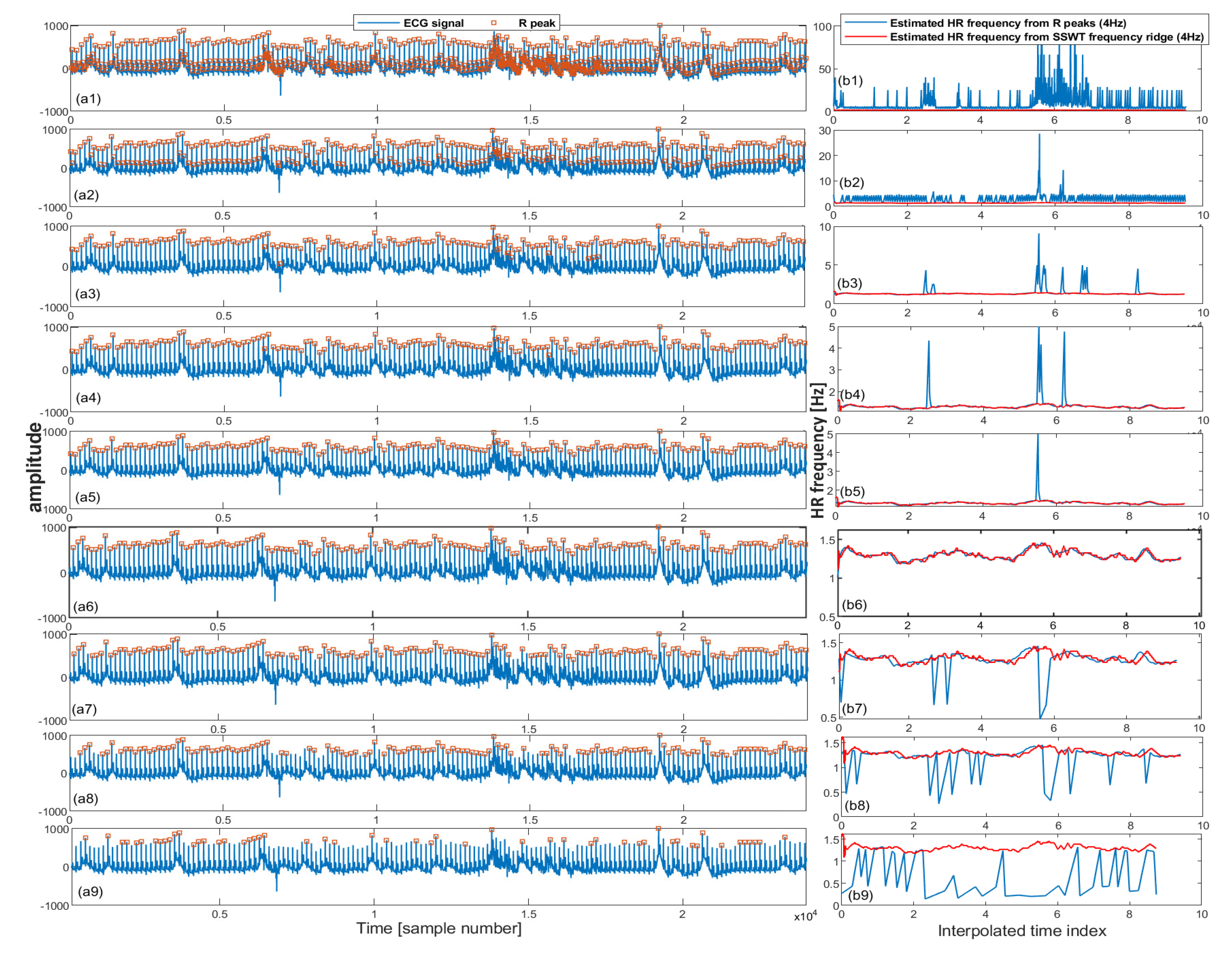
2.2.3. Iterative Pulse/R Peak Detection
| Algorithm 1: Iterative pulse peak detection. |
| - Input ECG: ← input ECG |
| - Set sampling frequency |
| - Set a vector of thresholds for detecting the pulse peaks |
| - = WSST(s, ) - SSWT transform |
| - = TFRIDGE(W) - frequency ridge in the range of Hz |
| - = INTERPRR(...4...) - Resample frequencies into 4 Hz |
| - for in [p] = PEAKDET(s, ) = /DIFF(p) - HR frequency = INTERPRR(...4...) - Resample frequencies into 4 Hz error() = ABS(-) |
| - Find set of pulse peaks associated with minimum error |
2.2.4. ECG Features
2.3. A Multimodal Deep Learning Neural Network for Sleep Disorder Detection
- Input Layer EMG: 5 × 3 tensor of the feature layer; the first dimension is the statistical parameters as presented in Table 1 and the second dimension is temporal window of the trial.
