Integration of a Thermoelectric Heating Unit with Ionic Wind-Induced Droplet Centrifugation Chip to Develop Miniaturized Concentration Device for Rapid Determination of Salmonella on Food Samples Using Antibody-Functionalized SERS Tags
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Preparations
2.2.1. Preparation of Antibody-Conjugated NAEBs
2.2.2. Preparation of Spiked Salmonella Samples in De-Ionized Water
2.2.3. Preparation of Spiked Salmonella Samples of Ice Cubes
2.3. Preparation of Spiked Salmonella Samples of Lettuce Leaves
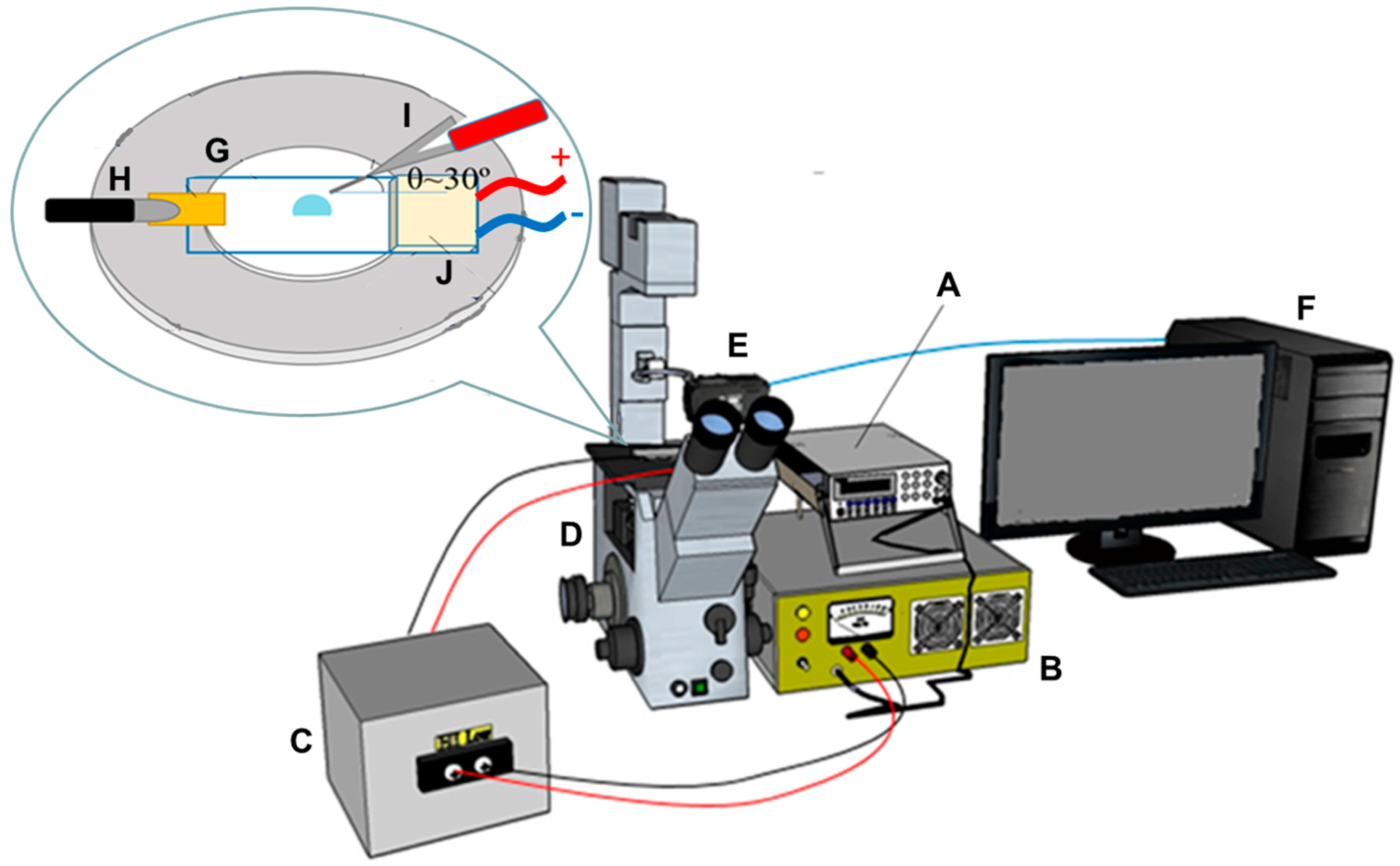
2.4. Equipmental Set-Up
2.5. Evaporation and Concentration of Salmonella Samples
3. Results and Discussion
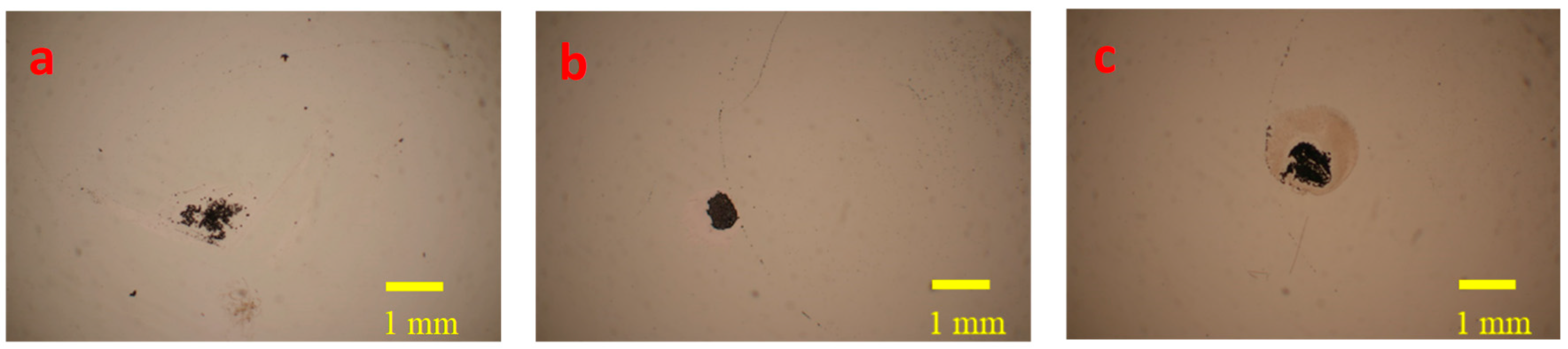
3.1. Droplet Shrinking and Drying Processes
3.2. Determination of Concentrated Salmonella Cells Bound with Antibody-Conjugated NAEBs
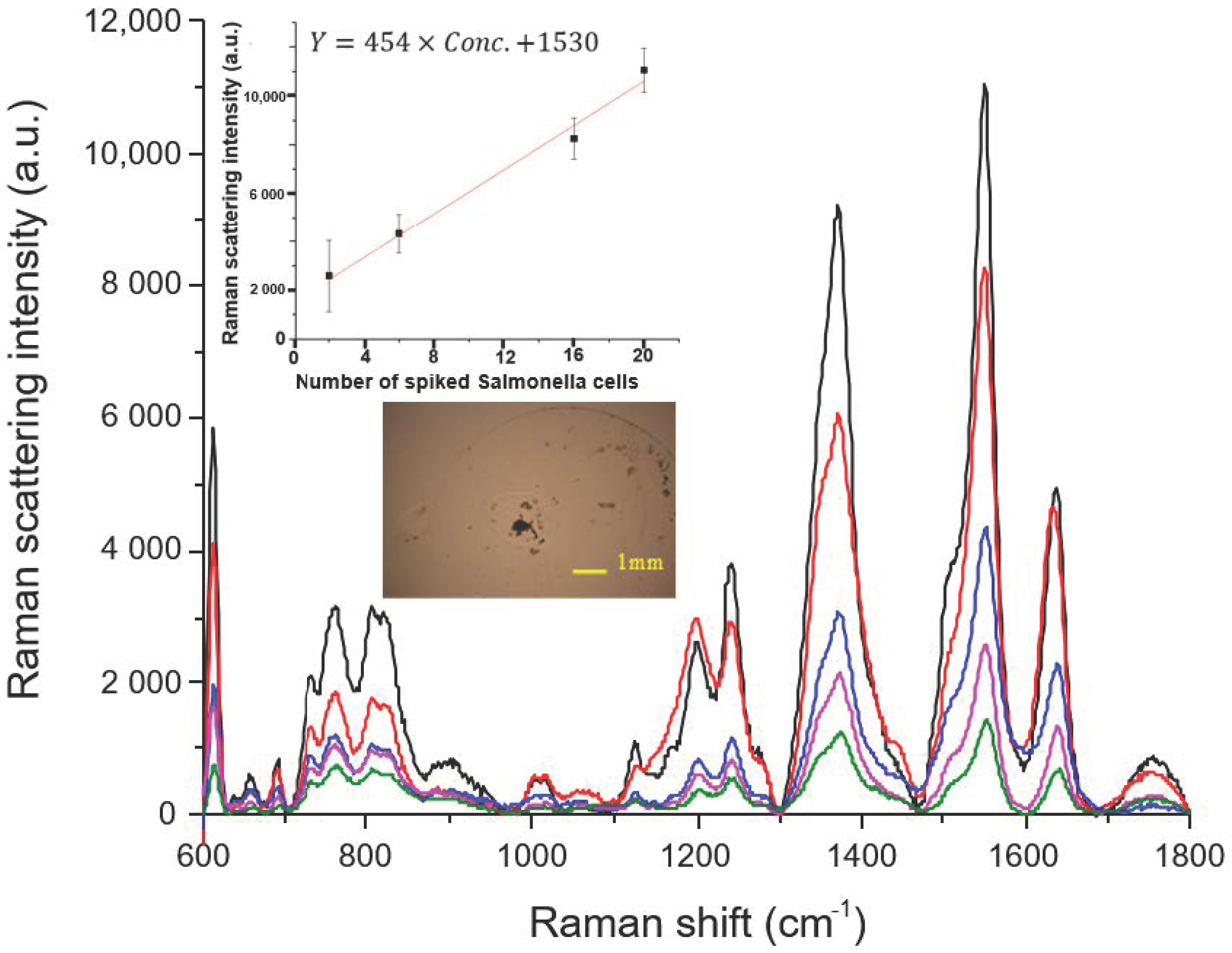
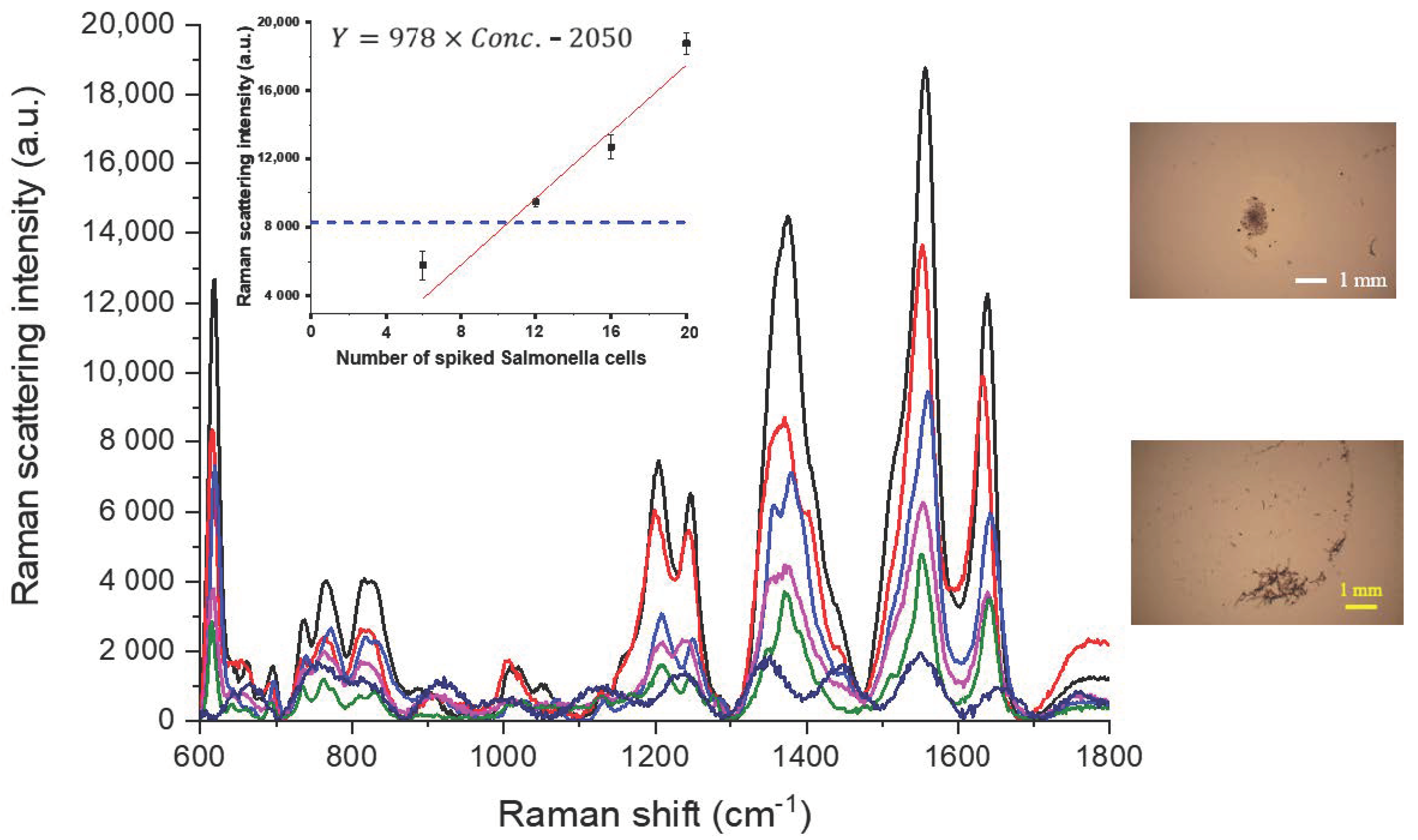
3.2.1. Determination of Salmonella in Spiked Water Samples
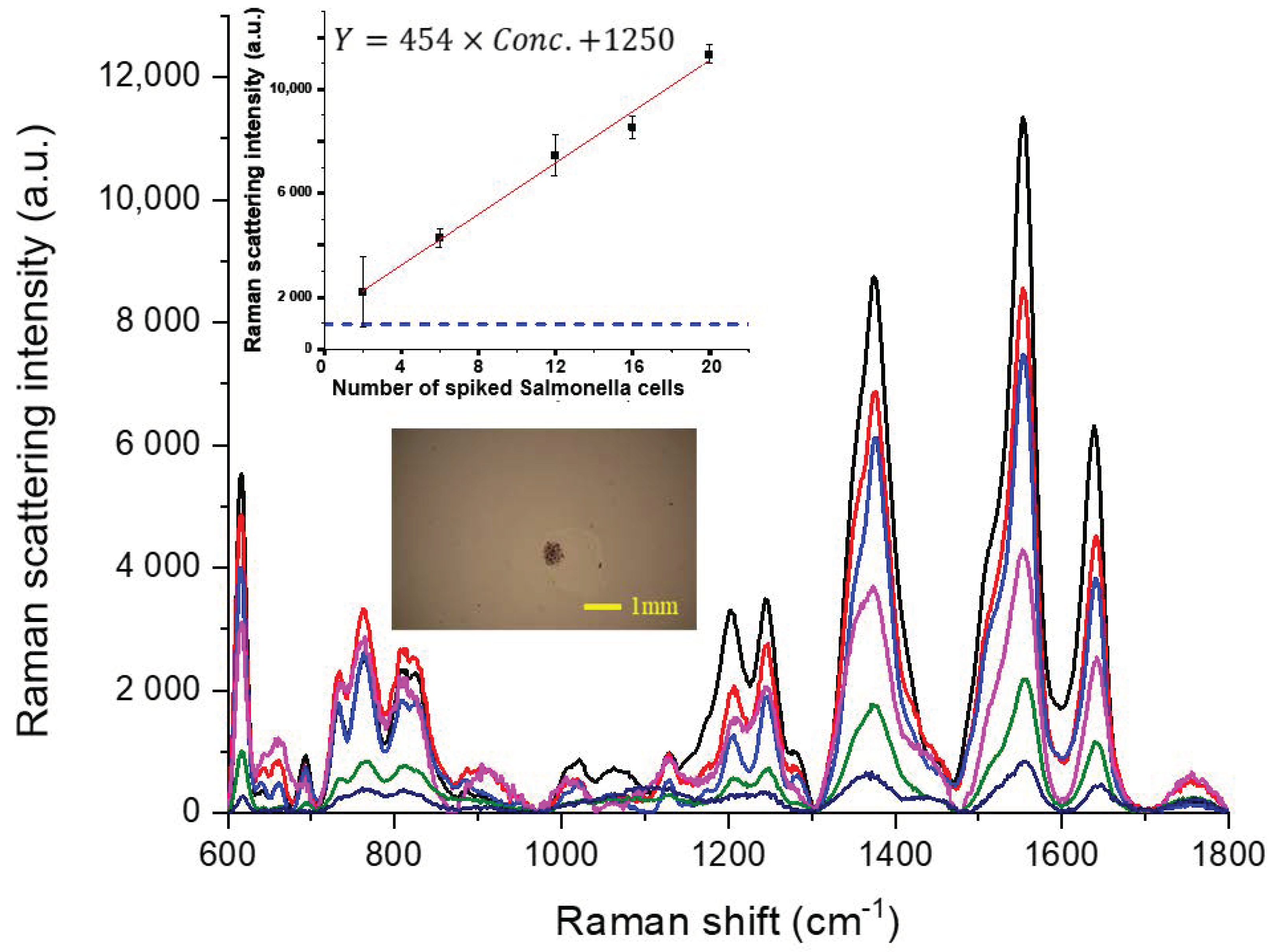
3.2.2. Determination of Salmonella in Spiked Ice Cube Samples
3.2.3. Determination of Salmonella in Spiked Lettuce Leaf Samples
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A

Appendix B
| SERS (cm−1) | Assignment |
|---|---|
| 615 | δ(C–C–C) |
| 759 | ω(NH2) + γ(C–C–C) |
| 1202 | υ(N–C) + δ(H-C-C) |
| 1377 | υ(C–C) |
| 1556 | υ(C–C) |
| 1641 | δ(NH2) + υ(C–C) |
References
- Compendium of Microbiological Criteria for Food; Food Standards Australia New Zealand (FSANZ): Kingston, Australia; Food Standards Australia New Zealand (FSANZ): Wellington, New Zealand, 2018. Available online: https://www.foodstandards.gov.au/publications/Documents/Compedium%20of%20Microbiological%20Criteria/Compendium_revised-Sep%202018.pdf (accessed on 12 December 2020).
