Pesticide Aptasensors—State of the Art and Perspectives
Abstract
1. Introduction
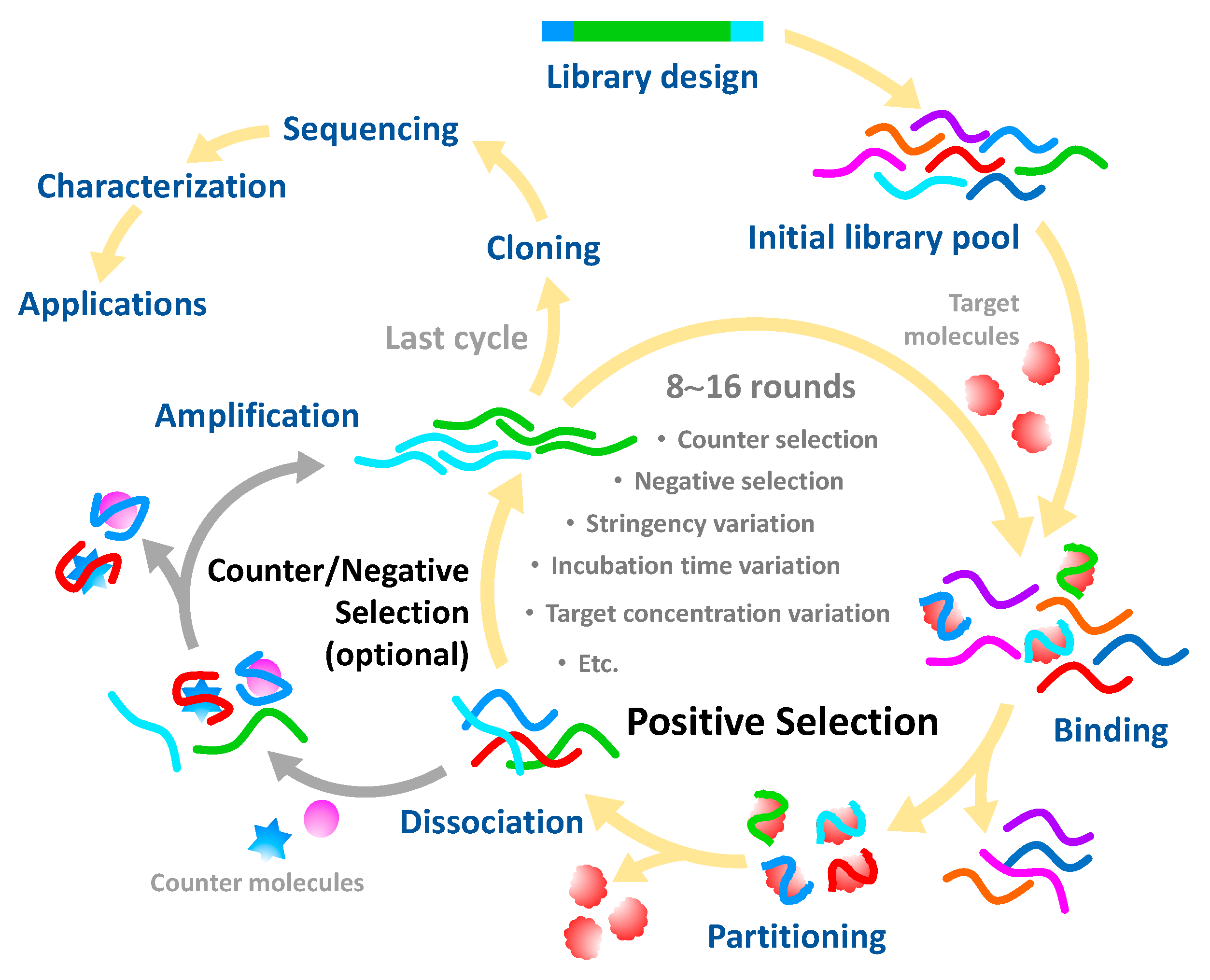
2. Generation of Aptamers against Pesticides
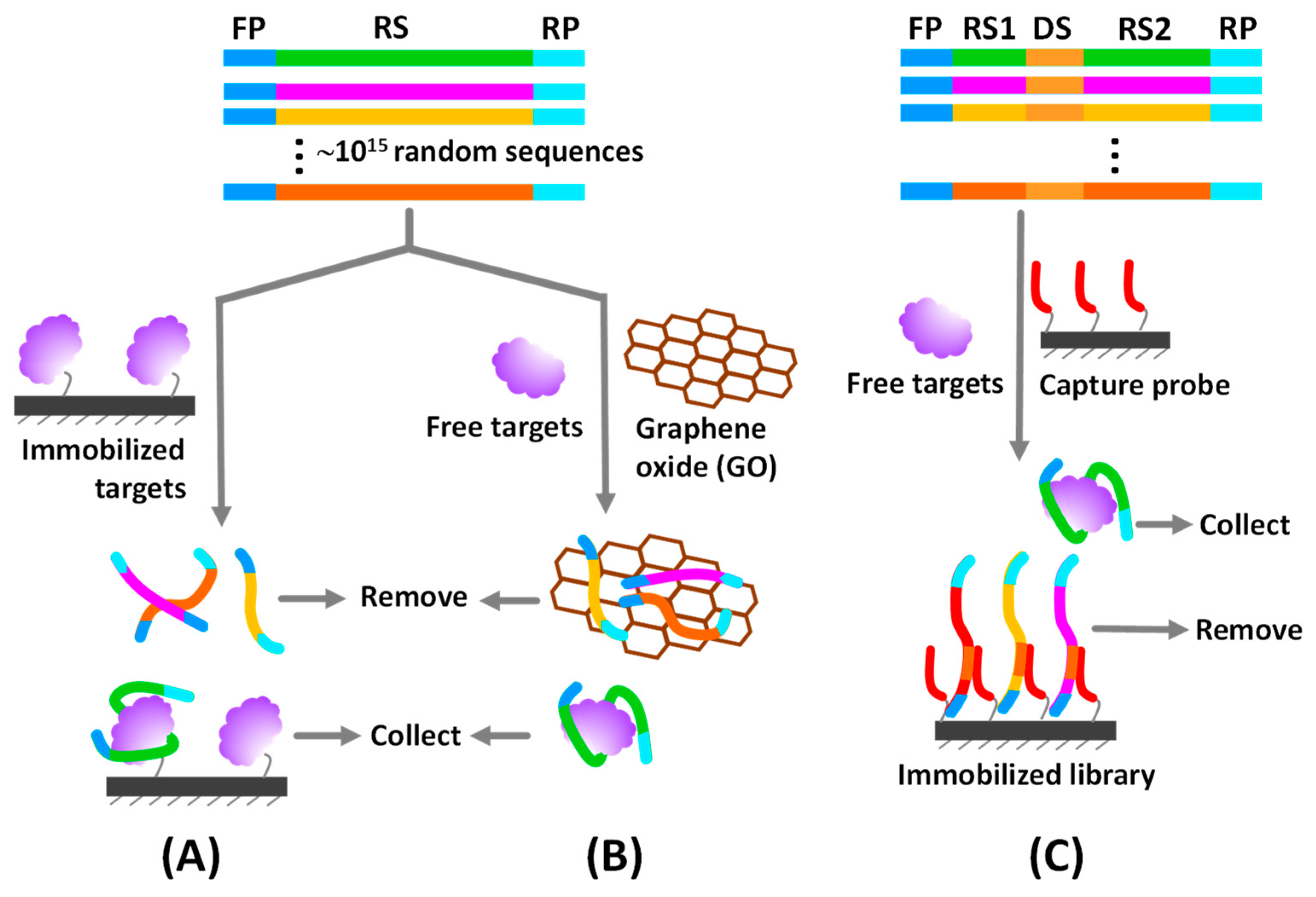
2.1. Immobilized SELEX Approach
2.1.1. Immobilization of Target Molecules
2.1.2. Immobilization of Random Library
2.2. Non-Immobilized SELEX Approach
2.2.1. Capillary Electrophoresis-Based SELEX Process (CE-SELEX)
2.2.2. Physical Adsorption-Based SELEX Process
3. Improvement of Aptamer Specificity and Affinity
3.1. Eliminating Non-Specificity with Selection Matrix and Counter Molecules
3.2. Enhancing Affinity with Stringency Modification
3.3. Trimming off Nonessential Sequences for Target Recognition
3.4. Understanding Molecular Interaction for Rational Design and Selection
4. Essential Characterization of Pesticide Aptamers
5. Current Advances in Aptasensors for Pesticide Determination
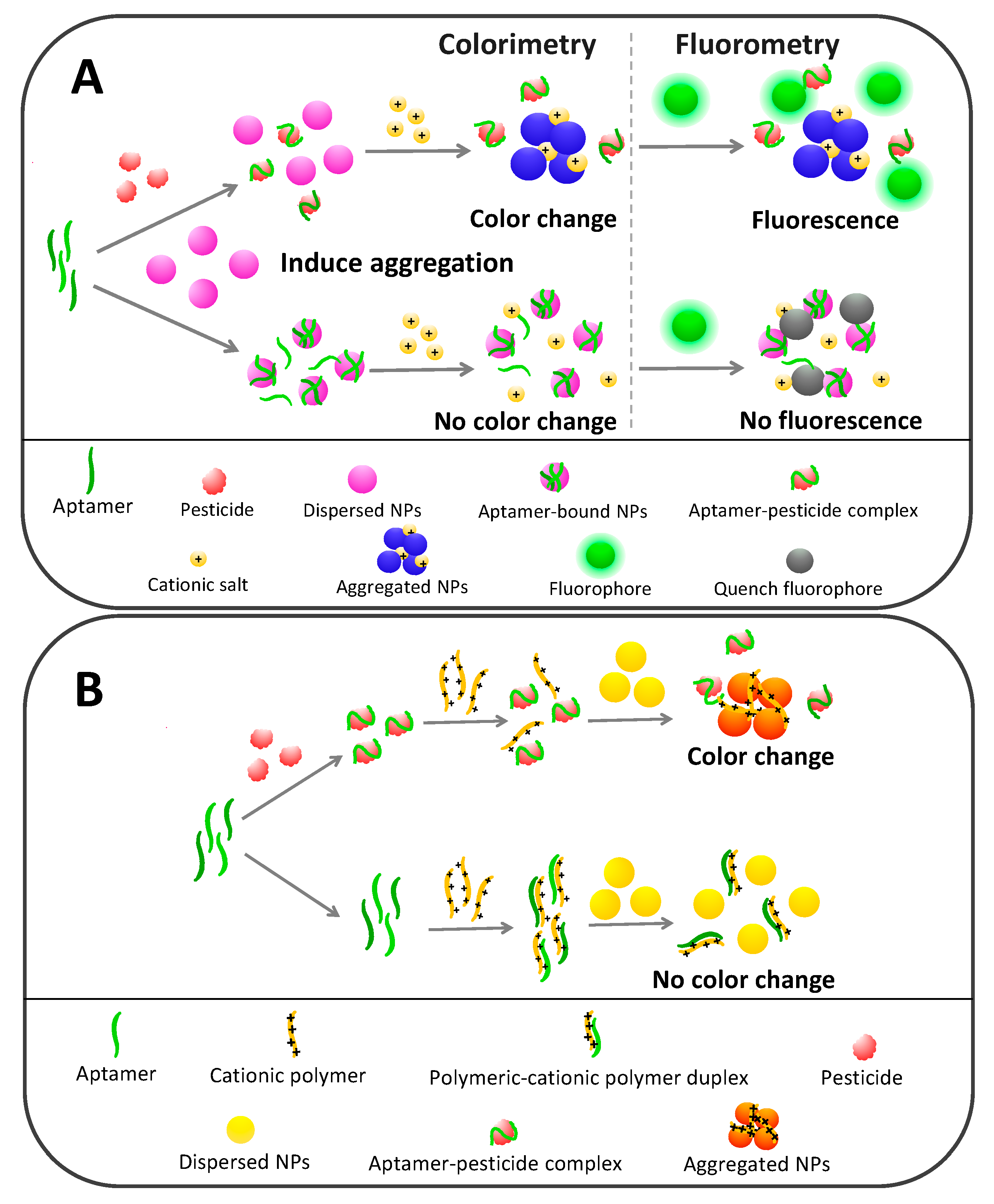
5.1. Colorimetric Pesticide Aptasensors Based on Nanoparticle Agglomeration
5.2. Fluorometric Aptasensors for Sensitive Agrochemical Detection
5.2.1. Dual Colorimetric and Fluorometric Aptasensors Based on Noble Metal Nanoparticles
5.2.2. Graphene Oxide- and Carbon Nanomaterial-Based Fluorescence Aptasensors
5.2.3. Complementary Fluorescent Probe-Based Aptasensors
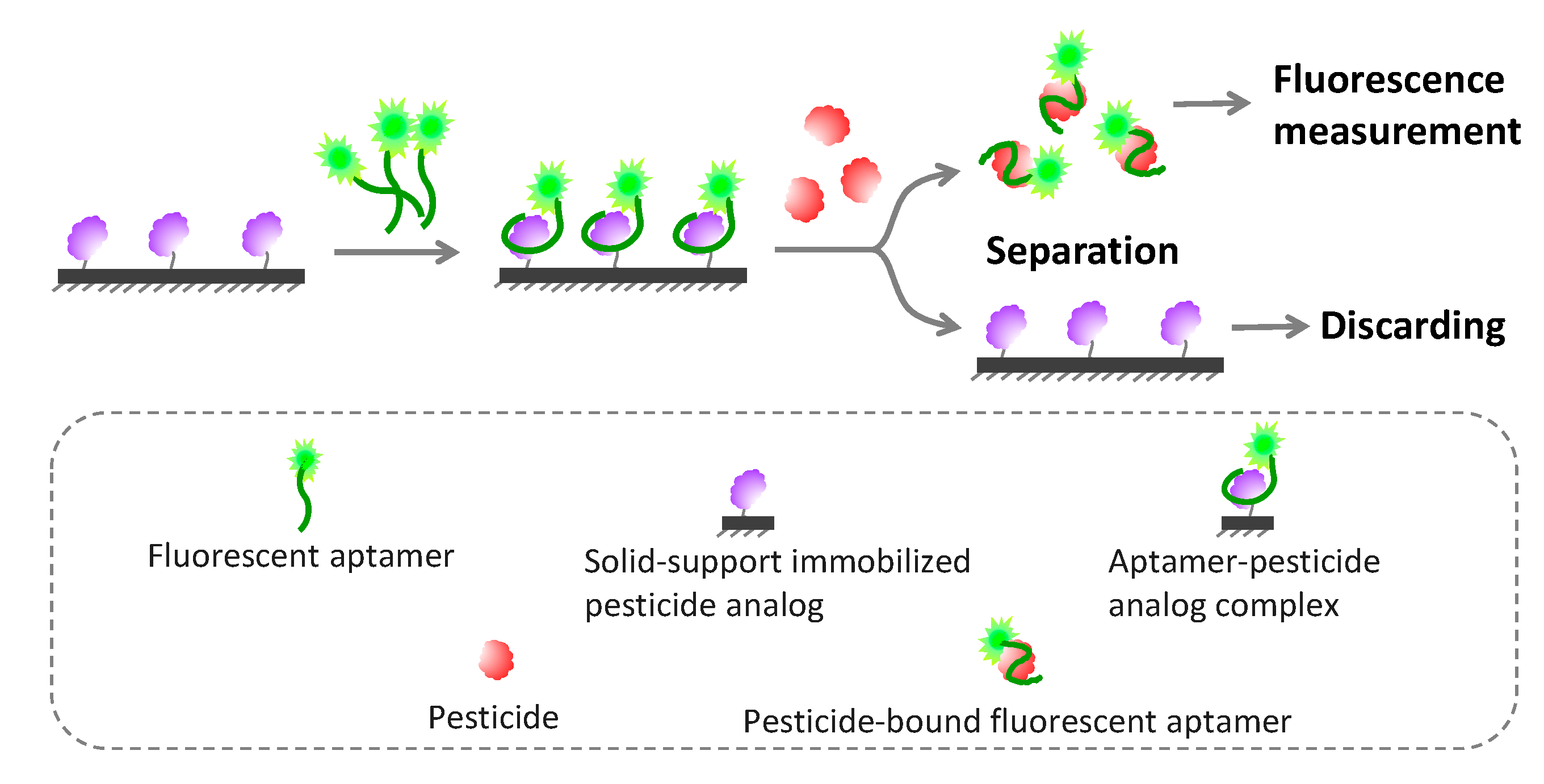
5.2.4. Solid Phase-Based Fluorometric Aptasensors
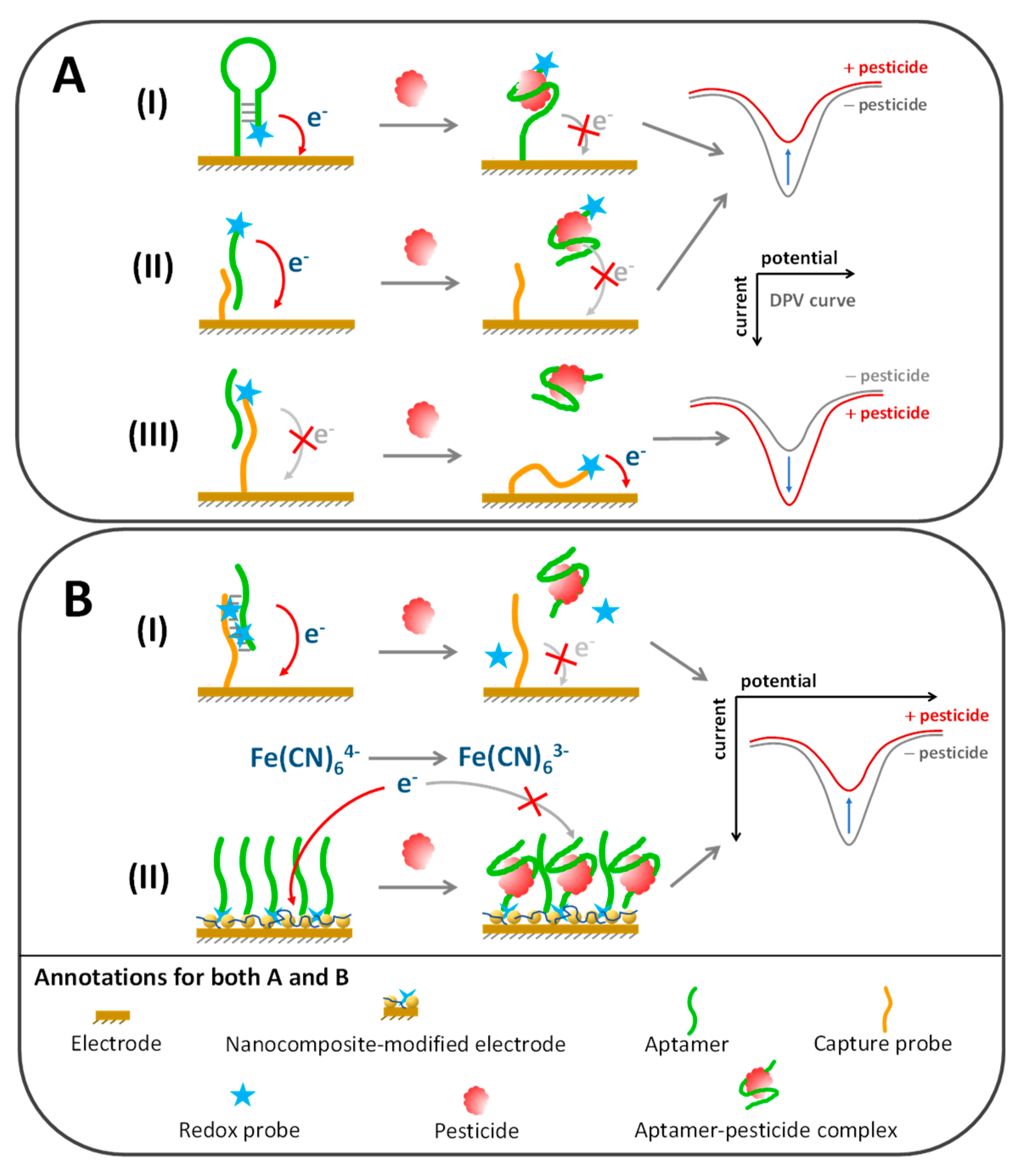
5.3. Electrochemical Aptasensors
5.3.1. Conjugation of Redox Probe to Aptamer or Molecular Probe
5.3.2. Noncovalent Incorporation of Aptamer Duplex with Redox Probe
5.3.3. Integration of Redox Probes in Aqueous or Nanocomposite Phase
5.4. Electrochemiluminescence-Based Aptasensors
5.5. Surface-Enhanced Raman Spectrometry-Based Aptasensors
5.6. Miscellaneous Approaches
6. Benchmarking with the ASSURED Criteria
7. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Zamora-Sequeira, R.; Starbird-Pérez, R.; Rojas-Carillo, O.; Vargas-Villalobos, S. What are the main sensor methods for quantifying pesticides in agricultural activities? A Review. Molecules 2019, 24, 2659. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Bhardwaj, A. Biosensor technology for pesticides—A review. Appl. Biochem. Biotechnol. 2015, 175, 3093–3119. [Google Scholar] [CrossRef] [PubMed]
- Pundir, C.S.; Malik, A.; Preety. Bio-sensing of organophosphorus pesticides: A review. Biosens. Bioelectron. 2019, 140, 111348. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Li, H.; Su, X. Review of optical sensors for pesticides. Trends Anal. Chem. 2018, 103, 1–20. [Google Scholar] [CrossRef]
- Apilux, A.; Ayudhya, C.I.N.; Tantimongcolwat, T.; Prachayasittikul, V. Paper-based acetylcholinesterase inhibition assay combining a wet system for organophosphate and carbamate pesticides detection. EXCLI J. 2015, 14, 307–319. [Google Scholar]
- Yang, N.; Shaheen, N.; Xie, L.; Yu, J.; Ahmad, H.; Mao, H. Pesticide residues identification by optical spectrum in the time-sequence of enzyme inhibitors performed on microfluidic paper-based analytical devices (µPADs). Molecules 2019, 24, 2428. [Google Scholar] [CrossRef]
- Cetrangolo, P.G.; Gori, C.; Rusko, J.; Terreri, S.; Manco, G.; Cimmino, A.; Febbraio, F. Determination of picomolar concentrations of paraoxon in human urine by fluorescence-based enzymatic assay. Sensors 2019, 19, 4852. [Google Scholar] [CrossRef]
- Singh, A.P.; Balayan, S.; Hooda, V.; Sarin, R.K.; Chauhan, N. Nano-interface driven electrochemical sensor for pesticides detection based on the acetylcholinesterase enzyme inhibition. Int. J. Biol. Macromol. 2020, 164, 3943–3952. [Google Scholar] [CrossRef]
- Cervera-Chiner, L.; March, C.; Arnau, A.; Jiménez, Y.; Montoya, Á. Detection of DDT and carbaryl pesticides in honey by means of immunosensors based on high fundamental frequency quartz crystal microbalance (HFF-QCM). J. Sci. Food Agric. 2020, 100, 2468–2472. [Google Scholar] [CrossRef]
- Sroysee, W.; Chunta, S.; Amatatongchai, M.; Lieberzeit, P.A. Molecularly imprinted polymers to detect profenofos and carbofuran selectively with QCM sensors. Phys. Med. 2019, 7, 100016. [Google Scholar] [CrossRef]
- Kant, R. Surface plasmon resonance based fiber–optic nanosensor for the pesticide fenitrothion utilizing Ta2O5 nanostructures sequestered onto a reduced graphene oxide matrix. Microchim. Acta 2019, 187, 8. [Google Scholar] [CrossRef] [PubMed]
- Özcan, A.; Hamid, F.; Özcan, A.A. Synthesizing of a nanocomposite based on the formation of silver nanoparticles on fumed silica to develop an electrochemical sensor for carbendazim detection. Talanta 2020, 222, 121591. [Google Scholar] [CrossRef] [PubMed]
- El-Moghazy, A.Y.; Huo, J.; Amaly, N.; Vasylieva, N.; Hammock, B.D.; Sun, G. An innovative nanobody-based electrochemical immunosensor using decorated nylon nanofibers for point-of-care monitoring of human exposure to pyrethroid insecticides. ACS Appl. Mater. Interfaces 2020, 12, 6159–6168. [Google Scholar] [CrossRef]
- Modh, H.; Scheper, T.; Walter, J.-G. Aptamer-modified magnetic beads in biosensing. Sensors 2018, 18, 1041. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zu, Y. A highlight of recent advances in aptamer technology and its application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef]
- Liu, M.; Khan, A.; Wang, Z.; Liu, Y.; Yang, G.; Deng, Y.; He, N. Aptasensors for pesticide detection. Biosens. Bioelectron. 2019, 130, 174–184. [Google Scholar] [CrossRef]
- Sekhon, S.S.; Park, G.-Y.; Park, D.-Y.; Kim, S.Y.; Wee, J.-H.; Ahn, J.-Y.; Kim, Y.-H. Aptasensors for pesticide detection. Toxicol. Environ. Health Sci. 2018, 10, 229–236. [Google Scholar] [CrossRef]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Reverdatto, S.; Burz, D.S.; Shekhtman, A. Peptide aptamers: Development and applications. Curr. Top. Med. Chem. 2015, 15, 1082–1101. [Google Scholar] [CrossRef]
- Beisel, C.L.; Smolke, C.D. Design principles for riboswitch function. PLoS Comput. Biol. 2009, 5, e1000363. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, H.; Wang, J.; Xu, L.; Chen, H.; Pei, R. Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd(II). Talanta 2016, 154, 498–503. [Google Scholar] [CrossRef]
