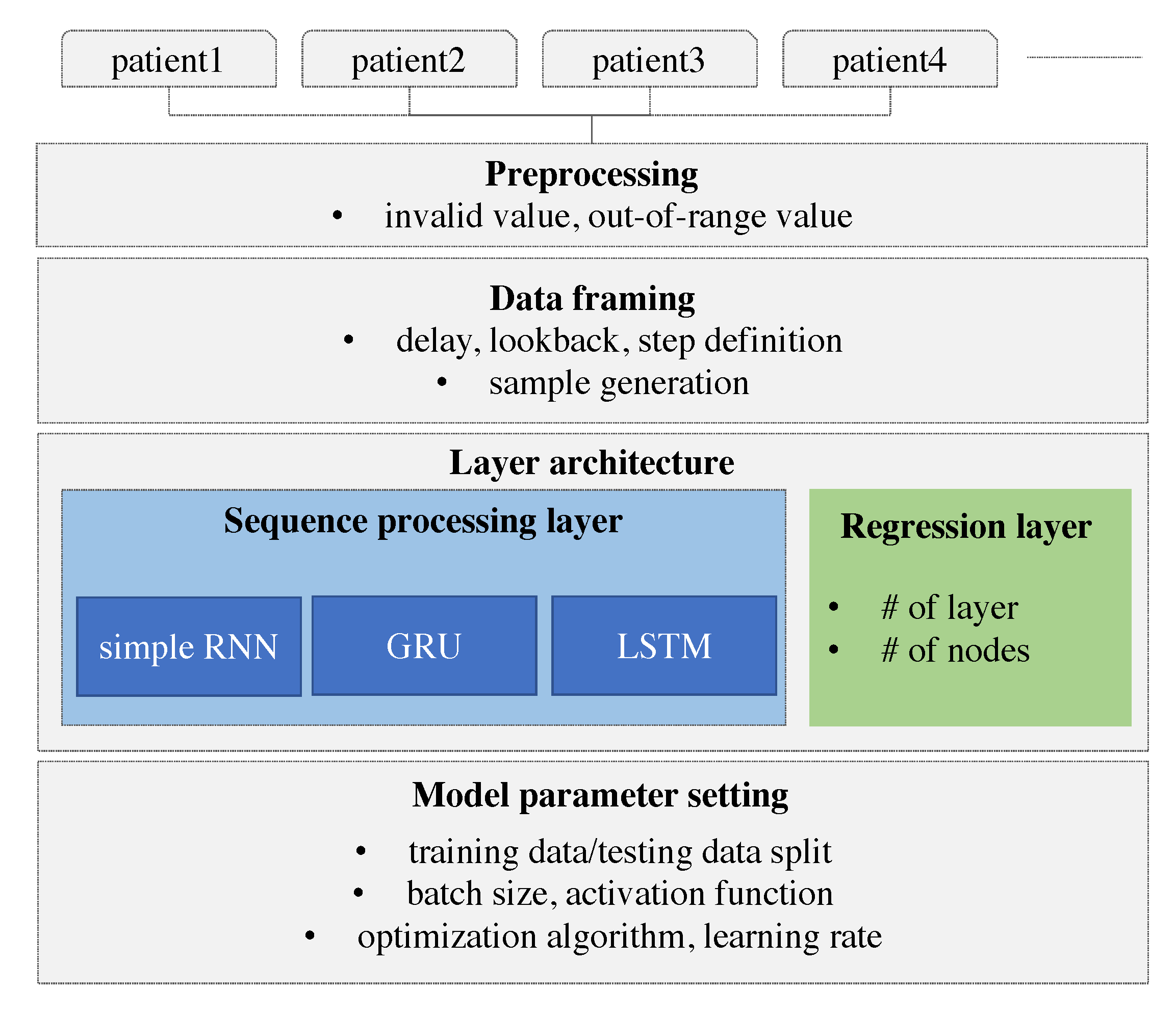
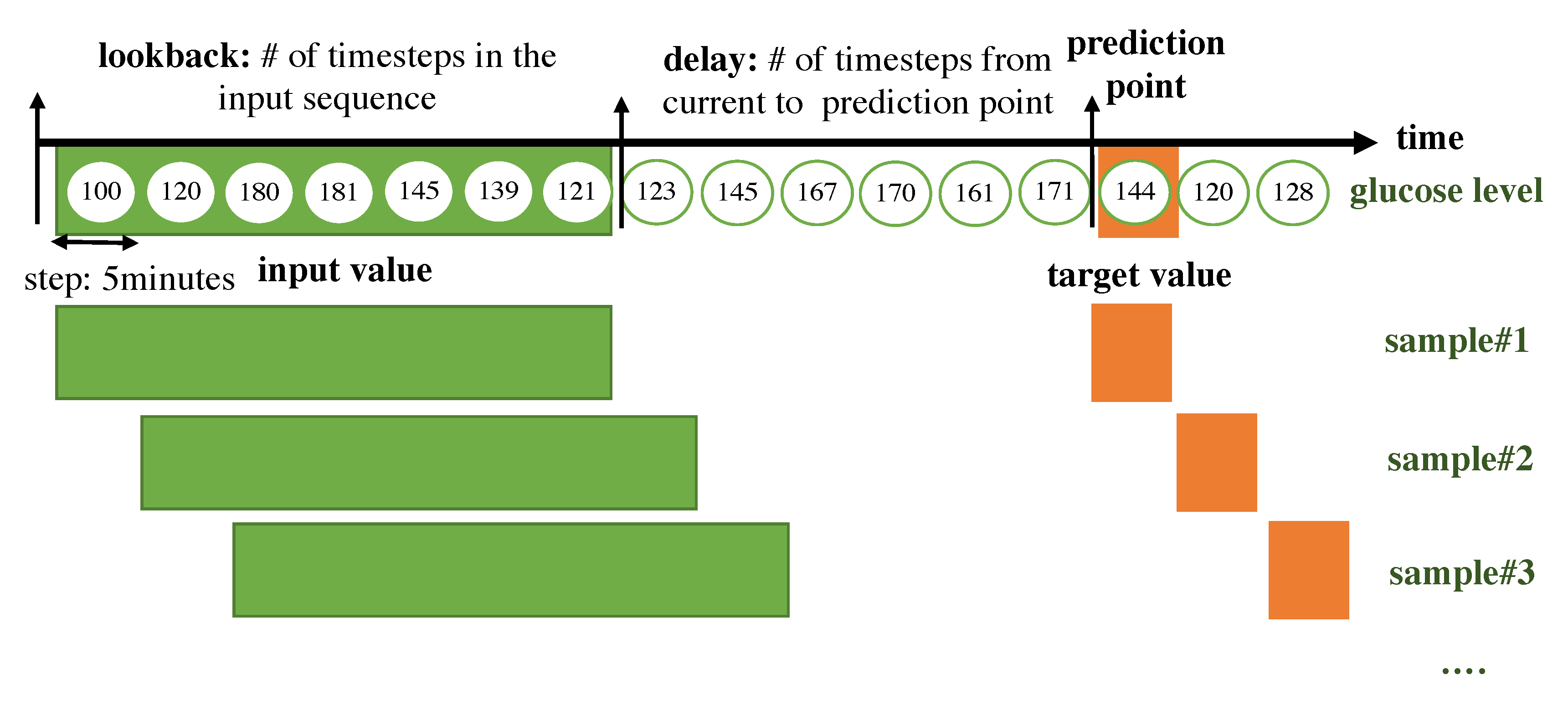
1. Introduction
Diabetes is a major risk factor for patients suffering from various diseases including cardiovascular disease [
1]. The socioeconomic losses resulting from acute and chronic complications and the burden of medical expenses for individual patients are enormous [
2,
3]. It is also widely known that diabetic patients experience longer hospitalization periods and have higher mortality rates from other diseases than general patients without diabetes [
4,
5]. Therefore, it is critical to thoroughly monitor, predict, and regulate blood glucose in diabetic patients.