- Du, X.-J.; Zhou, T.-J.; Li, P.; Wang, S. A rapid Salmonella detection method involving thermophilic helicase-dependent amplification and a lateral flow assay. Mol. Cell. Probes 2017, 34, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.N.T.; Yoon, J.; Jin, C.E.; Koo, B.; Han, K.; Shin, Y.; Lee, T.Y. Rapid and sensitive detection of Salmonella based on microfluidic enrichment with a label-free nanobiosensing platform. Sens. Actuators B Chem. 2018, 262, 588–594. [Google Scholar] [CrossRef]
- Hu, J.; Huang, R.; Sun, Y.; Wei, X.; Wang, Y.; Jiang, C.; Geng, Y.; Sun, X.; Jing, J.; Gao, H.; et al. Sensitive and rapid visual detection of Salmonella Typhimurium in milk based on recombinase polymerase amplification with lateral flow dipsticks. J. Microbiol. Methods 2019, 158, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tian, J.; Li, K.; Tian, H.; Xu, W. Label-free visual biosensor based on cascade amplification for the detection of Salmonella. Anal. Chim. Acta 2019, 1075, 144–151. [Google Scholar] [CrossRef]
- Rosen, D.K.; Gallardo, M.; Vail, M.; Hellberg, R.S. Microplate immunocapture coupled with the 3M molecular detection system and selective plating for the rapid detection of Salmonella infantis in dry dog food and treats. J. Microbiol. Methods 2020, 172, 105881. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S.; Volpe, G.; Piermarini, S.; Delibato, E.; Palleschi, G. Electrochemical Biosensors for Rapid Detection of Foodborne Salmonella: A Critical Overview. Sensors 2017, 17, 1910. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Magalhães, J.M.C.S.; Freire, C.; Delerue-Matos, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, Y.; He, Y.; Fang, W.; Ying, Y.; Li, Y.; Fu, Y. Cooperation Mode of Outer Surface and Inner Space of Nanochannel: Separation-Detection System Based on Integrated Nanochannel Electrode for Rapid and Facile Detection of Salmonella. Anal. Chem. 2020, 92, 1818–1825. [Google Scholar] [CrossRef]
- Hou, Y.; Tang, W.; Qi, W.; Guo, X.; Lin, J. An ultrasensitive biosensor for fast detection of Salmonella using 3D magnetic grid separation and urease catalysis. Biosens. Bioelectron. 2020, 157, 112160. [Google Scholar] [CrossRef]
- Dinshaw, I.J.; Muniandy, S.; Teh, S.J.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Development of an aptasensor using reduced graphene oxide chitosan complex to detect Salmonella. J. Electroanal. Chem. 2017, 806, 88–96. [Google Scholar] [CrossRef]
- Appaturi, J.N.; Pulingam, T.; Thong, K.L.; Muniandy, S.; Ahmad, N.; Leo, B.F. Rapid and sensitive detection of Salmonella with reduced graphene oxide-carbon nanotube based electrochemical aptasensor. Anal. Biochem. 2020, 589, 113489. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Pang, G.; Xie, J. A new phosphothreonine lyase electrochemical immunosensor for detecting Salmonella based on horseradish peroxidase/GNPs-thionine/chitosan. Biomed. Microdevices 2017, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Dai, G.; Li, Z.; Luo, F.; Ai, S.; Chen, B.; Wang, Q. Electrochemical determination of Salmonella typhimurium by using aptamer-loaded gold nanoparticles and a composite prepared from a metal-organic framework (type UiO-67) and graphene. Microchim. Acta 2019, 186, 620. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yan, W.; Liu, Y.; He, S.; Cao, X.; Xu, X.; Zheng, H.; Gunasekaran, S. Electrochemical detection of Salmonella using an invA genosensor on polypyrrole-reduced graphene oxide modified glassy carbon electrode and AuNPs-horseradish peroxidase-streptavidin as nanotag. Anal. Chim. Acta 2019, 1074, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huo, X.; Zheng, L.; Cai, G.; Wang, Y.; Liu, N.; Wang, M.; Lin, J. An ultrasensitive biosensor for colorimetric detection of Salmonella in large-volume sample using magnetic grid separation and platinum loaded zeolitic imidazolate Framework-8 nanocatalysts. Biosens. Bioelectron. 2020, 150, 111862. [Google Scholar] [CrossRef]
- Cheng, K.; Zhang, J.; Zhang, L.; Wang, L.; Chen, H. Aptamer biosensor for Salmonella typhimurium detection based on luminescence energy transfer from Mn2+-doped NaYF4:Yb, Tm upconverting nanoparticles to gold nanorods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 171, 168–173. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Huang, C.-H.; Hsieh, W.-H.; Liu, L.-H.; Lin, Y.-C.; Chu, C.-C.; Wang, S.-T.; Kuo, I.T.; Chau, L.-K.; Yang, C.-Y. On-line SERS Detection of Single Bacterium Using Novel SERS Nanoprobes and A Microfluidic Dielectrophoresis Device. Small 2014, 10, 4700–4710. [Google Scholar] [CrossRef]