- Ma, X.; Wang, W.; Chen, X.; Xia, Y.; Wu, S.; Duan, N.; Wang, Z. Selection, identification, and application of Aflatoxin B1 aptamer. Eur. Food Res. Technol. 2014, 238, 919–925. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, M.; Su, H.; Xiao, H.; Wu, S.; Qin, X.; Li, S.; Mi, H.; Lu, Z.; Shi, D.; et al. Selection and characterization of ssDNA aptamers specifically recognizing pathogenic Vibrio alginolyticus. J. Fish Dis. 2019, 42, 851–858. [Google Scholar] [CrossRef]
- Affinito, A.; Quintavalle, C.; Esposito, C.L.; Roscigno, G.; Vilardo, C.; Nuzzo, S.; Vitiani, L.R.; De Luca, G.; Pallini, R.; Kichkailo, A.S.; et al. The discovery of RNA aptamers that selectively bind glioblastoma stem cells. Mol. Ther. Nucleic Acids 2019, 18, 99–109. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Wu, Y.; Belmonte, I.; Sykes, K.S.; Xiao, Y.; White, R.J. Perspective on the future role of aptamers in analytical chemistry. Anal. Chem. 2019, 91, 15335–15344. [Google Scholar] [CrossRef]
- Yan, J.; Xiong, H.; Cai, S.; Wen, N.; He, Q.; Liu, Y.; Peng, D.; Liu, Z. Advances in aptamer screening technologies. Talanta 2019, 200, 124–144. [Google Scholar] [CrossRef] [PubMed]
- Mulchandani, A. DNA Apatmers-Based Detection of Atrazine in Water; Report; University of California Water Resources Center: Riverside County, CA, USA, 2004; pp. 69–70. [Google Scholar]
- He, J.; Liu, Y.; Fan, M.; Liu, X. Isolation and identification of the DNA aptamer target to acetamiprid. J. Agric. Food Chem. 2011, 59, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, C.; Han, B.; Luo, J.; Yin, G. An electrochemiluminescence aptasensor switch for aldicarb recognition via ruthenium complex-modified dendrimers on multiwalled carbon nanotubes. Microchim. Acta 2017, 184, 1669–1675. [Google Scholar] [CrossRef]
- Williams, R.M.; Crihfield, C.L.; Gattu, S.; Holland, L.A.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against atrazine. Int. J. Mol. Sci. 2014, 15, 14332–14347. [Google Scholar] [CrossRef]
- Abraham, K.M.; Roueinfar, M.; Ponce, A.T.; Lussier, M.E.; Benson, D.B.; Hong, K.L. In vitro selection and characterization of a single-stranded DNA aptamer against the herbicide atrazine. ACS Omega 2018, 3, 13576–13583. [Google Scholar] [CrossRef]
- Sanchez, P.E. DNA Aptamer Development for Detection of Atrazine and Protective Antigen Toxin Using Fluorescence Polarization; University of California Riverside: Riverside, CA, USA, 2012. [Google Scholar]
- Williams, R.M.; Kulick, A.R.; Yedlapalli, S.; Battistella, L.; Hajiran, C.J.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element specific for bromacil. J. Nucleic Acids 2014, 2014, 102968. [Google Scholar] [CrossRef]
- Eissa, S.; Zourob, M. Selection and characterization of DNA aptamers for electrochemical biosensing of carbendazim. Anal. Chem. 2017, 89, 3138–3145. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Luo, J.; Xu, Z.; Ma, X. A microfluidic chip containing a molecularly imprinted polymer and a DNA aptamer for voltammetric determination of carbofuran. Microchim. Acta 2018, 185, 295. [Google Scholar] [CrossRef]
- Li, S.; Wu, X.; Liu, C.; Yin, G.; Luo, J.; Xu, Z. Application of DNA aptamers as sensing layers for detection of carbofuran by electrogenerated chemiluminescence energy transfer. Anal. Chim. Acta 2016, 941, 94–100. [Google Scholar] [CrossRef]
- Xian-jin, L.; Cun-zheng, Z.; Yuan, L.; Li, W.; Qiu-hui, H.; Xian-jin, L. Selection of chlorpyrifos-binding ssDNA aptamer by SELEX. Jiangsu J. Agric. Sci. 2012, 1, 198–203. [Google Scholar]
- Bruno, J.G.; Chanpong, J. Methods of Producing Competitive Aptamer FRET Reagents and Assays. U.S. Patent No. 2006/0257915 A1, 16 November 2006. [Google Scholar]
- Jokar, M.; Safaralizadeh, M.H.; Hadizadeh, F.; Rahmani, F.; Kalani, M.R. Apta-nanosensor preparation and in vitro assay for rapid Diazinon detection using a computational molecular approach. J. Biomol. Struct. Dyn. 2017, 35, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, B.; Kalyani, N.; Anand, A.; Khan, E.; Das, S.; Bansal, V.; Kumar, A.; Sharma, T.K. GOLD SELEX: A novel SELEX approach for the development of high-affinity aptamers against small molecules without residual activity. Microchim. Acta 2020, 187, 618. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against the pesticide fipronil and sensitive detection in river water. Int. J. Mol. Sci. 2017, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Lu, X.; Hu, X.; Zhang, Y.; Zeng, L.; Chen, L.; Sun, M. In vitro selection of DNA aptamers binding pesticide fluoroacetamide. Biosci. Biotechnol. Biochem. 2016, 80, 823–832. [Google Scholar] [CrossRef]
- Bor, G.; Man, E.; Ugurlu, O.; Ceylan, A.E.; Balaban, S.; Durmus, C.; Pinar Gumus, Z.; Evran, S.; Timur, S. In vitro selection of aptamer for imidacloprid recognition as model analyte and construction of a water analysis platform. Electroanalysis 2020, 32, 1922–1929. [Google Scholar] [CrossRef]
- Barahona, F.; Bardliving, C.L.; Phifer, A.; Bruno, J.G.; Batt, C.A. An aptasensor based on polymer-gold nanoparticle composite microspheres for the detection of malathion using surface-enhanced raman spectroscopy. Ind. Biotechnol. 2013, 9, 42–50. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Nguyen, V.-T.; Park, J.G.; Gu, M.B. Detection of iprobenfos and edifenphos using a new multi-aptasensor. Anal. Chim. Acta 2015, 868, 60–66. [Google Scholar] [CrossRef]
- Wang, L.; Liu, X.; Zhang, Q.; Zhang, C.; Liu, Y.; Tu, K.; Tu, J. Selection of DNA aptamers that bind to four organophosphorus pesticides. Biotechnol. Lett. 2012, 34, 869–874. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, L.; Tu, Z.; Sun, X.; He, Q.; Lei, Z.; Xu, C.; Liu, Y.; Zhang, X.; Yang, J.; et al. Organophosphorus pesticides detection using broad-specific single-stranded DNA based fluorescence polarization aptamer assay. Biosens. Bioelectron. 2014, 55, 216–219. [Google Scholar] [CrossRef]
- Phan, H.T.M.; Bartelt-Hunt, S.; Rodenhausen, K.B.; Schubert, M.; Bartz, J.C. Investigation of bovine serum albumin (BSA) attachment onto self-assembled monolayers (SAMs) using combinatorial quartz crystal microbalance with dissipation (QCM-D) and spectroscopic ellipsometry (SE). PLoS ONE 2015, 10, e0141282. [Google Scholar] [CrossRef]