Most previous studies on the prediction of blood glucose were conducted on outpatients, and most of these patients were those with Type-1 diabetes [
6,
7,
8,
9,
10,
11]. As inpatients are hospitalized owing to various diseases, have different personal characteristics, and show dynamic changes during hospitalization, the fluctuation of blood glucose in these inpatients is more severe than in outpatients, and it is, therefore, difficult to predict blood glucose. Inpatients have higher risk of hyperglycemia and hypoglycemia for reasons such as instability of vital signs due to various diseases, stress, and inflammation, immune responses, and drugs administered for treatment [
12,
13].
The role of medical professionals, especially doctors, is the key to controlling blood glucose in hospitalized patients. On the basis of the blood glucose value measured in the patient’s ward, doctors empirically predict changes in blood glucose levels and adjust drugs including insulin. However, this system is very inefficient, particularly because the number of medical professionals who specialize in diabetes is small compared to the number of diabetic patients. This issue needs to be resolved because excessive labor of medical personnel is demanded in such cases.
Recently, automated systems using artificial intelligence (AI) were investigated in various engineering fields, and the results have been used as a basis for real-life considerations. Medical specialists have attempted to combine AI with medicine in line with this trend of the times. However, the amount of information generated by the human body is close to infinity, and signals from various organs are complexly affected. In the field of AI, the analysis of limited information can lead to an incorrect result; subsequently, the application of an incorrect result is applied to real patients, it can lead to fatal results. Therefore, research involving the application of AI in the medical field is lagging behind that in other fields. However, the utilization of AI is a trend that is difficult to resist, and it is likely to be used in the medical field while reducing errors. We, therefore, believed that the prediction of blood glucose was appropriate as a starting point for the study because such an avenue is less likely to cause immediate harm to patients. Our work has the following motivations. The research uses continuous glucose monitoring for Type-2 patients who make up a major portion in diabetics. Continuous glucose monitoring (CGM) has been performed for Type-1 patients who are usually children and need parents’ care. Parents who have a role to monitor their children’s glucose level often use a CGM device. Thus, previous works for glucose prediction were focused on Type-1 diabetes. Type-2 patients, who are usually adults, are somewhat able to control their glucose level and are reluctant to use CGM devices which are expensive. However, when the patients are in the hospital, their glucose level should be monitored because of its high variability. The prediction model will eventually help to automatically control the amount of insulin according to hypoglycemia, hyperglycemia, and nocturnal glucose. With these motivations, we collected blood glucose data from hospitalized patients with Type-2 diabetes via a CGM system. We then developed a personalized prediction model that applies a recurrent neural network (RNN) to the blood glucose data collected through the CGM. For this, we examined various types of models with different model architectures and hyperparameters, designed data frames to be fitted to the prediction model, and performed experiments. Our work has a contribution in three folds. First, we performed the continuous glucose monitoring for Type-2 inpatients. Second, we found the best architecture for the RNN-based model to capture the patterns from the time-series of the continuous glucose level. Third, our RNN-based model showed comparative results with outpatients with Type-1.
2. Related Work
Several studies on blood glucose prediction have been conducted. Type-2 diabetes accounts for 90% of all diabetes, but there are few related studies. It is difficult to predict blood glucose in the case of hospitalized patients with Type-2 diabetes since blood glucose fluctuations are high. There are a few studies on outbreak prediction in hospitalized patients with Type-2 diabetes. We investigated studies using AI on the glucose level prediction using CGM data in patients with Type-1 diabetes and on the outbreak prediction for Type-2 diabetes.
Most of the blood glucose prediction studies used CGM data and diet information of Type-1 diabetes patients [
6,
7,
8,
9,
10,
11,
14,
15,
16,
17,
18,
19,
20,
21,
22,
23,
24]. Pérez-Gandía et al. [
6] predicted blood glucose after 15, 30, and 45 min after a given time using an artificial neural network (ANN) model and used the CGM data from 15 Type-1 diabetic patients. The root mean square error (RMSE) was used to evaluate the accuracy of the model in a glucose prediction study, and a continuous glucose error grid analysis (CG-EMA) and mean absolute difference percentage (MAD) analysis were also conducted. In this study, RMSE values of 10, 18, and 27 mg/dL were found corresponding to 15, 30, and 45 min, respectively.
