DNAzyme Sensor for the Detection of Ca2+ Using Resistive Pulse Sensing
Abstract
:1. Introduction
2. Experimental
2.1. Chemicals and Reagents
2.2. Custom DNA Oligonucleotides
2.3. Particle Preparation
2.4. RPS Set Up
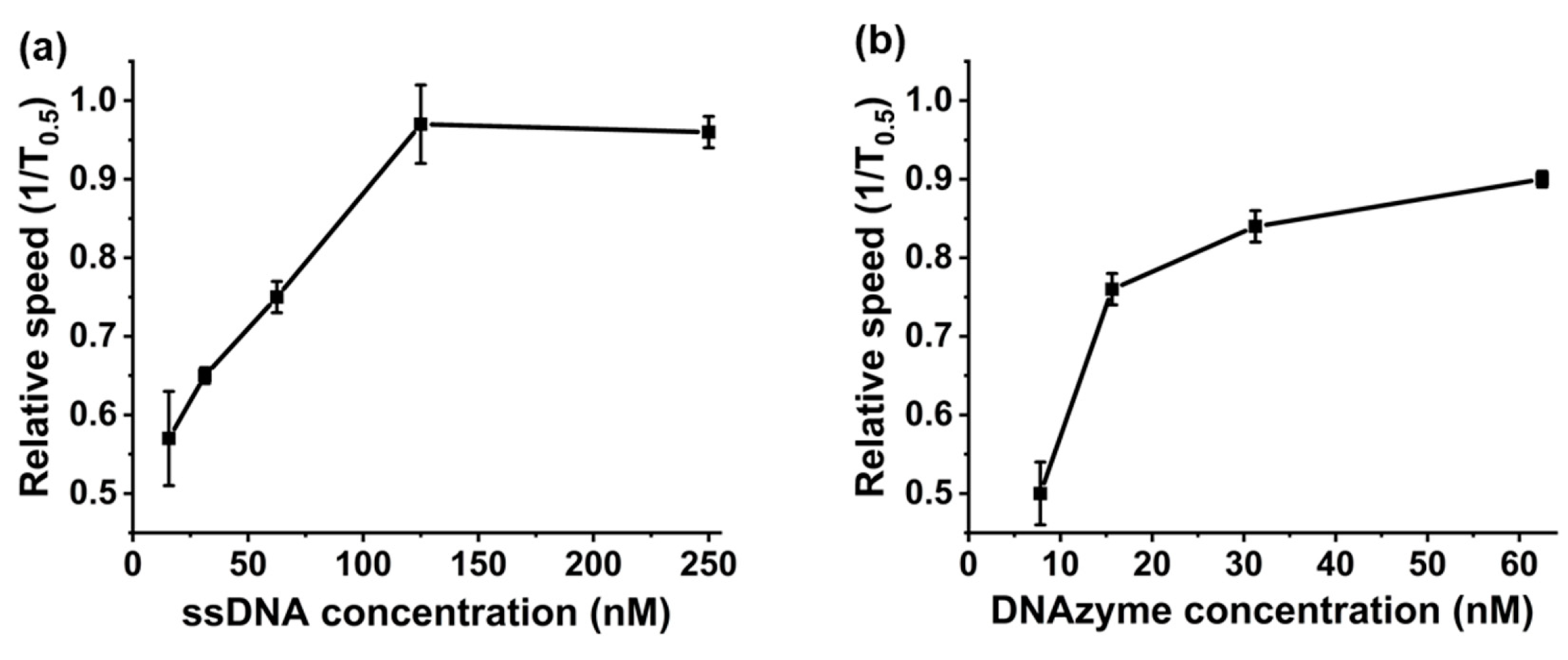
2.5. Saturation Point Testing
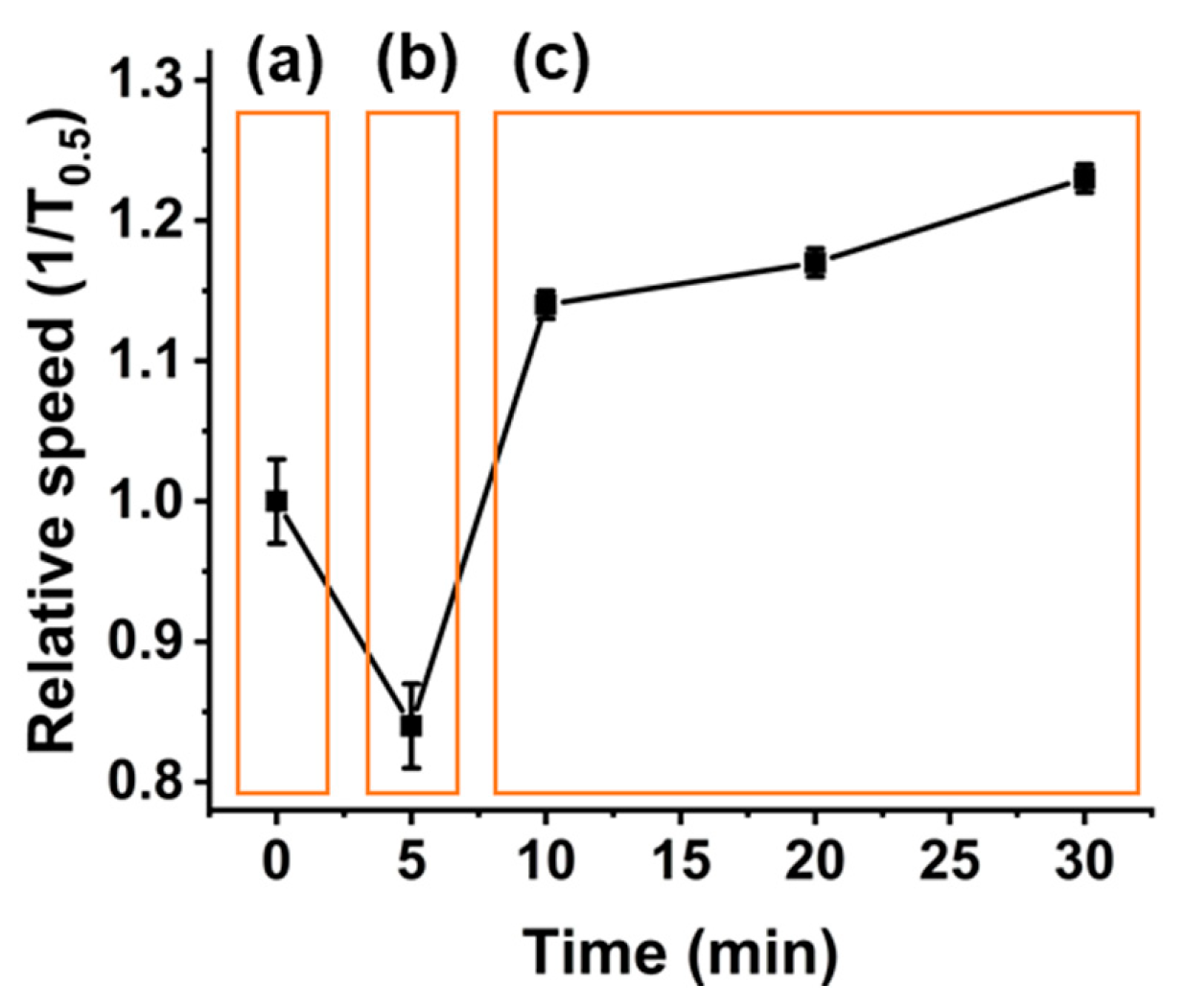
2.6. Binding Time
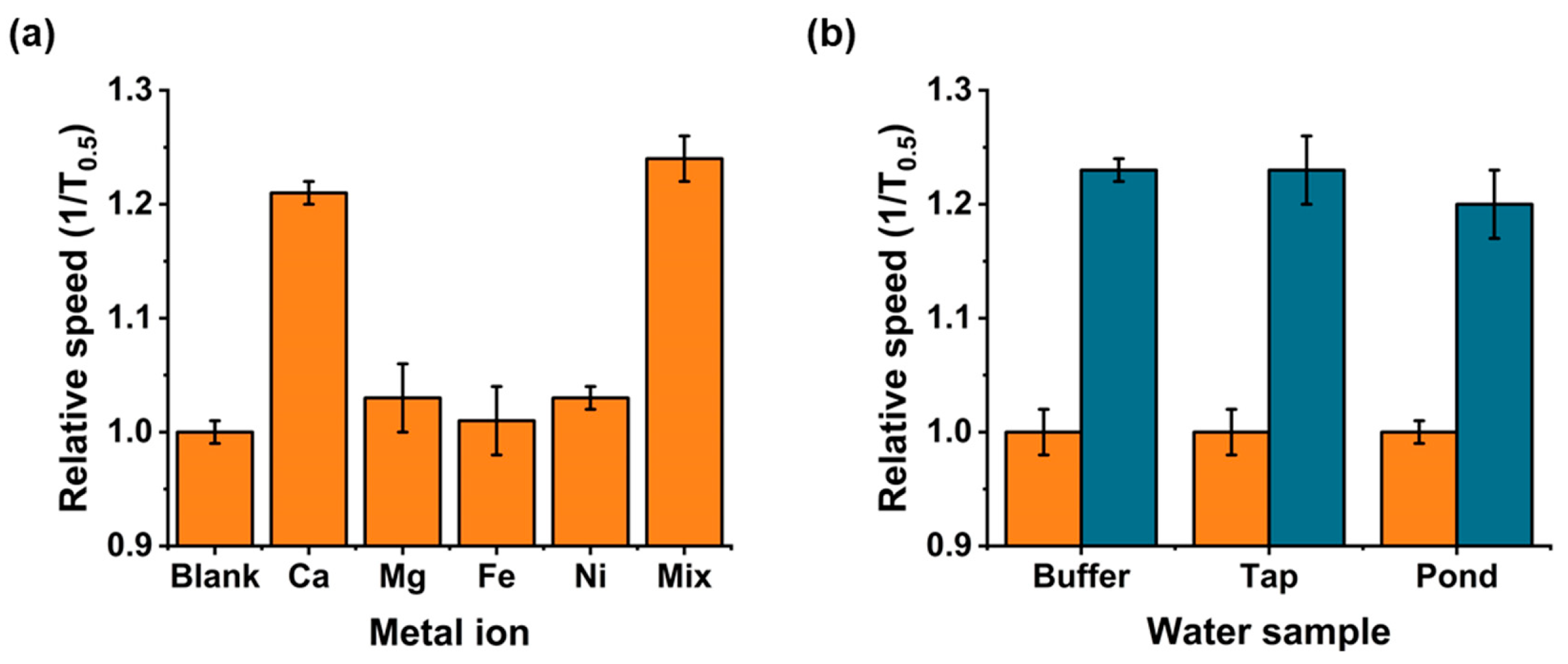
2.7. Metal Ion Interferences
2.8. Environmental Water Samples
2.9. Particle Speed
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sui, B.; Liu, X.; Wang, M.; Belfield, K.D. A Highly Selective Fluorescence Turn-On Sensor for Extracellular Calcium Ion Detection. Chem. A Eur. J. 2016, 22, 10351–10354. [Google Scholar] [CrossRef] [PubMed]
- He, H.Z.; Wang, M.; Chan, D.S.H.; Leung, C.H.; Lin, X.; Lin, J.M.; Ma, D.L. A parallel G-quadruplex-selective luminescent probe for the detection of nanomolar calcium(II) ion. Methods 2013, 64, 212–217. [Google Scholar] [CrossRef]
- Yu, T.; Zhou, W.; Liu, J. Screening of DNAzyme mutants for highly sensitive and selective detection of calcium in milk. Anal. Methods 2018, 10, 1740–1746. [Google Scholar] [CrossRef]
- Li, L.; Ma, X.; Dong, W.; Miao, P.; Tang, Y. Electrochemical Determination of Ca2+ Based On Recycling Formation of Highly Selective DNAzyme and Gold Nanoparticle-Mediated Amplification. Bioconjug. Chem. 2018, 29, 1021–1024. [Google Scholar] [CrossRef]
- Kozisek, F. Regulations for calcium, magnesium or hardness in drinking water in the European Union member states. Regul. Toxicol. Pharmacol. 2020, 112, 104589. [Google Scholar] [CrossRef] [PubMed]
- Hampe, D.; Gleisberg, B.; Akhmadaliev, S.; Rugel, G.; Merchel, S. Determination of 41Ca with LSC and AMS: Method development, modifications and applications. J. Radioanal. Nucl. Chem. 2013, 296, 617–624. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.W.; Kim, D.; Kim, D.; Lee, I.H.; Jon, S. Bioinspired colorimetric detection of calcium(II) ions in serum using calsequestrin-functionalized gold nanoparticles. Angew. Chem. Int. Ed. 2009, 48, 4138–4141. [Google Scholar] [CrossRef]