- Mendonsa, S.D.; Bowser, M.T. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal. Chem. 2004, 76, 5387–5392. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Goo, N.-I.; Kim, D.-E. Mechanism of DNA adsorption and desorption on graphene oxide. Langmuir 2014, 30, 12587–12595. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-W.; Tatavarty, R.; Kim, D.W.; Jung, H.-T.; Gu, M.B. Immobilization-free screening of aptamers assisted by graphene oxide. ChemComm 2012, 48, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.-T.; Kwon, Y.S.; Kim, J.H.; Gu, M.B. Multiple GO-SELEX for efficient screening of flexible aptamers. ChemComm 2014, 50, 10513–10516. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Macdonald, J.; Houghton, P.; Xiang, D.; Duan, W.; Shigdar, S. Truncation and mutation of a transferrin receptor aptamer enhances binding affinity. Nucleic Acid Ther. 2016, 26, 348–354. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, Y.; Sheng, Z.; Li, T.; Li, X. A colorimetric detection method of pesticide acetamiprid by fine-tuning aptamer length. Anal. Biochem. 2016, 513, 87–92. [Google Scholar] [CrossRef]
- Arvand, M.; Mirroshandel, A.A. An efficient fluorescence resonance energy transfer system from quantum dots to graphene oxide nano sheets: Application in a photoluminescence aptasensing probe for the sensitive detection of diazinon. Food Chem. 2019, 280, 115–122. [Google Scholar] [CrossRef]
- Hassani, S.; Akmal, M.R.; Salek-Maghsoudi, A.; Rahmani, S.; Ganjali, M.R.; Norouzi, P.; Abdollahi, M. Novel label-free electrochemical aptasensor for determination of Diazinon using gold nanoparticles-modified screen-printed gold electrode. Biosens. Bioelectron. 2018, 120, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Belinskaia, D.A.; Avdonin, P.V.; Avdonin, P.P.; Jenkins, R.O.; Goncharov, N.V. Rational in silico design of aptamers for organophosphates based on the example of paraoxon. Comput. Biol. Chem. 2019, 80, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Yan, J.; Xiong, H.; Liu, Y.; Peng, D.; Liu, Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 2018, 143, 5317–5338. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhao, G.; Liu, M.; Fan, L.; Cao, T. Aptamer-based colorimetric sensing of acetamiprid in soil samples: Sensitivity, selectivity and mechanism. J. Hazard. Mater. 2013, 260, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Weisshoff, H.; Wenzel, K.; Schulze-Rothe, S.; Nikolenko, H.; Davideit, H.; Becker, N.-P.; Göttel, P.; Srivatsa, G.S.; Dathe, M.; Müller, J.; et al. Characterization of aptamer BC 007 substance and product using circular dichroism and nuclear magnetic resonance spectroscopy. J. Pharm. Sci. 2018, 107, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.-W.; Kwok, J.; Law, A.W.L.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci.USA 2013, 110, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Oshannessy, D.J.; Brighamburke, M.; Soneson, K.K.; Hensley, P.; Brooks, I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: Use of nonlinear least squares analysis methods. Anal. Biochem. 1993, 212, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.L.; McKeague, M.; Smolke, C.D. Facile characterization of aptamer kinetic and equilibrium binding properties using surface plasmon resonance. Methods Enzymol 2014, 549, 451–466. [Google Scholar] [CrossRef]
- Mayer, K.M.; Hafner, J.H. Localized surface plasmon resonance sensors. Chem. Rev. 2011, 111, 3828–3857. [Google Scholar] [CrossRef]
- Hammond, J.L.; Bhalla, N.; Rafiee, S.D.; Estrela, P. Localized surface plasmon resonance as a biosensing platform for developing countries. Biosensors 2014, 4, 172–188. [Google Scholar] [CrossRef]
- Kelly, K.L.; Coronado, E.; Zhao, L.L.; Schatz, G.C. The optical properties of metal nanoparticles: The influence of size, shape, and dielectric environment. J. Phys. Chem. B 2003, 107, 668–677. [Google Scholar] [CrossRef]
- Unser, S.; Bruzas, I.; He, J.; Sagle, L. Localized surface plasmon resonance biosensing: Current challenges and approaches. Sensors 2015, 15, 15684–15716. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, M.; Ferreira Carlos, F.; Pedrosa, P.; Lopez, A.; Baptista, P.V. Gold nanoparticles for diagnostics: Advances towards points of care. Diagnostics 2016, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Saraji, M. Optical aptasensor based on silver nanoparticles for the colorimetric detection of adenosine. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 213, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Q.; Yuan, F.; Wang, L. Simple and sensitive turn-on luminescent detection of biothiols based on energy transfer between green-emitting upconversion nanocrystals and gold nanoparticles. Anal. Methods 2013, 5, 2873–2879. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2018, 10, 18–33. [Google Scholar] [CrossRef]
- Carnerero, J.M.; Jimenez-Ruiz, A.; Grueso, E.M.; Prado-Gotor, R. Understanding and improving aggregated gold nanoparticle/dsDNA interactions by molecular spectroscopy and deconvolution methods. Phys. Chem. Chem. Phys. 2017, 19, 16113–16123. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J. Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir 2012, 28, 3896–3902. [Google Scholar] [CrossRef]
- Bala, R.; Dhingra, S.; Kumar, M.; Bansal, K.; Mittal, S.; Sharma, R.K.; Wangoo, N. Detection of organophosphorus pesticide—Malathion in environmental samples using peptide and aptamer based nanoprobes. Chem. Eng. J. 2017, 311, 111–116. [Google Scholar] [CrossRef]
- Wang, S.; Su, L.; Wang, L.; Zhang, D.; Shen, G.; Ma, Y. Colorimetric determination of carbendazim based on the specific recognition of aptamer and the poly-diallyldimethylammonium chloride aggregation of gold nanoparticles. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 117809. [Google Scholar] [CrossRef]
- Luo, Y.; He, L.; Zhan, S.; Wu, Y.; Liu, L.; Zhi, W.; Zhou, P. Ultrasensitive resonance scattering (RS) spectral detection for trace tetracycline in milk using aptamer-coated nanogold (ACNG) as a catalyst. J. Agric. Food Chem. 2014, 62, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Bala, R.; Mittal, S.; Sharma, R.K.; Wangoo, N. A supersensitive silver nanoprobe based aptasensor for low cost detection of malathion residues in water and food samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 196, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Bala, R.; Kumar, M.; Bansal, K.; Sharma, R.K.; Wangoo, N. Ultrasensitive aptamer biosensor for malathion detection based on cationic polymer and gold nanoparticles. Biosens. Bioelectron. 2016, 85, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Sabela, M.; Balme, S.; Bechelany, M.; Janot, J.-M.; Bisetty, K. A review of gold and silver nanoparticle-based colorimetric sensing assays. Adv. Eng. Mater. 2017, 19, 1700270. [Google Scholar] [CrossRef]