- Input Layer ECG: 1 × 10 tensor of statistical parameters of the ECG extracted from Table 1.
- Two hidden RNN layers for EMG input: Two hidden RNN layers with five units each.
- Fully connected layers for ECG input (FC1): A fully connected layer (FC) that performs a linear transformation with a weight matrix and bias; an input length of 10 and a number of hidden units of 20 with a rectifier linear unit (ReLU) activation function.
- Merging Layer: A layer that concatenates output tensors from the RNN layers and FC1 as a 1 dimensional tensor 1 × 25.
- Drop-out layer: A drop out layer with attention weights set to 0.4.
- Fully connected layer (FC1): A fully connected layer that applies a linear transformation (i.e., weight matrix and bias) with 25 inputs and 15 hidden units with ReLU activation.
- Fully connected layer (FC2): A fully connected layer that applies a linear transformation with 15 inputs and 15 hidden units with ReLU activation.
- Fully connected layer (FC3): A fully connected layer that applies a linear transformation with 15 inputs and 10 hidden units with ReLU activation.
- Fully connected layer (FC4): A fully connected layer that applies a linear transformation with 15 inputs and four hidden units.
- Softmax layer (SF): This layer re-scales between 0 and 1, with each output tensor element () from the fully connected layer FC4 as .
3. Results
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fang, Y.; Jiang, Z.; Wang, H. A novel sleep respiratory rate detection method for obstructive sleep apnea based on characteristic moment waveform. J. Healthc. Eng. 2018, 2018, 1902176. [Google Scholar] [CrossRef]
- Khandoker, A.H.; Palaniswami, M.; Karmakar, C.K. Support vector machines for automated recognition of obstructive sleep apnea syndrome from ECG recordings. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 37–48. [Google Scholar] [CrossRef]
- de Weerd, A.W.; Rijsman, R.M. Activity patterns of leg muscles in periodic limb movement disorder. J. Neurol. Neurosurg. Psychiatry 2004, 75, 317–319. [Google Scholar] [PubMed]
- Roux, F.J. Restless legs syndrome: Impact on sleep-related breathing disorders. Respirology 2013, 18, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Ferreri, F.; Rossini, P.M. Neurophysiological investigations in restless legs syndrome and other disorders of movement during sleep. Sleep Med. 2004, 5, 397–399. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, G.; Meredith, I.; Walker, A.; Solin, P. Obstructive sleep apnea leads to transient uncoupling of coronary blood flow and myocardial work in humans. Sleep 2009, 32, 263–270. [Google Scholar] [CrossRef]
- Takama, N.; Kurabayashi, M. Influence of untreated sleep disordered breathing on the long-term prognosis of patients with cardiovascular disease. Am. J. Cardiol. 2009, 103, 730–734. [Google Scholar] [CrossRef]