- Zhou, W.; Saran, R.; Liu, J. Metal Sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. [Google Scholar] [CrossRef] [Green Version]
- Ankireddy, S.R.; Kim, J. Highly selective and sensitive detection of calcium (II) ions in human serum using novel fluorescent carbon dots. Sens. Actuators B Chem. 2018, 255, 3425–3433. [Google Scholar] [CrossRef]
- Yue, J.; Li, L.; Cao, L.; Zan, M.; Yang, D.; Wang, Z.; Chang, Z.; Mei, Q.; Miao, P.; Dong, W.F. Two-Step Hydrothermal Preparation of Carbon Dots for Calcium Ion Detection. ACS Appl. Mater. Interfaces 2019, 11, 44566–44572. [Google Scholar] [CrossRef]
- Araújo, A.N.; Costa, R.C.C.; Lima, J.L.F.C.; Reis, B.F. Sequential injection system in flame atomic absorption spectrometry for the determination of calcium and magnesium in mineral waters. Anal. Chim. Acta 1998, 358, 111–119. [Google Scholar] [CrossRef]
- Szymczycha-Madeja, A.; Welna, M.; Pohl, P. Method Validation for Multi-Elemental Analysis of Dialyzable and Non-dialyzable Fractions of Coffee Brews by F AAS and ICP OES: A Bioaccessibility Study. Food Anal. Methods 2019, 12, 198–216. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.L.S.; Fritz, J. Rapid determination of magnesium and calcium hardness in water by ion chromatography. Anal. Chim. Acta 1988, 204, 87–93. [Google Scholar] [CrossRef]
- García-Fernández, R.; García-Alonso, J.I.; Sanz-Medel, A. Simultaneous determination of inorganic anions, calcium and magnesium by suppressed ion chromatography. J. Chromatogr. A 2004, 1033, 127–133. [Google Scholar] [CrossRef]
- Paull, B.; MacKa, M.; Haddad, P.R. Determination of calcium and magnesium in water samples by high- performance liquid chromatography on a graphitic stationary phase with a mobile phase containing o-cresolphthalein complexone. J. Chromatogr. A 1997, 789, 329–337. [Google Scholar] [CrossRef]