- Chávez, J.L.; MacCuspie, R.I.; Stone, M.O.; Kelley-Loughnane, N. Colorimetric detection with aptamer–gold nanoparticle conjugates: Effect of aptamer length on response. J. Nanopart. Res. 2012, 14, 1166. [Google Scholar] [CrossRef]
- Qi, Y.; Chen, Y.; Xiu, F.-R.; Hou, J. An aptamer-based colorimetric sensing of acetamiprid in environmental samples: Convenience, sensitivity and practicability. Sens. Actuators B Chem. 2020, 304, 127359. [Google Scholar] [CrossRef]
- Weerathunge, P.; Behera, B.K.; Zihara, S.; Singh, M.; Prasad, S.N.; Hashmi, S.; Mariathomas, P.R.D.; Bansal, V.; Ramanathan, R. Dynamic interactions between peroxidase-mimic silver NanoZymes and chlorpyrifos-specific aptamers enable highly-specific pesticide sensing in river water. Anal. Chim. Acta 2019, 1083, 157–165. [Google Scholar] [CrossRef]
- Wang, R.-H.; Zhu, C.-L.; Wang, L.-L.; Xu, L.-Z.; Wang, W.-L.; Yang, C.; Zhang, Y. Dual-modal aptasensor for the detection of isocarbophos in vegetables. Talanta 2019, 205, 120094. [Google Scholar] [CrossRef]
- Li, X.; Jiang, X.; Liu, Q.; Liang, A.; Jiang, Z. Using N-doped carbon dots prepared rapidly by microwave digestion as nanoprobes and nanocatalysts for fluorescence determination of ultratrace isocarbophos with label-free aptamers. Nanomaterials 2019, 9, 223. [Google Scholar] [CrossRef]
- Abnous, K.; Danesh, N.M.; Ramezani, M.; Alibolandi, M.; Emrani, A.S.; Lavaee, P.; Taghdisi, S.M. A colorimetric gold nanoparticle aggregation assay for malathion based on target-induced hairpin structure assembly of complementary strands of aptamer. Microchim. Acta 2018, 185, 216. [Google Scholar] [CrossRef]
- Wang, P.; Wan, Y.; Aldalbahi, A.; Deng, S.; Su, Y.; Fan, C.; Yang, S. Aptamer-wrapped gold nanoparticles for the colorimetric detection of omethoate. Sci. China Chem. 2015, 59. [Google Scholar] [CrossRef]
- Li, X.; Shi, J.; Chen, C.; Li, W.; Han, L.; Lan, L.; Guo, Y.; Chang, Y.; Cai, J.; Ding, Y. One-step, visual and sensitive detection of phorate in blood based on a DNA–AgNC aptasensor. New J. Chem. 2018, 42, 6293–6298. [Google Scholar] [CrossRef]
- Hu, W.; Chen, Q.; Li, H.; Ouyang, Q.; Zhao, J. Fabricating a novel label-free aptasensor for acetamiprid by fluorescence resonance energy transfer between NH2-NaYF4: Yb, Ho@SiO2 and Au nanoparticles. Biosens. Bioelectron. 2016, 80, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Wang, S.; Wang, L.; Yan, Z.; Yi, H.; Zhang, D.; Shen, G.; Ma, Y. Fluorescent aptasensor for carbendazim detection in aqueous samples based on gold nanoparticles quenching Rhodamine B. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 225, 117511. [Google Scholar] [CrossRef] [PubMed]
- Arvand, M.; Mirroshandel, A.A. Highly-sensitive aptasensor based on fluorescence resonance energy transfer between l-cysteine capped ZnS quantum dots and graphene oxide sheets for the determination of edifenphos fungicide. Biosens. Bioelectron. 2017, 96, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, H.; Han, P.; Feng, X. Fluorescent aptasensing of chlorpyrifos based on the assembly of cationic conjugated polymer-aggregated gold nanoparticles and luminescent metal–organic frameworks. Analyst 2019, 144, 6025–6032. [Google Scholar] [CrossRef]
- Kim, T.-Y.; Lim, J.W.; Lim, M.-C.; Song, N.-E.; Woo, M.-A. Aptamer-based fluorescent assay for simple and sensitive detection of fipronil in liquid eggs. Biotechnol. Bioprocess Eng. 2020, 25, 246–254. [Google Scholar] [CrossRef]
- Li, X.; Tang, X.; Chen, X.; Qu, B.; Lu, L. Label-free and enzyme-free fluorescent isocarbophos aptasensor based on MWCNTs and G-quadruplex. Talanta 2018, 188, 232–237. [Google Scholar] [CrossRef]
- Bala, R.; Swami, A.; Tabujew, I.; Peneva, K.; Wangoo, N.; Sharma, R.K. Ultra-sensitive detection of malathion using quantum dots-polymer based fluorescence aptasensor. Biosens. Bioelectron. 2018, 104, 45–49. [Google Scholar] [CrossRef]
- Xiong, J.e.; Li, S.; Li, Y.; Chen, Y.; Liu, Y.; Gan, J.; Ju, J.; Xian, Y.; Xiong, X. Fluorescent aptamer-polyethylene glycol functionalized graphene oxide biosensor for profenofos detection in food. Chem. Res. Chin. Univ. 2019. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, B.; Cao, Y.; Guo, M.; Yu, Y. Fluorescence determination of omethoate based on a dual strategy for improving sensitivity. J. Agric. Food Chem. 2017, 65, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ye, H.; Sang, H.-Q.; Wang, D.-D. Aptamer-Based fluorescence assay for detection of isocarbophos and profenofos. Chin. J. Anal. Chem. 2016, 44, 799–803. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, C.; He, J.; Zhang, H.; Xu, Z. Fluorescence assay for three organophosphorus pesticides in agricultural products based on Magnetic-Assisted fluorescence labeling aptamer probe. Food Chem. 2020, 307, 125534. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Song, Y.; Fu, Q.; Du, D.; Luo, Y.; Wang, Y.; Xu, W.; Lin, Y. Aptasensor based on fluorophore-quencher nano-pair and smartphone spectrum reader for on-site quantification of multi-pesticides. Biosens. Bioelectron. 2018, 117, 75–83. [Google Scholar] [CrossRef]
- Dou, X.; Chu, X.; Kong, W.; Luo, J.; Yang, M. A gold-based nanobeacon probe for fluorescence sensing of organophosphorus pesticides. Anal. Chim. Acta 2015, 891, 291–297. [Google Scholar] [CrossRef]
- Swierczewska, M.; Lee, S.; Chen, X. The design and application of fluorophore–gold nanoparticle activatable probes. Phys. Chem. Chem. Phys. 2011, 13, 9929–9941. [Google Scholar] [CrossRef]
- Guo, J.; Liu, X.; Gao, H.; Bie, J.; Zhang, Y.; Liu, B.; Sun, C. Highly sensitive turn-on fluorescent detection of cartap via a nonconjugated gold nanoparticle–quantum dot pair mediated by inner filter effect. RSC Adv. 2014, 4, 27228–27235. [Google Scholar] [CrossRef]