- Abdulla, S.; Diykh, M.; Luaibi Laft, L.; Saleh, K.; Deo, R.C. Sleep EEG signal analysis based on correlation graph similarity coupled with an ensemble extreme machine learning algorithm. Expert Syst. Appl. 2019, 138, 112790. [Google Scholar] [CrossRef]
- Diykh, M.; Li, Y.; Abdulla, S. EEG sleep stages identification based on weighted undirected complex networks. Comput. Methods Programs Biomed. 2020, 184, 105116. [Google Scholar] [CrossRef]
- Saha, S.; Bhattacharjee, A.; Fattah, S.A. Automatic detection of sleep apnea events based on inter-band energy ratio obtained from multi-band EEG signal. Healthc. Technol. Lett. 2019, 6, 82–86. [Google Scholar] [CrossRef]
- Sugi, T.; Kawana, F.; Nakamura, M. Automatic EEG arousal detection for sleep apnea syndrome. Biomed. Signal Process. Control 2009, 4, 329–337. [Google Scholar] [CrossRef]
- Moridani, M.K.; Heydar, M.; Jabbari Behnam, S.S. A Reliable Algorithm Based on Combination of EMG, ECG and EEG Signals for Sleep Apnea Detection: (A Reliable Algorithm for Sleep Apnea Detection). In Proceedings of the 2019 5th Conference on Knowledge Based Engineering and Innovation (KBEI), Tehran, Iran, 28 February–1 March 2019; pp. 256–262. [Google Scholar]
- Mosquera-Lopez, C.; Leitschuh, J.; Condon, J.; Hagen, C.C.; Rajhbeharrysingh, U.; Hanks, C.; Jacobs, P.G. Design and Evaluation of a Non-Contact Bed-Mounted Sensing Device for Automated In-Home Detection of Obstructive Sleep Apnea: A Pilot Study. Biosensors 2019, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Andreu-Perez, J.; Cao, F.; Hagras, H.; Yang, G.Z. A self-adaptive online brain-machine interface of a humanoid robot through a general type-2 fuzzy inference system. IEEE Trans. Fuzzy Syst. 2016, 26, 101–116. [Google Scholar] [CrossRef]
- Bsoul, M.; Minn, H.; Tamil, L. Apnea MedAs-sist: Real-time sleep apnea monitor using single-lead ECG. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 416–427. [Google Scholar] [CrossRef]
- Jarchi, D.; Sanei, S.; Prochazka, A. Detection of sleep apnea/hypopnea events using synchrosqueezed wavelet transform. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1199–1203. [Google Scholar]
- Jarchi, D.; Sanei, S. Derivation of respiratory effort from photoplethysmography. In Proceedings of the 2019 27th European Signal Processing Conference (EUSIPCO), A Coruna, Spain, 2–6 September 2019; pp. 1–5. [Google Scholar]
- Shokrollahi, M.; Krishnan, S. A Review of Sleep Disorder Diagnosis by Electromyogram Signal Analysis. Crit. Rev. Biomed. Eng. 2015, 43, 1–20. [Google Scholar] [CrossRef]
- Podlipnik, M.; Sarc, I.; Ziherl, K. Restless leg syndrome is common in patients with obstructive sleep apnoea. ERJ Open Res. 2017, 3, 20. [Google Scholar]
- Prochazka, A.; Kuchynka, J.; Vysata, O.; Cejnar, P.; Valis, M.; Marik, V. Multi-class sleep stage analysis and adaptive pattern recognition. Appl. Sci. 2018, 8, 697. [Google Scholar] [CrossRef]
- Prochazka, A.; Kuchynka, J.; Vysata, O.; Yadollahi, M.; Sanei, S.; Marik, V.; Valis, M. Sleep scoring using polysomnography data features. Signal Image Video Process. 2018, 12, 1043–1051. [Google Scholar] [CrossRef]