- Ding, Y.; Ling, J.; Qiao, Y.; Li, Z.; Sun, Z.; Cai, J.; Guo, Y.; Wang, H. A high-Throughput fluorimetric microarray with enhanced fluorescence and suppressed “coffee-ring” effects for the detection of calcium ions in blood. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Atanasijevic, T.; Shusteff, M.; Fam, P.; Jasanoff, A. Calcium-sensitive MRI contrast agents based on superparamagnetic iron oxide nanoparticles and calmodulin. Natl. Acad. Sci. 2006, 103, 14707–14712. [Google Scholar] [CrossRef] [Green Version]
- Miyawaki, A.; Llopis, J.; Heim, R.; Michael McCaffery, J.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Taktak, S.; Weissleder, R.; Josephson, L. Electrode chemistry yields a nanoparticle-based NMR sensor for calcium. Langmuir 2008, 24, 7596–7598. [Google Scholar] [CrossRef] [Green Version]
- Rojas, T.C.; Rojo, J.; Can, J.; Ferna, Â. Glycosphingolipid clustering and interactions at the cell membrane can be modeled by gold glyconanoparticles prepared with biologically significant oligosaccharides. For more information see the following pages. Angew. Chemie Int. Ed. English 2001, 40, 2257–2261. [Google Scholar] [CrossRef]
- Sigel, R.K.O.; Pyle, A.M. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 2007, 107, 97–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y. New transition-metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chem. A Eur. J. 2002, 8, 4588–4596. [Google Scholar] [CrossRef]
- Lan, T.; Furuya, K.; Lu, Y. A highly selective lead sensor based on a classic lead DNAzyme. Chem. Commun. 2010, 46, 3896–3898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Lu, Y. A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J. Am. Chem. Soc. 2000, 122, 10466–10467. [Google Scholar] [CrossRef]