- Srisantitham, S.; Sukwattanasinitt, M.; Unarunotai, S. Effect of pH on fluorescence quenching of organic dyes by graphene oxide. Colloids Surf. Physicochem. Eng. Asp. 2018, 550, 123–131. [Google Scholar] [CrossRef]
- Das, R.; Rajender, G.; Giri, P.K. Anomalous fluorescence enhancement and fluorescence quenching of graphene quantum dots by single walled carbon nanotubes. Phys. Chem. Chem. Phys. 2018, 20, 4527–4537. [Google Scholar] [CrossRef]
- Kim, M.S.; Cho, S.; Joo, S.H.; Lee, J.; Kwak, S.K.; Kim, M.I.; Lee, J. N- and B-Codoped graphene: A strong candidate to replace natural peroxidase in sensitive and selective bioassays. ACS Nano 2019, 13, 4312–4321. [Google Scholar] [CrossRef]
- Pokhrel, S.; Yadav, P.N. Functionalization of chitosan polymer and their applications. J. Macromol. Sci. Part A 2019, 56, 450–475. [Google Scholar] [CrossRef]
- Prabhakar, N.; Thakur, H.; Bharti, A.; Kaur, N. Chitosan-iron oxide nanocomposite based electrochemical aptasensor for determination of malathion. Anal. Chim. Acta 2016, 939, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Khosropour, H.; Rezaei, B.; Rezaei, P.; Ensafi, A.A. Ultrasensitive voltammetric and impedimetric aptasensor for diazinon pesticide detection by VS2 quantum dots-graphene nanoplatelets/carboxylated multiwalled carbon nanotubes as a new group nanocomposite for signal enrichment. Anal. Chim. Acta 2020, 1111, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, J.; Yao, Y.; Liu, H.; Huang, J.; Guo, Y.; Sun, X. Novel electrochemical aptasensor with dual signal amplification strategy for detection of acetamiprid. Sci. Total Environ. 2020, 705, 135905. [Google Scholar] [CrossRef]
- Roushani, M.; Nezhadali, A.; Jalilian, Z. An electrochemical chlorpyrifos aptasensor based on the use of a glassy carbon electrode modified with an electropolymerized aptamer-imprinted polymer and gold nanorods. Microchim. Acta 2018, 185, 551. [Google Scholar] [CrossRef]
- Fu, J.; Dong, H.; Zhao, Q.; Cheng, S.; Guo, Y.; Sun, X. Fabrication of refreshable aptasensor based on hydrophobic screen-printed carbon electrode interface. Sci. Total Environ. 2020, 712, 136410. [Google Scholar] [CrossRef]
- Hayat, A.; Marty, J.L. Aptamer based electrochemical sensors for emerging environmental pollutants. Front. Chem. 2014, 2, 41. [Google Scholar] [CrossRef]
- Selvolini, G.; Băjan, I.; Hosu, O.; Cristea, C.; Săndulescu, R.; Marrazza, G. DNA-Based sensor for the detection of an organophosphorus pesticide: Profenofos. Sensors 2018, 18, 2035. [Google Scholar] [CrossRef]
- Fan, L.; Zhang, C.; Yan, W.; Guo, Y.; Shuang, S.; Dong, C.; Bi, Y. Design of a facile and label-free electrochemical aptasensor for detection of atrazine. Talanta 2019, 201, 156–164. [Google Scholar] [CrossRef]
- Trotter, M.; Borst, N.; Thewes, R.; von Stetten, F. Review: Electrochemical DNA sensing—Principles, commercial systems, and applications. Biosens. Bioelectron. 2020, 154, 112069. [Google Scholar] [CrossRef]
- Li, F.; Yu, Z.; Han, X.; Lai, R.Y. Electrochemical aptamer-based sensors for food and water analysis: A review. Anal. Chim. Acta 2019, 1051, 1–23. [Google Scholar] [CrossRef]
- Yi, J.; Liu, Z.; Liu, J.; Liu, H.; Xia, F.; Tian, D.; Zhou, C. A label-free electrochemical aptasensor based on 3D porous CS/rGO/GCE for acetamiprid residue detection. Biosens. Bioelectron. 2020, 148, 111827. [Google Scholar] [CrossRef]
- Yao, Y.; Wang, G.X.; Shi, X.J.; Li, J.S.; Yang, F.Z.; Cheng, S.T.; Zhang, H.; Dong, H.W.; Guo, Y.M.; Sun, X.; et al. Ultrasensitive aptamer-based biosensor for acetamiprid using tetrahedral DNA nanostructures. J. Mater. Sci. 2020, 55, 15975–15987. [Google Scholar] [CrossRef]
- Jiao, Y.; Hou, W.; Fu, J.; Guo, Y.; Sun, X.; Wang, X.; Zhao, J. A nanostructured electrochemical aptasensor for highly sensitive detection of chlorpyrifos. Sens. Actuators B Chem. 2017, 243, 1164–1170. [Google Scholar] [CrossRef]
- Xu, G.; Huo, D.; Hou, C.; Zhao, Y.; Bao, J.; Yang, M.; Fa, H. A regenerative and selective electrochemical aptasensor based on copper oxide nanoflowers-single walled carbon nanotubes nanocomposite for chlorpyrifos detection. Talanta 2018, 178, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Jia, H.; Guo, Y.; Zhang, H.; Wang, Z.; Sun, X.; Zhao, J. An ultrasensitive aptasensor for chlorpyrifos based on ordered mesoporous carbon/ferrocene hybrid multiwalled carbon nanotubes. RSC Adv. 2016, 6, 58541–58548. [Google Scholar] [CrossRef]
- Xu, G.; Hou, J.; Zhao, Y.; Bao, J.; Yang, M.; Fa, H.; Yang, Y.; Li, L.; Huo, D.; Hou, C. Dual-signal aptamer sensor based on polydopamine-gold nanoparticles and exonuclease I for ultrasensitive malathion detection. Sens. Actuators B Chem. 2019, 287, 428–436. [Google Scholar] [CrossRef]
- Kaur, N.; Thakur, H.; Prabhakar, N. Multi walled carbon nanotubes embedded conducting polymer based electrochemical aptasensor for estimation of malathion. Microchem. J. 2019, 147, 393–402. [Google Scholar] [CrossRef]
- Jiao, Y.; Fu, J.; Hou, W.; Shi, Z.; Guo, Y.; Sun, X.; Yang, Q.; Li, F. Homogeneous electrochemical aptasensor based on a dual amplification strategy for sensitive detection of profenofos residues. New J. Chem. 2018, 42, 14642–14647. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, J.; Cheng, S.; Liu, H.; Li, F.; Guo, Y.; Sun, X. A Dual-Amplification electrochemical aptasensor for profenofos detection. J. Electrochem. Soc. 2020, 167, 027515. [Google Scholar] [CrossRef]
- Fu, J.; An, X.; Yao, Y.; Guo, Y.; Sun, X. Electrochemical aptasensor based on one step co-electrodeposition of aptamer and GO-CuNPs nanocomposite for organophosphorus pesticide detection. Sens. Actuators B Chem. 2019, 287, 503–509. [Google Scholar] [CrossRef]
- Fu, J.; Yao, Y.; An, X.; Wang, G.; Guo, Y.; Sun, X.; Li, F. Voltammetric determination of organophosphorus pesticides using a hairpin aptamer immobilized in a graphene oxide-chitosan composite. Microchim. Acta 2019, 187, 36. [Google Scholar] [CrossRef] [PubMed]
- Madianos, L.; Skotadis, E.; Tsekenis, G.; Patsiouras, L.; Tsigkourakos, M.; Tsoukalas, D. Ιmpedimetric nanoparticle aptasensor for selective and label free pesticide detection. Microelectron. Eng. 2018, 189, 39–45. [Google Scholar] [CrossRef]