- Faulds, K.; Jarvis, R.; Smith, W.E.; Graham, D.; Goodacre, R. Multiplexed detection of six labelled oligonucleotides using surface enhanced resonance Raman scattering (SERRS). Analyst 2008, 133, 1505–1512. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, X.; Liu, Y.; Duan, N.; Wu, S.; Wang, Z.; Xu, B. Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Biosens. Bioelectron. 2015, 74, 872–877. [Google Scholar] [CrossRef]
- Kearns, H.; Goodacre, R.; Jamieson, L.E.; Graham, D.; Faulds, K. SERS Detection of Multiple Antimicrobial-Resistant Pathogens Using Nanosensors. Anal. Chem. 2017, 89, 12666–12673. [Google Scholar] [CrossRef]
- Sánchez-Purrà, M.; Carré-Camps, M.; de Puig, H.; Bosch, I.; Gehrke, L.; Hamad-Schifferli, K. Surface-Enhanced Raman Spectroscopy-Based Sandwich Immunoassays for Multiplexed Detection of Zika and Dengue Viral Biomarkers. ACS Infect. Dis. 2017, 3, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Choi, N.; Wang, R.; Lee, S.; Moon, K.C.; Yoon, S.-Y.; Chen, L.; Choo, J. Simultaneous Detection of Dual Prostate Specific Antigens Using Surface-Enhanced Raman Scattering-Based Immunoassay for Accurate Diagnosis of Prostate Cancer. ACS Nano 2017, 11, 4926–4933. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Fang, Y.-C.; Lin, S.-Y.; Lin, Y.-J.; Yen, S.-Y.; Huang, C.-H.; Yang, C.-Y.; Chau, L.-K.; Wang, S.-C. Corona-induced micro-centrifugal flows for concentration of Neisseria and Salmonella bacteria prior to their quantitation using antibody-functionalized SERS-reporter nanobeads. Microchim. Acta 2017, 184, 1021–1028. [Google Scholar] [CrossRef]