- Peng, D.; Li, Y.; Huang, Z.; Liang, R.P.; Qiu, J.D.; Liu, J. Efficient DNA-Catalyzed Porphyrin Metalation for Fluorescent Ratiometric Pb2+ Detection. Anal. Chem. 2019, 91, 11403–11408. [Google Scholar] [CrossRef]
- Li, J.; Zheng, W.; Kwon, A.; Lu, Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000, 28, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Li, G.; Shao, X.; Huang, K.; Luo, Y.; Xu, W. A colorimetric zinc(II) assay based on the use of hairpin DNAzyme recycling and a hemin/G-quadruplex lighted DNA nanoladder. Microchim. Acta 2020, 187, 26. [Google Scholar] [CrossRef]
- Carmi, N.; Shultz, L.A.; Breaker, R.R. In vitro selection of self-cleaving DNAs. Chem. Biol. 1996, 3, 1039–1046. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Lu, Y. A DNAzyme catalytic beacon sensor for paramagnetic Cu2+ ions in aqueous solution with high sensitivity and selectivity. J. Am. Chem. Soc. 2007, 129, 9838–9839. [Google Scholar] [CrossRef]
- Liu, J.; Brown, A.K.; Meng, X.; Cropek, D.M.; Istok, J.D.; Watson, D.B.; Lu, Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. USA 2007, 104, 2056–2061. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Lu, Y. Metal-ion-dependent folding of a uranyl-specific DNAzyme: Insight into function from fluorescence resonance energy transfer studies. Chem. A Eur. J. 2011, 17, 13732–13742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willner, I.; Shlyahovsky, B.; Zayats, M.; Willner, B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008, 37, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, J. Functional DNA nanotechnology: Emerging applications of DNAzymes and aptamers. Curr. Opin. Biotechnol. 2006, 17, 580–588. [Google Scholar] [CrossRef]