Pappada et al. [
7] collected and analyzed data on 27 Type-1 diabetic patients using the CGM data. They used a feedforward neural network (FNN) model to predict blood glucose levels 75 min after a given time. The FNN model yielded an RMSE of 43.9 mg/dL and MAD of 22.1. A majority of the predicted values (92.3%) were contained within zones A (62.3%) and B (30.0%) of the CG-EMA and were therefore considered clinically acceptable.
Zarkogianni et al. [
8] evaluated four glucose prediction models for patients with Type-1 diabetes via the CGM data. The four models were based on an FNN, a self-organizing map (SOM), a neuro-fuzzy network with wavelets as activation functions (WFNN), and a linear regression model (LRM). Each model predicted blood glucose 30, 60, and 120 min after a given time, and the predictive performance was compared. The model using SOM showed better results than using the FNN, WFNN, and LRM. The RMSE values for 30, 60, and 120 min corresponded to 12.29, 21.06, and 33.68 mg/dL. In addition, according to the CG-EMA, the model using SOM showed relatively high accuracy for not only euglycemia but also hypoglycemia and hyperglycemia.
Mhaskar et al. [
9] analyzed clinical data from 25 Type-1 diabetic patients using the CGM data and predicted blood glucose 30 min after a given time using deep neural networks (DNNs), especially deep convolutional neural networks (DCNNs). While most studies dealt with one patient at a time, these scholars considered the data of only a certain percentage of patients in the dataset as the training data and tested the remainder of the patients. Based on the CG-EGA, for 50% of the data used for training, the percentage of accurate prediction and predictions with benign consequences was 96.43% in the hypoglycemic range, 97.96% in the euglycemic range, and 85.29% in the hyperglycemic range.
Sun et al. [
10] trained and tested on 26 datasets from 20 Type-1 diabetic patients using long-short term memory (LSTM) and bi-directional LSTM (Bi-LSTM)-based deep neural network. RMSE values for 15, 30, 45, 60 min were 11.633, 21.747, 30.215, 36.918 mg/dL.
Li et al. [
11] analyzed the data from 10 patients with Type-1 diabetes and proposed a deep learning algorithm using a multi-layer convolutional recurrent neural network (CRNN) model. RMSE values for 30 and 60 min were 21.07 and 33.27 mg/dL.
Sparacino et al. [
14] predicted the hypoglycemic threshold 20–25 min ahead using a first-order autoregressive (AR) model. They collected glucose for 48 h using a CGM device that was monitored every 3 min on 28 Type-1 diabetes volunteers. The RMSE values are 18.78 and 34.64 mg/dL for 30 and 45 min. They showed that even using this simple method, glucose can be predicted in advance.
Mougiakakou et al. [
15] developed models for simulating glucose–insulin metabolism. They used CGM data, insulin, and food intake from four children with Type-1 diabetes as the input to the models. The FNN, RNN/Free-Run (FR), and RNN/Teacher-Forcing (TF) models were compared. RNN/FR ignores glucose measurements available during training. RNN/TF replaces the actual output during training with the corresponding available glucose measurement. They have shown that models using RNN trained with the Real-Time Recurrent Learning (RTRL) algorithm can more accurately simulate metabolism in children with Type-1 diabetes.
Turksoy et al. [
16] used linear ARMAX models composed of autoregressive (AR), moving average (MA), and external inputs (X). They proposed the hypoglycemia alarm system for preventing hypoglycemia before it happens with glucose concentration, insulin, and physical activity information. CGM data from 14 Type-1 diabetic young adults were used for the prediction and alarm algorithm. In the real-time case, the optimal performance was the RMSE value of 11.7 mg/dL for 30 min.