- Rostaghi, M.; Azami, H. Dispersion Entropy: A measure for time-series analysis. IEEE Signal Process. Lett. 2016, 23, 610–614. [Google Scholar] [CrossRef]
- Sanei, S.; Lee, T.K.M.; Abolghasemi, V. A new adaptive line enhancer based on singular spectrum analysis. IEEE Trans. Biomed. Eng. 2012, 59, 428–434. [Google Scholar] [CrossRef]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Bandt, C.; Pompe, B. Permutation entropy: A natural complexity measure for time series. Phys. Rev. Lett. 2002, 88, 1–4. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Daubechies, I.; Lu, J.; Wu, H. Synchrosqueezed wavelet transforms: An empirical mode decomposition-like tool. Appl. Comput. Harmon. Anal. 2011, 30, 243–261. [Google Scholar] [CrossRef]
- Thakur, G.; Brevdo, E.; Fuckar, N.S.; Wu, H.T. The Synchrosqueezing algorithm for time-varying spectral analysis: Robustness properties and new paleoclimate applications. IEEE Trans. Signal Process. 2011, 93, 1094–1097. [Google Scholar] [CrossRef]
- Carmona, R.A.; Wang, W.L.; Torresani, B. Characterization of signals by the ridges of their wavelet transforms. IEEE Trans. Signal Process. 1997, 45, 2586–2590. [Google Scholar] [CrossRef]
- Charlton, P.H.; Bonnici, T.; Tarassenko, L.; Clifton, D.A.; Beale, R.; Watkinson, P.J. An assessment of algorithms to estimate respiratory rate from the electrocardiogram and photoplethysmogram. Physiol. Meas. 2016, 37, 610–626. [Google Scholar] [CrossRef]
- Charlton, P.H.; Birrenkott, D.A.; Bonnici, T.; Pimentel, M.A.; Johnson, A.E.; Alastruey, J.; Tarassenko, L.; Watkinson, P.J.; Beale, R.; Clifton, D.A. Breathing rate estimation from the electrocardiogram and photoplethysmogram: A Review. IEEE Rev. Biomed. Eng. 2018, 11, 2–20. [Google Scholar] [CrossRef]
- Varoquaux, G.; Buitinck, L.; Louppe, G.; Grisel, O.; Pedregosa, F.; Mueller, A. Scikit-learn: Machine learning without learning the machinery. GetMob. Mob. Comput. Commun. 2015, 19, 29–33. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 6, 1–27. [Google Scholar] [CrossRef]
- Fan, R.E.; Chang, K.W.; Hsieh, C.J.; Wang, X.R.; Lin, C.J. Liblinear: A library for large linear classification. J. Mach. Learn. Res. 2008, 9, 1871–1874. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Short, R.; Fukunaga, K. The optimal distance measure for nearest neighbor classification. IEEE Trans. Inf. Theory 1981, 27, 622–627. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Jin, H.; Song, Q.; Hu, X. Auto-keras: An efficient neural architecture search system. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 1946–1956. [Google Scholar]
- Nielsen, D. Tree Boosting with XGBoost-Why does Xgboost Win “Every” Machine Learning Competition? Master’s Thesis, NTNU, Trondheim, Norway, 2016. [Google Scholar]