- Zhou, W.; Ding, J.; Liu, J. Theranostic dnazymes. Theranostics 2017, 7, 1010–1025. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Saran, R.; Chen, Q.; Ding, J.; Liu, J. A New Na + -Dependent RNA-Cleaving DNAzyme with over 1000-fold Rate Acceleration by Ethanol. ChemBioChem 2016, 17, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Blundell, E.L.C.J.; Vogel, R.; Platt, M. Particle-by-Particle Charge Analysis of DNA-Modified Nanoparticles Using Tunable Resistive Pulse Sensing. Langmuir 2016, 32, 1082–1090. [Google Scholar] [CrossRef] [Green Version]
- Mayne, L.; Lin, C.Y.; Christie, S.D.R.; Siwy, Z.S.; Platt, M. The Design and Characterization of Multifunctional Aptamer Nanopore Sensors. ACS Nano 2018, 12, 4844–4852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willmott, G.R.; Vogel, R.; Yu, S.S.C.; Groenewegen, L.G.; Roberts, G.S.; Kozak, D.; Anderson, W.; Trau, M. Use of tunable nanopore blockade rates to investigate colloidal dispersions. J. Phys. Condens. Matter 2010, 22, 454116. [Google Scholar] [CrossRef]
- Kozak, D.; Anderson, W.; Vogel, R.; Trau, M. Advances in resistive pulse sensors: Devices bridging the void between molecular and microscopic detection. Nano Today 2011, 6, 531–545. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; German, S.R.; Lan, W.-J.; Holden, D.A.; Mega, T.L.; White, H.S. Resistive-Pulse Analysis of Nanoparticles. Annu. Rev. Anal. Chem. 2014, 7, 513–535. [Google Scholar] [CrossRef]
- Maugi, R.; Hauer, P.; Bowen, J.; Ashman, E.; Hunsicker, E.; Platt, M. A methodology for characterising nanoparticle size and shape using nanopores. Nanoscale 2020, 12, 262–270. [Google Scholar] [CrossRef]
- Billinge, E.R.; Platt, M. Multiplexed, label-free detection of biomarkers using aptamers and Tunable Resistive Pulse Sensing (AptaTRPS). Biosens. Bioelectron. 2015, 68, 741–748. [Google Scholar] [CrossRef] [Green Version]
- Kohli, P.; Harrell, C.C.; Cao, Z.; Gasparac, R.; Tan, W.; Martin, C.R. DNA-functionalized nanotube, membranes with single-base mismatch selectivity. Science 2004, 305, 984–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sexton, L.T.; Horen, L.P.; Martin, C.R. Biosensing with Nanopores and Nanotubes. In Molecular- and Nano-Tubes SE-6; Hayden, O., Nielsch, K., Eds.; Springer: Boston, MA USA, 2011; pp. 165–207. [Google Scholar]