- Madianos, L.; Tsekenis, G.; Skotadis, E.; Patsiouras, L.; Tsoukalas, D. A highly sensitive impedimetric aptasensor for the selective detection of acetamiprid and atrazine based on microwires formed by platinum nanoparticles. Biosens. Bioelectron. 2018, 101, 268–274. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, D.; Chen, Z.; Li, L.; You, T. An ultra-sensitive aptasensor based on carbon nanohorns/gold nanoparticles composites for impedimetric detection of carbendazim at picogram levels. J. Colloid Interface Sci. 2019, 546, 92–100. [Google Scholar] [CrossRef]
- Zhang, J.; Fang, X.; Wu, J.; Hu, Z.; Jiang, Y.; Qi, H.; Zheng, L.; Xuan, X. An interdigitated microelectrode based aptasensor for real-time and ultratrace detection of four organophosphorus pesticides. Biosens. Bioelectron. 2020, 150, 111879. [Google Scholar] [CrossRef]
- Villalonga, A.; Pérez-Calabuig, A.M.; Villalonga, R. Electrochemical biosensors based on nucleic acid aptamers. Anal. Bioanal. Chem. 2020, 412, 55–72. [Google Scholar] [CrossRef]
- Erdem, A.; Kerman, K.; Meric, B.; Ozsoz, M. Methylene blue as a novel electrochemical hybridization indicator. Electroanalysis 2001, 13, 219–223. [Google Scholar] [CrossRef]
- Ding, L.; Jiang, D.; Wen, Z.; Xu, Y.; Guo, Y.; Ding, C.; Wang, K. Ultrasensitive and visible light-responsive photoelectrochemical aptasensor for edifenphos based on Zinc phthalocyanine sensitized MoS2 nanosheets. Biosens. Bioelectron. 2020, 150, 111867. [Google Scholar] [CrossRef]
- Nie, Y.; Teng, Y.; Li, P.; Liu, W.; Shi, Q.; Zhang, Y. Label-free aptamer-based sensor for specific detection of malathion residues by surface-enhanced Raman scattering. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 191, 271–276. [Google Scholar] [CrossRef]
- Pang, S.; Labuza, T.P.; He, L. Development of a single aptamer-based surface enhanced Raman scattering method for rapid detection of multiple pesticides. Analyst 2014, 139, 1895–1901. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, D.; Wang, Q.; Wang, Q. Aptamer-based resonance light scattering for sensitive detection of acetamiprid. Anal. Sci. 2016, 32, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Zargar, T.; Khayamian, T.; Jafari, M.T. Immobilized aptamer paper spray ionization source for ion mobility spectrometry. J. Pharm. Biomed. Anal. 2017, 132, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Nobukawa, A.; Osaki, T.; Morimoto, Y.; Kamiya, K.; Misawa, N.; Takeuchi, S. Pesticide vapor sensing using an aptamer, nanopore, and agarose gel on a chip. Lab Chip 2017, 17, 2421–2425. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, G.; Wu, S.; Zhang, Q. Aptamer-based microcantilever-array biosensor for profenofos detection. Anal. Chim. Acta 2018, 1020, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Deng, J.; Zhang, M.; Shi, G.; Zhou, T. Quantum dot-DNA aptamer conjugates coupled with capillary electrophoresis: A universal strategy for ratiometric detection of organophosphorus pesticides. Talanta 2016, 146, 55–61. [Google Scholar] [CrossRef]
- Xu, K.; Zhou, R.; Takei, K.; Hong, M. Toward flexible surface-enhanced raman scattering (SERS) sensors for point-of-care diagnostics. Adv. Sci. 2019, 6, 1900925. [Google Scholar] [CrossRef]
- Blackie, E.J.; Le Ru, E.C.; Etchegoin, P.G. Single-Molecule surface-enhanced raman spectroscopy of nonresonant molecules. J. Am. Chem. Soc. 2009, 131, 14466–14472. [Google Scholar] [CrossRef]
- Kettler, H.; White, K.; Hawkes, S. Mapping the Landscape of Diagnostics for Sexually Transmitted Infections: Key Findings and Recommandations; Special Programme for Research and Training in Tropical Diseases; WHO: Geneva, Switzerland, 2004. [Google Scholar]
- Mabey, D.; Peeling, R.W.; Ustianowski, A.; Perkins, M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004, 2, 231–240. [Google Scholar] [CrossRef]
- Martínez-del-Río, J.; Martínez Vidal, J.L.; Garrido Frenich, A. Economic evaluation of pesticide-residue analysis of vegetables. Trends Anal. Chem. 2013, 44, 90–97. [Google Scholar] [CrossRef]
- Peeling, R.W.; Mabey, D. Point-of-care tests for diagnosing infections in the developing world. Clin. Microbiol. Infect. 2010, 16, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Xiao, M.; Fu, Q.; Yu, S.; Shen, H.; Bian, H.; Tang, Y. A portable smart-phone readout device for the detection of mercury contamination based on an aptamer-assay nanosensor. Sensors 2016, 16, 1871. [Google Scholar] [CrossRef] [PubMed]
- Hoilett, O.S.; Walker, J.F.; Balash, B.M.; Jaras, N.J.; Boppana, S.; Linnes, J.C. KickStat: A coin-sized potentiostat for high-resolution electrochemical analysis. Sensors 2020, 20, 2407. [Google Scholar] [CrossRef]
- Dryden, M.D.M.; Wheeler, A.R. DStat: A versatile, open-source potentiostat for electroanalysis and integration. PLoS ONE 2015, 10, e0140349. [Google Scholar] [CrossRef] [PubMed]
- Boucaud-Maitre, D.; Rambourg, M.O.; Sinno-Tellier, S.; Puskarczyk, E.; Pineau, X.; Kammerer, M.; Bloch, J.; Langrand, J. Human exposure to banned pesticides reported to the French Poison Control Centers: 2012–2016. Environ. Toxicol. Pharmacol. 2019, 69, 51–56. [Google Scholar] [CrossRef]
- Nguyen Quang, N.; Perret, G.; Ducongé, F. Applications of high-throughput sequencing for in vitro selection and characterization of aptamers. Pharmaceuticals 2016, 9, 76. [Google Scholar] [CrossRef]
- Cole, K.H.; Lupták, A. Chapter eighteen—High-throughput methods in aptamer discovery and analysis. In Methods Enzymol; Shukla, A.K., Ed.; Academic Press: New York, NY, USA, 2019; Volume 621, pp. 329–346. [Google Scholar]
- Kinghorn, A.B.; Fraser, L.A.; Lang, S.; Shiu, S.C.-C.; Tanner, J.A. Aptamer bioinformatics. Int. J. Mol. Sci. 2017, 18, 2516. [Google Scholar] [CrossRef]
- Wu, D.; Feagin, T.; Mage, P.; Rangel, A.; Wan, L.; Li, A.; Coller, J.; Eisenstein, M.; Pitteri, S.; Soh, H.T. Automated platform for high-throughput screening of base-modified aptamers for affinity and specificity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ruscito, A.; DeRosa, M.C. Small-Molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef]
- Presnell, K.V.; Alper, H.S. Thermodynamic and first-principles biomolecular simulations applied to synthetic biology: Promoter and aptamer designs. Mol. Syst. Des. Eng. 2018, 3, 19–37. [Google Scholar] [CrossRef]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar] [CrossRef] [PubMed]