- Andreu-Perez, J.; Garcia-Gancedo, L.; McKinnell, J.; Van der Drift, A.; Powell, A.; Hamy, V.; Keller, T.; Yang, G.Z. Developing fine-grained Actigraphies for rheumatoid arthritis patients from a single accelerometer using machine learning. Sensors 2017, 17, 2113. [Google Scholar] [CrossRef] [PubMed]


| EMG Features | |
| Feature | Feature Type |
| Mean | Raw |
| Standard deviation | Raw |
| Skewness | Raw |
| Kurtosis | Raw |
| Dispersion Entropy | Raw |
| ECG Features | |
| Feature | Feature type |
| Maximum difference of pulse peaks | Time-domain |
| Minimum difference of pulse peaks | Time-domain |
| Mean difference of pulse peaks | Time-domain |
| Maximum amplitude of respiratory amplitude modulation | Time-domain |
| Minimum amplitude of respiratory amplitude modulation | Time-domain |
| Mean amplitude of respiratory amplitude modulation | Time-domain |
| Maximum of instantaneous frequencies of respiratory amplitude modulation | Frequency-domain |
| Minimum of instantaneous frequencies of respiratory amplitude modulation | Frequency-domain |
| Mean of instantaneous frequencies of respiratory amplitude modulation | Frequency-domain |
| Standard deviation of instantaneous frequencies of respiratory amplitude modulation | Frequency-domain |
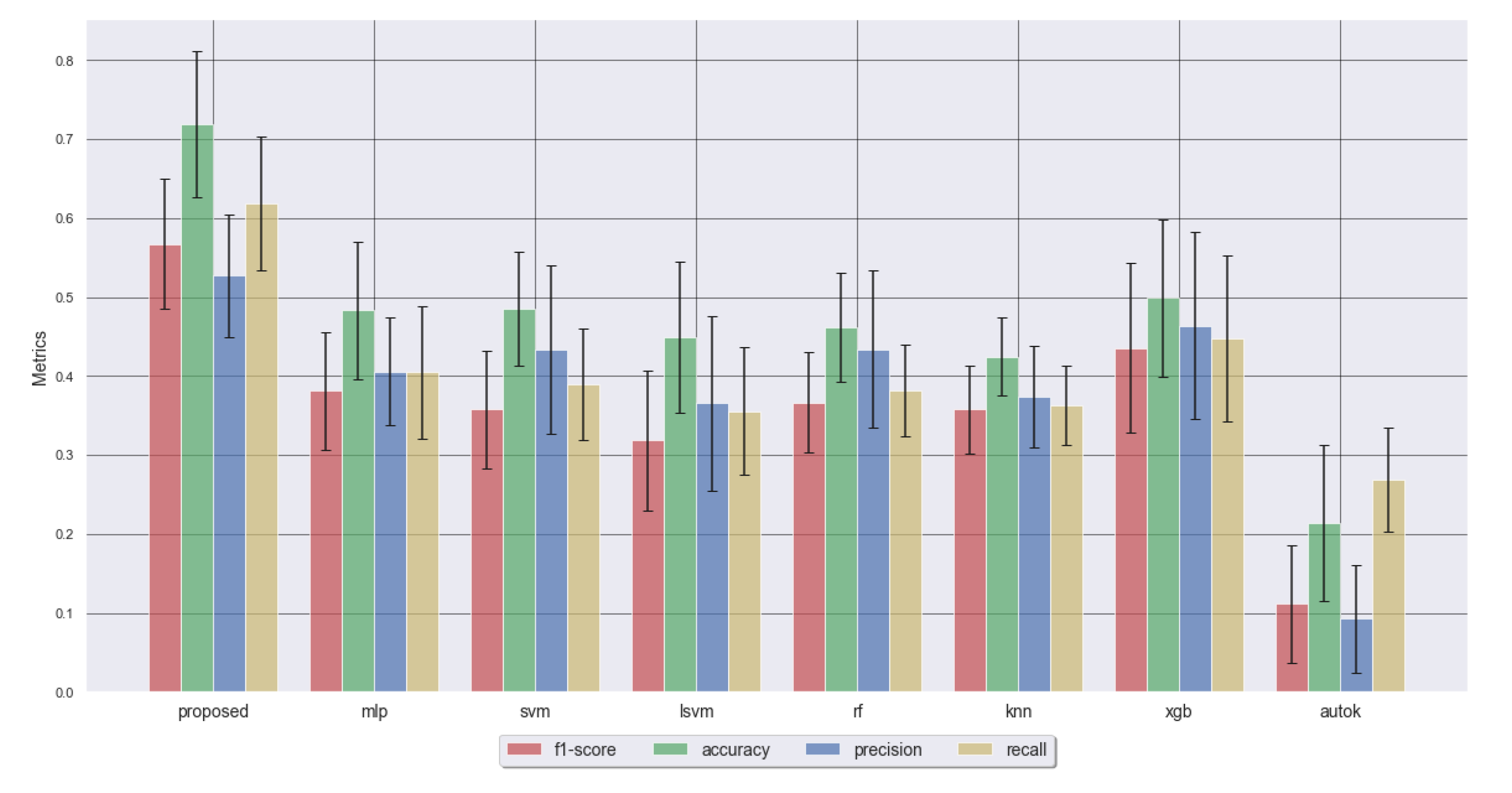
| Metric | Proposed | MLP | SVM | LSVM | RF | KNN | XGB | AUTOK |
|---|---|---|---|---|---|---|---|---|
| F1 Score | 0.57 ± 0.08 | 0.38 ± 0.07 | 0.36 ± 0.07 | 0.32 ± 0.09 | 0.37 ± 0.06 | 0.36 ± 0.06 | 0.44 ± 0.11 | 0.11 ± 0.07 |
| Accuracy | 0.72 ± 0.09 | 0.48 ± 0.09 | 0.48 ± 0.07 | 0.45 ± 0.1 | 0.46 ± 0.07 | 0.42 ± 0.05 | 0.5 ± 0.1 | 0.21 ± 0.1 |
| Precision | 0.53 ± 0.08 | 0.41 ± 0.07 | 0.43 ± 0.11 | 0.37 ± 0.11 | 0.43 ± 0.1 | 0.37 ± 0.06 | 0.46 ± 0.12 | 0.09 ± 0.07 |
| Recall | 0.62 ± 0.09 | 0.4 ± 0.08 | 0.39 ± 0.07 | 0.36 ± 0.08 | 0.38 ± 0.06 | 0.36 ± 0.05 | 0.45 ± 0.1 | 0.27 ± 0.07 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarchi, D.; Andreu-Perez, J.; Kiani, M.; Vysata, O.; Kuchynka , J.; Prochazka, A.; Sanei, S. Recognition of Patient Groups with Sleep Related Disorders using Bio-signal Processing and Deep Learning. Sensors 2020, 20, 2594. https://doi.org/10.3390/s20092594
Jarchi D, Andreu-Perez J, Kiani M, Vysata O, Kuchynka J, Prochazka A, Sanei S. Recognition of Patient Groups with Sleep Related Disorders using Bio-signal Processing and Deep Learning. Sensors. 2020; 20(9):2594. https://doi.org/10.3390/s20092594
Chicago/Turabian StyleJarchi, Delaram, Javier Andreu-Perez, Mehrin Kiani, Oldrich Vysata, Jiri Kuchynka , Ales Prochazka, and Saeid Sanei. 2020. "Recognition of Patient Groups with Sleep Related Disorders using Bio-signal Processing and Deep Learning" Sensors 20, no. 9: 2594. https://doi.org/10.3390/s20092594
APA StyleJarchi, D., Andreu-Perez, J., Kiani, M., Vysata, O., Kuchynka , J., Prochazka, A., & Sanei, S. (2020). Recognition of Patient Groups with Sleep Related Disorders using Bio-signal Processing and Deep Learning. Sensors, 20(9), 2594. https://doi.org/10.3390/s20092594