- Blundell, E.L.C.J.; Mayne, L.J.; Billinge, E.R.; Platt, M. Emergence of tunable resistive pulse sensing as a biosensor. Anal. Methods 2015, 7, 7055–7066. [Google Scholar] [CrossRef] [Green Version]
- Bayley, H.; Martin, C.R. Resistive-pulse sensing—From microbes to molecules. Chem. Rev. 2000, 100, 2575–2594. [Google Scholar] [CrossRef]
- Mayne, L.J.; Christie, S.D.R.; Platt, M. A tunable nanopore sensor for the detection of metal ions using translocation velocity and biphasic pulses. Nanoscale 2016, 8, 19139–19147. [Google Scholar] [CrossRef] [Green Version]
- Billinge, E.R.; Broom, M.; Platt, M. Monitoring aptamer-protein interactions using tunable resistive pulse sensing. Anal. Chem. 2014, 86, 1030–1037. [Google Scholar] [CrossRef] [Green Version]
- Maugi, R.; Salkenova, Z.; Platt, M. Incorporating Peptide Aptamers into Resistive Pulse Sensing. Med. Devices Sens. 2020, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Xu, Y.; Ali, I.; Liu, L.; Wu, H.; Lu, Z.; Liu, Q. Solid-State Nanopore Single-Molecule Sensing of DNAzyme Cleavage Reaction Assisted with Nucleic Acid Nanostructure. ACS Appl. Mater. Interfaces 2018, 10, 26555–26565. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, L.; Dong, D.; Liu, Y.; Li, J. A label-free DNAzyme-based nanopore biosensor for highly sensitive and selective lead ion detection. Anal. Methods 2016, 8, 7040–7046. [Google Scholar] [CrossRef]
- Liu, N.; Hou, R.; Gao, P.; Lou, X.; Xia, F. Sensitive Zn2+ sensor based on biofunctionalized nanopores: Via combination of DNAzyme and DNA supersandwich structures. Analyst 2016, 141, 3626–3629. [Google Scholar] [CrossRef] [PubMed]