Zecchin et al. [
17] proposed a prediction algorithm combined with an NN model with a first-order polynomial extrapolation algorithm for short-time glucose prediction using CGM data and carbohydrate intake. They monitored 15 Type-1 diabetic patients for 7 days using a CGM system that returns glucose values every minute. They showed that using carbohydrate intake information improves the accuracy of short-term prediction of glucose concentration.
Robertson et al. [
18] performed blood glucose level (BGL) prediction using an Elman RNN model with the AIDA freeware diabetes simulator. CGM data, meal intake, and insulin injections were used as inputs to the model. The most accurate predictions in their study were in the nocturnal period of the 24-h day cycle. The results were an RMSE value of 0.15 ± 0.04 SD mmol/L for short-term (15, 30, 45, and 60 min) and an RMSE value of 0.14 ± 0.16 SD mmol/L for long-term predictions (8 and 10 h).
Georga et al. [
19] proposed the application of random forests (RF) and RReliefF feature evaluation algorithms on Type-1 diabetes data. RReliefF is a feature ranking algorithm for regression problems. They evaluated several features, CGM data, food intake, plasma insulin concentration, energy expenditure, and time of the day, extracted from medical and lifestyle self-monitoring data. They showed that the information on physical activities is able to improve performance.
Jaouher et al. [
20] proposed an ANN-based method for predicting blood glucose levels in Type-1 diabetes using CGM data as inputs. They investigated real 13 Type-1 diabetic patients to validate their ANN model. The RMSE values were 6.43 mg/dL, 7.45 mg/dL, 8.13 mg/dL, and 9.03 mg/dL for 15, 30, 45, and 60 min. They suggested that using only CGM data as inputs and limiting human intervention for improving the quality of life of Type-1 diabetic patients.
Martinsson et al. [
21] proposed an RNN model to predict blood glucose levels. CGM data from 6 Type-1 diabetic patients (OhioT1DM [
22]) were used for training and evaluation. OhioT1DM dataset consists of blood glucose level values for two men and four women. The RMSE values were 18.867 mg/dL and 31.403 mg/dL for 30 and 60 min. They pointed out that larger data sets and standards are needed.
Aliberti et al. [
23] developed a patient-specialized prediction model based on LSTM. They used OhioT1DM dataset for training and validation. Among the 6 patients, the RMSE values of the patient with the best predicted result were 11.55 mg/dL, 19.86 mg/dL, 25 mg/dL, and 30.95 mg/dL for 30, 45, 60, and 90 min. The worst results were 11.52 mg/dL, 19.58 mg/dL, 27.67 mg/dL, and 43.99 mg/dL for 30, 45, 60, and 90 min.
Carrillo-Moreno et al. [
24] proposed an LSTM model using historical glucose levels, insulin units, and carbohydrate intake data. They used CGM data from 3 Type-1 diabetic patients and created 12 models with various combinations of patient-specific, prediction horizon (PH), LSTM layers, and the number of neurons for performance comparison. They found that the predictor with a PH of 30 min is the best performance.
As far as we know, no study on blood glucose prediction has been conducted in patients with Type-2 diabetes. The current state is that the prediction model for Type-2 diabetes outbreak has been proposed [
25,
26]. Wu et al. [
25] proposed a new model using Weka open source machine learning software to predict Type-2 diabetes mellitus. Personal health data and medical examination results from 768 patients were used to train and test whether patients had diabetes. The number of times pregnant, plasma glucose concentration at 2 h in an oral glucose tolerance test, diastolic blood pressure, triceps skin fold thickness, 2-h serum insulin, body mass index, diabetes pedigree function, age, class variable were used as features. Diabetes prediction accuracy is 96%.
Kazerouni et al. [
26] proposed Type-2 diabetes mellitus prediction using data mining algorithms based on the long-noncoding RNAs’ (lncRNA) expression. They used data from 100 Type-2 diabetic patients and 100 healthy individuals. As features, 6 lncRNA expressions, gender, age, weight, height, BMI and fetal bovine serum (FBS) were used. They applied four classification models of K-nearest neighbor (KNN), support vector machine (SVM), logistic regression, and ANN and compared their diagnostic performance. Logistic regression and SVM showed 95% of mean area under the receiver operating characteristic (ROC) curve. These studies are in patients with Type-2 diabetes, but differs from our study, which predicts blood glucose with CGM data.