- Su, S.-R.; Chen, Y.-Y.; Li, K.-Y.; Fang, Y.-C.; Wang, C.-H.; Yang, C.-Y.; Chau, L.-K.; Wang, S.-C. Electrohydrodynamically enhanced drying droplets for concentration of Salmonella bacteria prior to their detections using antibody-functionalized SERS-reporter submicron beads. Sens. Actuators B Chem. 2019, 283, 384–389. [Google Scholar] [CrossRef]
- Huang, P.-J.; Chau, L.-K.; Yang, T.-S.; Tay, L.-L.; Lin, T.-T. Nanoaggregate-Embedded Beads as Novel Raman Labels for Biodetection. Adv. Funct. Mater. 2009, 19, 242–248. [Google Scholar] [CrossRef]
- Huang, P.-J.; Tay, L.-L.; Tanha, J.; Ryan, S.; Chau, L.-K. Single-Domain Antibody-Conjugated Nanoaggregate-Embedded Beads for Targeted Detection of Pathogenic Bacteria. Chem. Eur. J. 2009, 15, 9330–9334. [Google Scholar] [CrossRef]
- Tay, L.-L.; Huang, P.-J.; Tanha, J.; Ryan, S.; Wu, X.; Hulse, J.; Chau, L.-K. Silica encapsulated SERS nanoprobe conjugated to the bacteriophage tailspike protein for targeted detection of Salmonella. Chem. Commun. 2012, 48, 1024–1026. [Google Scholar] [CrossRef]
- Pathak, B.; Hatte, S.; Basu, S. Evaporation Dynamics of Mixed-Nanocolloidal Sessile Droplets. Langmuir 2017, 33, 14123–14129. [Google Scholar] [CrossRef]
- Lofrumento, C.; Arci, F.; Carlesi, S.; Ricci, M.; Castellucci, E.; Becucci, M. Safranin-O dye in the ground state. A study by density functional theory, Raman, SERS and infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 137, 677–684. [Google Scholar] [CrossRef]
- Sylvestre, S.; Sebastian, S.; Oudayakumar, K.; Jayavarthanan, T.; Sundaraganesan, N. Experimental (FT-IR, FT-Raman and UV–Vis) spectra and theoretical DFT investigations of 2,3-diaminophenazine. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 96, 401–412. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-J.; Chen, Y.-Y.; Wang, K.-H.; Wang, C.-H.; Yang, C.-Y.; Chau, L.-K.; Wang, S.-C. Integration of a Thermoelectric Heating Unit with Ionic Wind-Induced Droplet Centrifugation Chip to Develop Miniaturized Concentration Device for Rapid Determination of Salmonella on Food Samples Using Antibody-Functionalized SERS Tags. Sensors 2020, 20, 7177. https://doi.org/10.3390/s20247177
Chen Y-J, Chen Y-Y, Wang K-H, Wang C-H, Yang C-Y, Chau L-K, Wang S-C. Integration of a Thermoelectric Heating Unit with Ionic Wind-Induced Droplet Centrifugation Chip to Develop Miniaturized Concentration Device for Rapid Determination of Salmonella on Food Samples Using Antibody-Functionalized SERS Tags. Sensors. 2020; 20(24):7177. https://doi.org/10.3390/s20247177
Chicago/Turabian StyleChen, Yi-Jhen, Yuan-Yu Chen, Kai-Hao Wang, Chih-Hsien Wang, Chiou-Ying Yang, Lai-Kwan Chau, and Shau-Chun Wang. 2020. "Integration of a Thermoelectric Heating Unit with Ionic Wind-Induced Droplet Centrifugation Chip to Develop Miniaturized Concentration Device for Rapid Determination of Salmonella on Food Samples Using Antibody-Functionalized SERS Tags" Sensors 20, no. 24: 7177. https://doi.org/10.3390/s20247177
APA StyleChen, Y.-J., Chen, Y.-Y., Wang, K.-H., Wang, C.-H., Yang, C.-Y., Chau, L.-K., & Wang, S.-C. (2020). Integration of a Thermoelectric Heating Unit with Ionic Wind-Induced Droplet Centrifugation Chip to Develop Miniaturized Concentration Device for Rapid Determination of Salmonella on Food Samples Using Antibody-Functionalized SERS Tags. Sensors, 20(24), 7177. https://doi.org/10.3390/s20247177