| Pesticide | Group | Selection Approach | Aptamer Sequence | KD | Ref. |
|---|---|---|---|---|---|
| Acetamiprid | Insecticide | Immobilization of library on beads via capture probe (Capture-SELEX) | TGTAATTTGTCTGCAGCGATTCTTGATCGCTGACACCATATTATGAAGA | 4.98 µM | [33] |
| Aldicarb | Insecticide | N.A. | CCGGTGGGTGGTCAGCACCTGGGGGAGTCCGGATATGGCCCAGCGCATCACCAGTTCGCAAGC | N.A. | [34] |
| Atrazine | Herbicide | Immobilization of biotinylated-target on streptavidin-modified magnetic beads | TGTACCGTCTGAGCGATTCGTACGAACGGCTTTGTACTGTTTGCACTGGCGGATTTAGCCAGTCAGTGTTAAGGAGTGC | 0.62 nM | [35] |
| Capture-SELEX | TGTACCGTCTGAGCGATTCGTACTTTATTCGGGAAGGGTATCAGCGGGGTTCAACAAGCCAGTCAGTGTTAAGGAGTGC | N.A. | [36] | ||
| Truncation of sequence obtained from capture-SELEX | ACCGTCTGAGCGATTCGTACTTTATTCGGGAAGGGTATCAGCGGGG | 3.7 nM | |||
| Capillary electrophoresis-SELEX | CTACGCTAGCTTGTATGCCCATCTGACCTCTGTGCTGCTA | 890 nM | [37] | ||
| Bromacil | Herbicide | Immobilization of biotinylated-target on streptavidin-modified magnetic beads | TGTACCGTCTGAGCGATTCGTACTGTGGGCACCAATCGTACCCAATACTTGCGAATCAGCCAGTCAGTGTTAAGGAGTGC | 9.6 nM | [38] |
| Carbendazim | Fungicide | Immobilization of target-conjugated BSA on microplate | CGACACAGCGGAGGCCACCCGCCCACCAGCCCCTGCAGCTCCTGTACCTGTGTGTGTG | 60.2 nM | [39] |
| GGGCACACAACAACCGATGGTCCAGCCACCCGAATGACCAGCCCACCCGCCACCCCGCG | 65 nM | ||||
| Carbofuran | Insecticide | N.A. | CACCTGGGGGAGTATTGCGGAGGAAAGAGAACACTGGGGCAGATATGGGCCAGCAGGTC | N.A. | [40,41] |
| Chlorpyrifos | Insecticide | Immobilization of biotinylated-target on streptavidin-resin beads | CCTGCCACGCTCCGCAAGCTTAGGGTTACGCCTGCAGCGATTCTTGATCGCGCTGCTGGTAATCCTTCTTTAAGCTTGGCACCCGCATCGT | N.A. | [42] |
| Diazinon | Insecticide | N.A. | ATCCGTCACACCTGCTCTAATATAGAGGTATTGCTCTTGGACAAGGTACAGGGATGGTGTTGGCTCCCGTAT | N.A. | [43] |
| Computational screening of Bruno’s reported sequences [43] | ATCCGTCACACCTGCTCTAATATAGAGGTATTGCTCTTGGACAAGGTACAGGGATGGTGTTGGCTCCCGTAT | 55.51 µM * | [44] | ||
| Dichlorvos | Insecticide | GOLD SELEX | GGAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCG | 0.85 µM | [45] |
| Fipronil | Insecticide | Immobilization of target on magnetic beads via covalent coupling | TGTACCGTCTGAGCGATTCGTACAGTTTCTGGAGGACTGGGCGGGGTGACGGTTATAAGCCAGTCAGTGTTAAGGAGTGC | 48 nM | [46] |
| Fluoroacetamide | Rodenticide | Immobilization of target-conjugated BSA on microplate | ACCTGCAGGCGCGAGTTTCAGATCAAAACTTGTCTGGCGT | 1 µM | [47] |
| Imidacloprid | Insecticide | GO-SELEX | TGTCGTCTACGGTTTTGGTTGTTGTTTGTTGGTGGGTGTA | −2.86 kcal/mol † | [48] |
| GGTGTGTTTGTTGTTGTTCTTGGCTGGTTTTTCTTCCTG | −6.41 kcal/mol † | ||||
| Malathion | Insecticide | Immobilization of target on PharmaLink affinity column | ATCCGTCACACCTGCTCTTATACACAATTGTTTTTCTCTTAACTTCTTGACTGCTGGTGTTGGCTCCCGTAT | N.A. | [49] |
| Dual targets: edifenphos (ED) and iprobenfos (IP) | Insecticide | GO-SELEX | CGTACGGAATTCGCTAGCTAAGGGATTCCTGTAGAAGGAGCAGTCTGGATCCGAGCTCCACGTG | ED = 38 nM | [50] |
| IP = 1.67 µM | |||||
| Multiple targets (4OPs): phorate (PR), profenofos (PF), isocarbophos (IS) and omethoate (OM) | Insecticide | Capture-SELEX | AAGCTTTTTTGACTGACTGCAGCGATTCTTGATCGCCACGGTCTGGAAAAAGAG | PR = 1.43 µM | [51] |
| PF = 1.25 µM | |||||
| IS = 0.9 µM | |||||
| OM = 2 µM | |||||
| AAGCTTGCTTTATAGCCTGCAGCGATTCTTGATCGGAAAAGGCTGAGAGCTACGC | PR = 1.11 µM | ||||
| PF = 1 µM | |||||
| IS = 0.83 µM | |||||
| OM = 2.5 µM | |||||
| In silico design and truncation of above-mentioned sequence | AGCTTGCTGCAGCGATTCTTGATCGCCACAGAGCT | N.A. | [52] |
| Pesticide | Method | LOD | Linear Range | Sample | Ref. |
|---|---|---|---|---|---|
| Acetamiprid | Colorimetry | 5 nM | 75 nM–7.5 µM | Soil | [67] |
| Acetamiprid | Colorimetry | 0.56 nM | 8.7–920 nM | Waste water, soil and cucumber | [89] |
| Carbendazim | Colorimetry | 2.2 nM | 2.2–500 nM | Water | [83] |
| Chlorpyrifos | Colorimetry | 11.3 ppm | 35–210 ppm | River water | [90] |
| Isocarbophos | Colorimetry | 7.1 µg/L | 50–500 µg/L | Chinese cabbage, brassica rape and lettuce leaves | [91] |
| Isocarbophos | Colorimetry | 0.015 µg/L | 0.25–1.5 µg/L | Sewage, farm land water and pond water | [92] |
| Diazinon | Colorimetry | 17.903 nM | 0.141–0.65 nM | N.A. | [44] |
| Malathion | Colorimetry | 0.06 pM | 0.5-1000 pM | Lake water and apple | [86] |
| Malathion | Colorimetry | 1.94 pM | 0.01–0.75 nM | Lake water and apple | [82] |
| Malathion | Colorimetry | 1:00 pM 3 pM (in serum) | 5 pM–10 nM | Spiked human serum | [93] |
| Malathion | Colorimetry | 0.5 pM | 0.01 nM–0.75 nM | Lake water, tap water and apple | [85] |
| Omethoate | Colorimetry | 0.1 µM | 0.1 µM–10 µM | Soil | [94] |
| Phorate | Colorimetry | 0.012 ng/mL | 0–25 µg/mL | Human blood | [95] |
| Iprobenfos (IP) and edifenphos (ED) | Colorimetry | 10 nM (IB) and 5 nM (ED) | 10–100 nM (IP), 5–25 nM (ED) | Paddy and polished rice | [50] |
| Pesticide | Method | LOD | Linear Range | Sample | Ref. |
|---|---|---|---|---|---|
| Acetamiprid | Fluorometry | 3.2 nM | 50 nM–1000 nM | Adulterated tea | [96] |
| Carbendazim | Fluorometry | 2.33 nM | 2.33–800 nM | Water | [97] |
| Diazinon | Fluorometry | 0.13 nM | 1.05–206 nM | River water, apple and cucumber | [63] |
| Edifenphos | Fluorometry | 1.3 × 10−4 mg/L | 0.5–6 µg/mL | Surface water and rice | [98] |
| Carbofuran | Fluorometry | 3.8 nM | 5–600 nM | Tap water, cucumber, cabbage, kiwifruit and apple | [99] |
| Fipronil | Fluorometry | 105 nM | 5 nM–500 nM | River water | [46] |
| Fipronil | Fluorometry | 53.8 ppb | 25–300 ppb | Liquid egg | [100] |
| Isocarbophos | Fluorometry | 10 nM | 10–500 nM | Cabbage | [101] |
| Isocarbophos | Fluorometry | 0.11 µg/L | 0.25–1.5 µg/L | Sewage, farm land water and pond water | [92] |
| Isocarbophos | Phosphorescence | 0.57 µg/L | 5–160 µg/L | Chinese cabbage, brassica rape and lettuce leaves | [91] |
| Isocarbophos | Fluorometry | 0.11 µg/L | 0.25–1.5 µg/L | Sewage, farm land water and pond water | [92] |
| Malathion | Fluorometry | 4:00 p.m. | 0.01 nM–1 µM | Tap water, lake water, soil and orange juice | [102] |
| Profenofos | Fluorometry | 0.21 ng/mL | 0.5–100 ng/mL | Tap water, cabbage and milk | [103] |
| Omethoate | Fluorometry | 0.041 µM and | 0.1–17 nM and | Cabbage and river water | [104] |
| 0.029 pM by unpolarized and polarized fluorometry | 0.1 pM–1 µM by unpolarized and polarized fluorometry | ||||