Previous works developed from statistical models, AR species to deep learning, RNN. However, they limit the experiment to Type-1 diabetics, outpatients, and the limited number of patients and the short period. In this work, we set the experiment venue to an in-hospital patient with Type-2 diabetes.
6. Discussion
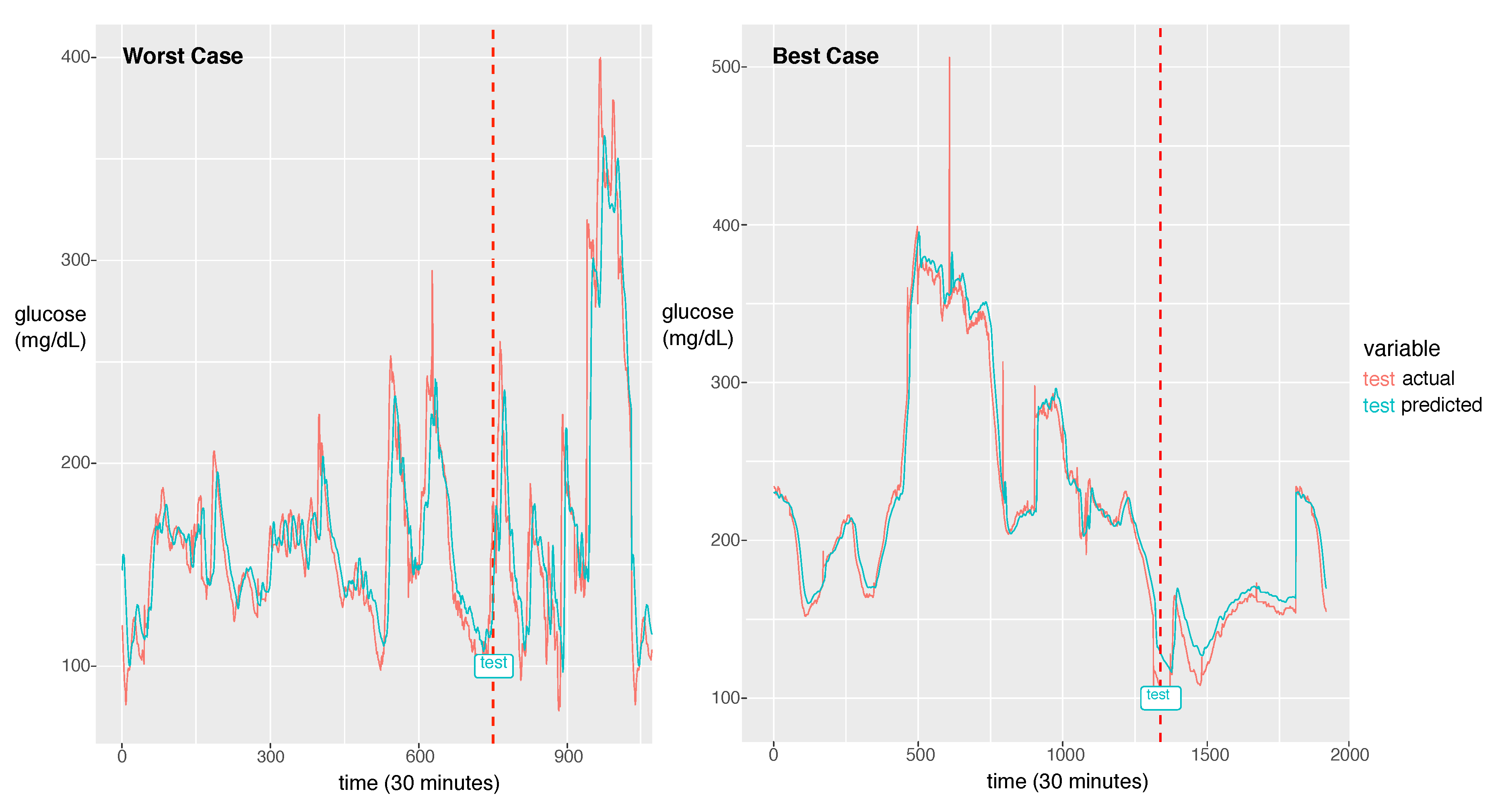
For the best model, the RMSE was 21.46 and the MAPE was calculated as 11.11%. The actual glucose value and the prediction value obtained using the best model for a patient with a 7:3 sampling split are shown in
Figure 6.