- Healey, M.J.; Sivakumaran, M.; Platt, M. Rapid quantification of prion proteins using resistive pulse sensing. Analyst 2020, 145, 2595–2601. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhou, W.; Liu, J. Ultrasensitive DNAzyme-Based Ca2+ Detection Boosted by Ethanol and a Solvent-Compatible Scaffold for Aptazyme Design. ChemBioChem 2018, 19, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Saran, R.; Huang, P.J.J.; Ding, J.; Liu, J. An Exceptionally Selective DNA Cooperatively Binding Two Ca2+ Ions. ChemBioChem 2017, 18, 518–522. [Google Scholar] [CrossRef]
- Healey, M.J.; Rowe, W.; Siati, S.; Sivakumaran, M.; Platt, M. Rapid Assessment of Site Specific DNA Methylation through Resistive Pulse Sensing. ACS Sens. 2018, 3, 655–660. [Google Scholar] [CrossRef] [Green Version]
- Kozak, D.; Anderson, W.; Vogel, R.; Chen, S.; Antaw, F.; Trau, M. Simultaneous size and ζ-potential measurements of individual nanoparticles in dispersion using size-tunable pore sensors. ACS Nano 2012, 6, 6990–6997. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridisation prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
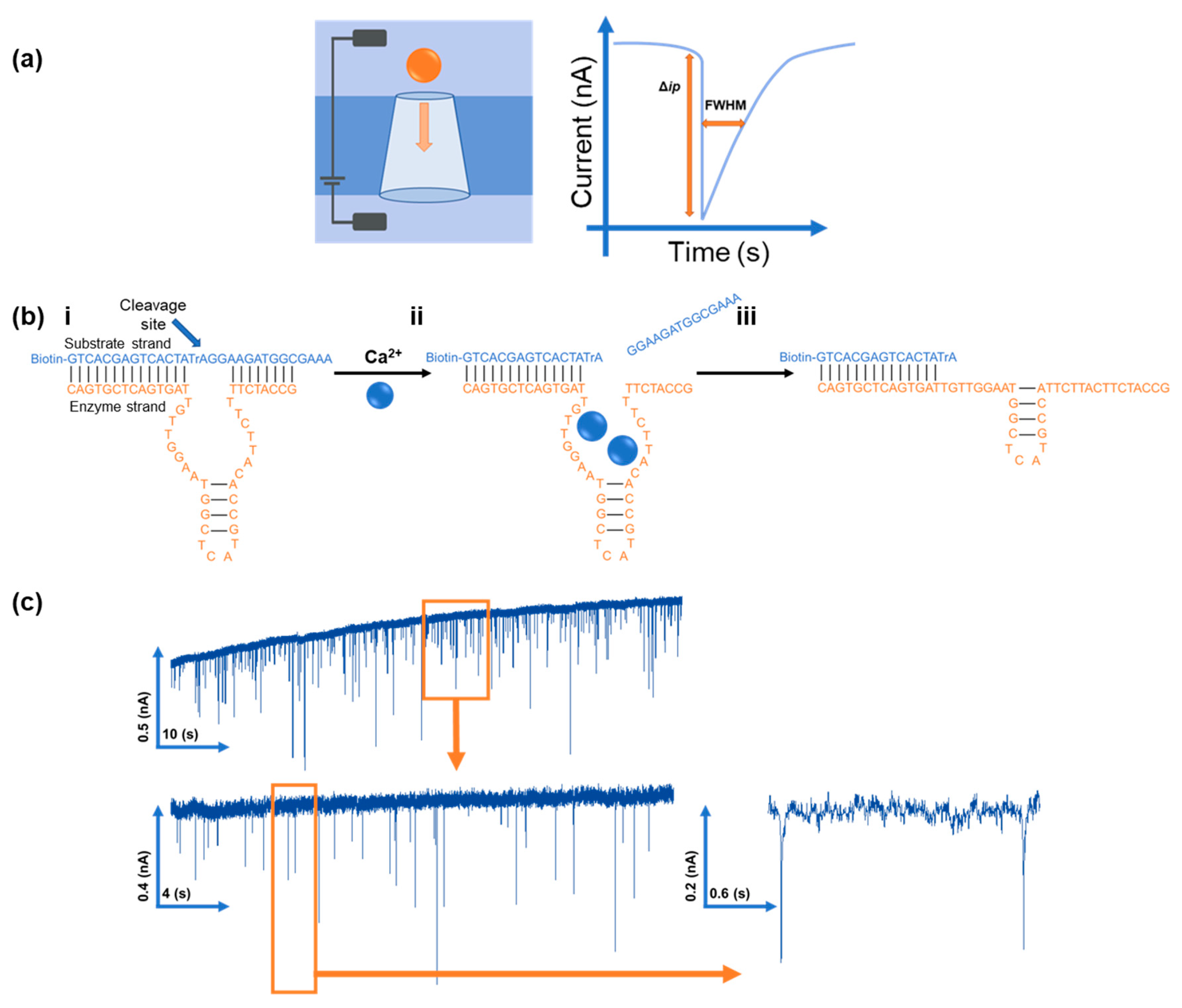

| DNA Sequence | ||
|---|---|---|
| Substrate strand | [Btn]GTCACGAGTCACTATrAGGAAGATGGCGAAA | 31 mer |
| Enzyme Strand | GCCATCTTTTCTCACAGCGTACTCGCTAAGGTTGTTAGTGACTCGTGAC | 49 mer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heaton, I.; Platt, M. DNAzyme Sensor for the Detection of Ca2+ Using Resistive Pulse Sensing. Sensors 2020, 20, 5877. https://doi.org/10.3390/s20205877
Heaton I, Platt M. DNAzyme Sensor for the Detection of Ca2+ Using Resistive Pulse Sensing. Sensors. 2020; 20(20):5877. https://doi.org/10.3390/s20205877
Chicago/Turabian StyleHeaton, Imogen, and Mark Platt. 2020. "DNAzyme Sensor for the Detection of Ca2+ Using Resistive Pulse Sensing" Sensors 20, no. 20: 5877. https://doi.org/10.3390/s20205877
APA StyleHeaton, I., & Platt, M. (2020). DNAzyme Sensor for the Detection of Ca2+ Using Resistive Pulse Sensing. Sensors, 20(20), 5877. https://doi.org/10.3390/s20205877