| Isocarbophos (IS) and profenofos (PF) | Fluorometry | 11.4 µM (IS) and 14 µM (PF) | 50–500 µM | Water | [105] |
| Trichorfon (TC), glyphosate (GP) and malathion (ML) | Fluorometry | 72.20 ng/L (TC), 88.80 ng/L (GP) and 195.37 ng/L (ML) | 0.1 µg/L–10 mg/L | Lettuce and carrot | [106] |
| Chlorpyrifos (CP), diazinon (DA) and malathion (ML) | Fluorometric-lateral flow strip | 0.73 ng/mL (CP), 6.7 ng/mL (DA) and 0.74 ng/mL (ML) | 1–5 ng/mL (CP), 2–4 ng/mL (DA), and 1–3 ng/mL (ML) | Maize, long bean, cauliflower, eggplant, oyster mushroom, shiitake mush-room, apple, orange, tomato, blueberry, spinach, lettuce and cabbage | [107] |
| 4OPs | Fluorometry | LOQ values are 19.2 nM (PR), 13.4 nM (PF), 17.2 nM (IS) and 23.4 nM (OM) | 0.01–10 mg/kg | Cabbage | [52] |
| 4OPs | Fluorometry | 0.384 µM (PR), 0.134 µM (PF), 0.035 µM (IS) and 2.35 µM (OM) | 0.268–26.8 µM (PF) and 0.346–34.6 µM (IS); no obvious relationship for PR and OM. | Dried tangerine peel | [108] |
| Pesticide | Method | LOD | Linear Range | Sample | Ref. |
|---|---|---|---|---|---|
| Acetamiprid | Voltammetry | 71.2 fM | 0.1 pM–0.1 µM | Tea | [125] |
| Acetamiprid | Voltammetry | 0.3 pM | 1 pM–1 µM | Lettuce and rape | [117] |
| Acetamiprid | Voltammetry | 0.077 pM | 0.1 pM–10 nM | Lettuce, cabbage and spinach | [126] |
| Aldicarb | Voltammetry | 0.1 pM | 0.25–250 pM | Lake and river water | [122] |
| Carbofuran | Voltammetry | 67 pM | 0.2–50 nM | Chinese cabbage, chili, lettuce, tomato, apple, banana, tangerine and watermelon | [40] |
| Carbofuran | Voltammetry | 0.033 ng/mL | 0.1 ng/mL–100 µg/mL | Cabbage, lettuce, leek and pakchoi | [127] |
| Carbofuran | Voltammetry | 70 pg/mL | 0.1–150 ng/mL | Apple, celery and cabbage | [128] |
| Carbofuran | Voltammetry | 0.35 fM | 1 fM–0.4 pM | Apple and lettuce | [118] |
| Carbofuran | Voltammetry | 0.33 ng/mL | 1 ng/mL–100 µg/mL | Lettuce, leek and pakchoi | [129] |
| Diazinon | Voltammetry | 0.0169 nM | 0.1–1000 nM | Plasma of Wistar rat | [64] |
| Diazinon | Voltammetry | 0.11 fM (DPV) and 2 fM (EIS) | 0.5 fM–10 nM (DPV) | Human serum, river water, soil, apple and lettuce | [116] |
| and EIS | 0.1 fM–10 nM (EIS) | ||||
| Malathion | Voltammetry | 0.001 ng/mL | 0.001–10 ng/mL | Lettuce and soil | [115] |
| Malathion | Voltammetry | 0.5 ng/mL | 0.5–600 ng/mL | Cauliflower and cabbage | [130] |
| Malathion | Voltammetry | 0.5 fM | 0.1 fM–0.1 µM | Lettuce | [131] |
| Profenofos | Voltammetry | 0.01 ng/mL | 0 to 6.5 ng/mL | Rape | [132] |
| Profenofos | Voltammetry | 0.27 µM | 0.1–10 µM | Pear juice | [121] |
| Profenofos | Voltammetry | 0.052 ng/mL | 0.1 ng/mL–100 µg/mL | Spinach, lettuce and cabbage | [133] |
| 4OPs | Voltammetry | 0.003 nM (PF), 0.3 nM (PR), 0.03 nM (IS), and 0.3 nM (OM) | 1–1000 nM (PR), 0.01–100 nM (PF), 0.1–1000 nM (IS), and 1–500 nM (OM) | Rape and spinach | [134] |
| 4OPs | Voltammetry | 0.1 nM (PR), 0.01 nM (PF), 0.01 nM (IS), and 0.1 nM (OM) | 0.01–1,000 nM (PF), 0.1–800 nM (PR), 0.01–1,000 nM (IS), and 0.1–100 nM (OM) | Rape, cabbage, spinach and baby cabbage | [135] |
| 4OPs | Refreshable voltammetry | N.A. | Qualitative detection above 1 µM | Baby cabbage | [119] |
| Acetamiprid | EIS | 6:00 pM | 40 pM–1 µM | N.A. | [136] |
| Acetamiprid | EIS | 1:00 pM | 10 pM–100 nM | Water | [137] |
| Aldicarb | EIS | 10:00 pM | 100 pM–1 µM | Water | [137] |
| Aldicarb | EIS | 40 pM | 0.6 nM–1 µM | N.A. | [136] |
| Carbendazim | EIS | 8.2 pg/mL | 10 pg/mL–10 ng/mL | Mango juice, soya milk, tomato and plum | [39] |
| Carbendazim | EIS | 0.5 pg/mL | 1–1000 pg/mL | Lettuce and orange juice | [138] |
| 4OPs | Capacitance | 0.24 fM (PR), 0.34 fM (PF), 0.38 fM (IS) and 1.67 fM (OM) | 3.84 fM–3.84 nM (PR), 2.68 fM–2.68 nM (PF), 3.46 fM–3.46 nM (IS) and 4.69 fM–4.69 nM (OM) | Lettuce | [139] |
| Pesticide | Method | LOD | Linear Range | Sample | Ref. |
|---|---|---|---|---|---|
| Aldicarb | ECL | 9.6 pM | 40 pM–4 nM | Turnip, cabbage, potato, banana, celery and irrigation water | [34] |
| Carbofuran | ECL | 0.88 pM | 20 pM–8 nM | Cowpea, cabbage, chili, tomato, lettuce, banana, celery, carrot, capsicum and apple | [41] |
| Edifenphos | ECL | 1.667 ng/L | 5 ng/L–10 µg/L | Rice | [142] |
| Malathion | SERS | 3.3 µg/mL | 3.3–33.3 µg/mL | N.A. | [49] |
| Malathion | SERS | N.A. | 0.5–10 µM | Tap water | [143] |
| 4OPs | SERS | 0.4 µM (PR), 14 µM (PF), 3.4 µM (IS), and 24 µM (OM) | 0–3.8 mM (PR): not available for other pesticides | Apple juice | [144] |
| Acetamiprid | Resonance Light Scattering (RLS) | 1.2 nM | 0–100 nM | Lake water | [145] |
| Acetamiprid | Sample extraction in ion mobility spectrometry | 1.8 ng/mL | 5–300 ng/ml | Wastewater, tomato and cucumber | [146] |
| Omethoate | Ionic current measurement | 4.8 nM (in solution) and 100 ppb (in vapor) | N.A. | N.A. | [147] |
| Profenofos | Microcantilever | 1.3 ng/mL (3.5 nM) | 5–1000 ng/mL | Chinese chive | [148] |
| 4OPs | Fluorometric-capillary electrophoresis (CE) | 0.20 µM (PR), 0.10 µM (PF), 0.17 µM (IS) and 0.23 µM (OM) | 0.6–10 µM (PR), 0.3–10 µM (PF), 0.5–10 µM (IS) and 0.7–10 µM (OM) | Apple | [149] |
| ASSURED | Sensing Principle | |||||
|---|---|---|---|---|---|---|
| Colorimetry | Fluorometry | Electrochemistry | ECL | SERS | Others | |
| Affordable | +++ | ++ | ++ | + | + | + |
| Sensitive | ++ | ++ | +++ | ++ | + | + |
| Specific | +++ | +++ | +++ | +++ | +++ | +++ |
| User-friendly | ++ | ++ | ++ | ++ | + | + |
| Rapid and robust | ++ | ++ | +++ | ++ | ++ | + |
| Equipment-free | +++ | ++ | + | + | + | + |
| Deliverable to end user | + | + | + | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phopin, K.; Tantimongcolwat, T. Pesticide Aptasensors—State of the Art and Perspectives. Sensors 2020, 20, 6809. https://doi.org/10.3390/s20236809
Phopin K, Tantimongcolwat T. Pesticide Aptasensors—State of the Art and Perspectives. Sensors. 2020; 20(23):6809. https://doi.org/10.3390/s20236809
Chicago/Turabian StylePhopin, Kamonrat, and Tanawut Tantimongcolwat. 2020. "Pesticide Aptasensors—State of the Art and Perspectives" Sensors 20, no. 23: 6809. https://doi.org/10.3390/s20236809
APA StylePhopin, K., & Tantimongcolwat, T. (2020). Pesticide Aptasensors—State of the Art and Perspectives. Sensors, 20(23), 6809. https://doi.org/10.3390/s20236809