The model could learn the sharp peak in the training phase and generate a larger error in the sharp peak zone compared to the smooth zone. The glucose prediction is expected to be sensitive to low values (hypoglycemia), but our model is not sensitive to high values (hyperglycemia). Thus, this problem can be overlooked when this algorithm is implemented in practice. As a future work, we will investigate the solution for the large error in hyperglycemia.
In
Figure 6, we demonstrate two cases of the best and the worst of the prediction performance. We could notice a big difference between the two. The case that the fluctuation with high frequency exhibits bad performance had an RMSE of 44.15 and the best case of the fluctuation with low frequency exhibits the best performance with an RMSE of 8.35. As a future work, we will update the model to capture the high frequent fluctuation. A probable solution is to train the model using the change rate of glucose as data instead of the glucose value itself. Another difference between the two cases is that the worst case has a much shorter experimental period than the best case. From this point of view, to achieve a good performance, enough training data should be provided. The other solution would be data augmentation to provide enough data for the fluctuation with high frequency.
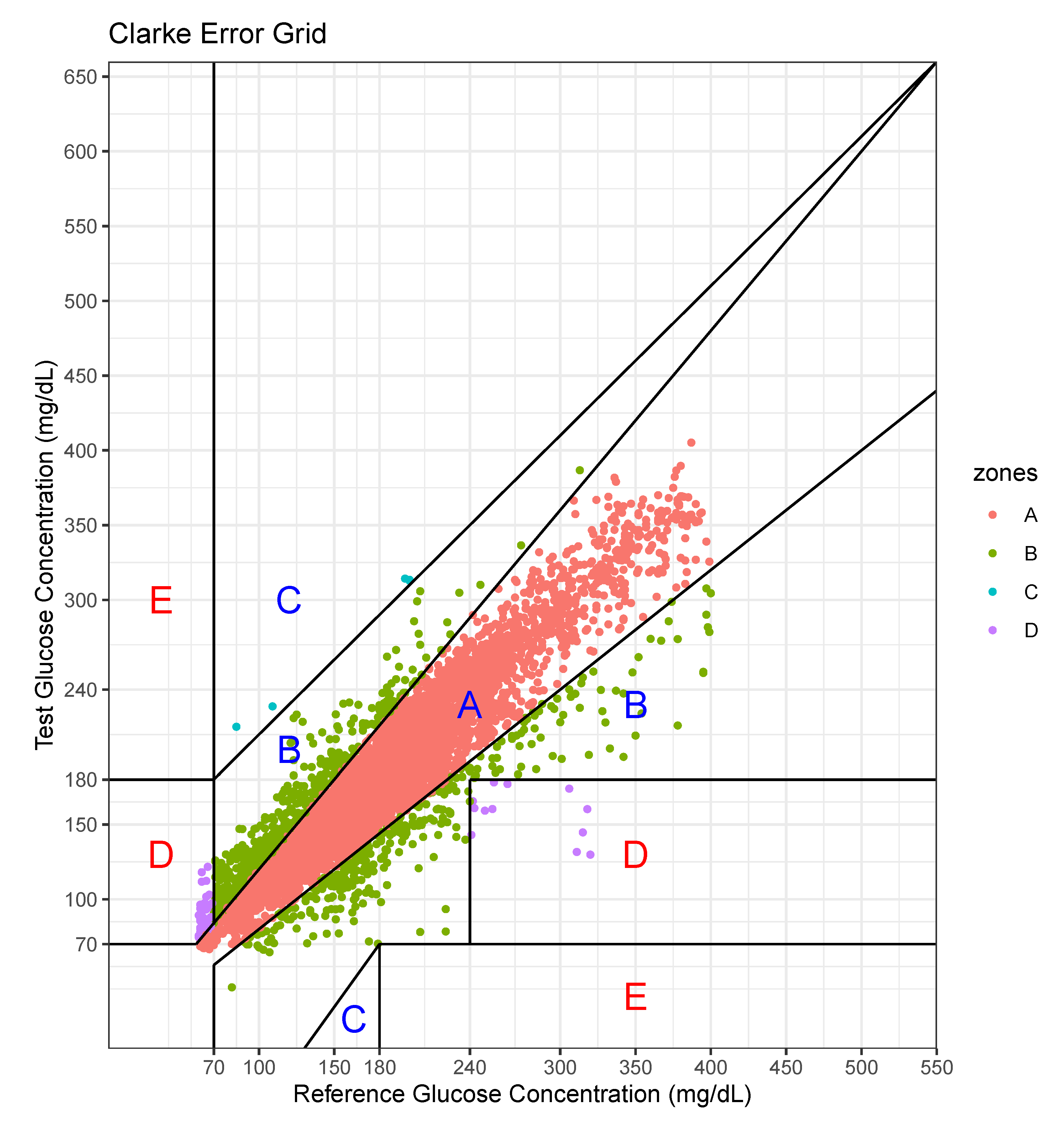
We performed an error analysis for the predicted values, as shown in
Figure 7. The error analysis follows the Clarke error grid lines according to the criteria [
30]. This graph confirms the very good clinical agreement between measured and predicted glucose values. Most of predicted values were located in the A zone. Occasional pairs of points fall in the B zone which is the slight overestimation/underestimation zone. Only a few points fall in the erroneous C and D zones. No point falls in the E zone which is a significantly erroneous zone. The percentage of data points in each zone is shown in
Table 7.
For this case, the model tended to overestimate the glucose level, especially at low levels of glucose.
In hospitals, the meals are served regularly and other food is prohibited. However, patients can take meals at different times and every person’s daily caloric intake is individual. Furthermore, the intake of other food is hard to be tracked unless patients voluntarily record their meal intake. Thus, in reality, it is difficult to record the exact event information. The event information including meal and insulin inject are important, but we could not afford to collect these data. However, our model is still working due to the following reasons. We collected the data from a hospital, so the same condition is applied to all patients in our experiments. Even in the absence of the event information, the model still can learn the sequence patterns reflecting event effects from the past history including the results of the event. Second, even if the event information becomes available, the stochastic event cannot be incorporated into the forecasting model because the model is required to predict the stochastic event as well. Only periodically deterministic events can be employed in the model because this information can be recorded in the input prior. Nevertheless, the event information would increase the model performance. As a future work, we will utilize the predefined and circadian event such as the meal intake, medicine intake, shot injection and treatment. In future work, we will further develop the model, incorporating more information regarding aspects such as insulin doses and food intake, which significantly affect the glucose level. Thus far, we have not recorded such information, but we plan to collect these details. Second, we developed an individual model that uses the patient’s own data. The number of patients involved for data collection was limited to 20, so it was difficult to leverage others’ data. However, when a large-scale data set becomes available, all patient glucose data can be used as the input. Then, the input will be in the form of vectors composed of patients’ own glucose data and others’ glucose data. The drawback in the current work, however, is that a significant portion of individual data are required for prediction; however, this can be resolved when the model exploits other patients’ data. Third, we will employ more advanced models such as a hybrid model incorporating a CNN and an RNN, a 1D CNN for a long sequence, and an attention-based model for achieving better performance. Dendritic neuron model considering the nonlinearity of synapses can be a good candidate for improving performance [
31,
32]. Neural network models including RNN or CNN only consider the linearity of synapses in the model. We will study how to incorporate Dendritic neuron model and RNN to derive the sequence pattern based on the nonlinearity.