Development of the Correction Algorithm to Limit the Deformation of Bacterial Colonies Diffraction Patterns Caused by Misalignment and Its Impact on the Bacteria Identification in the Proposed Optical Biosensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Sample Preparation
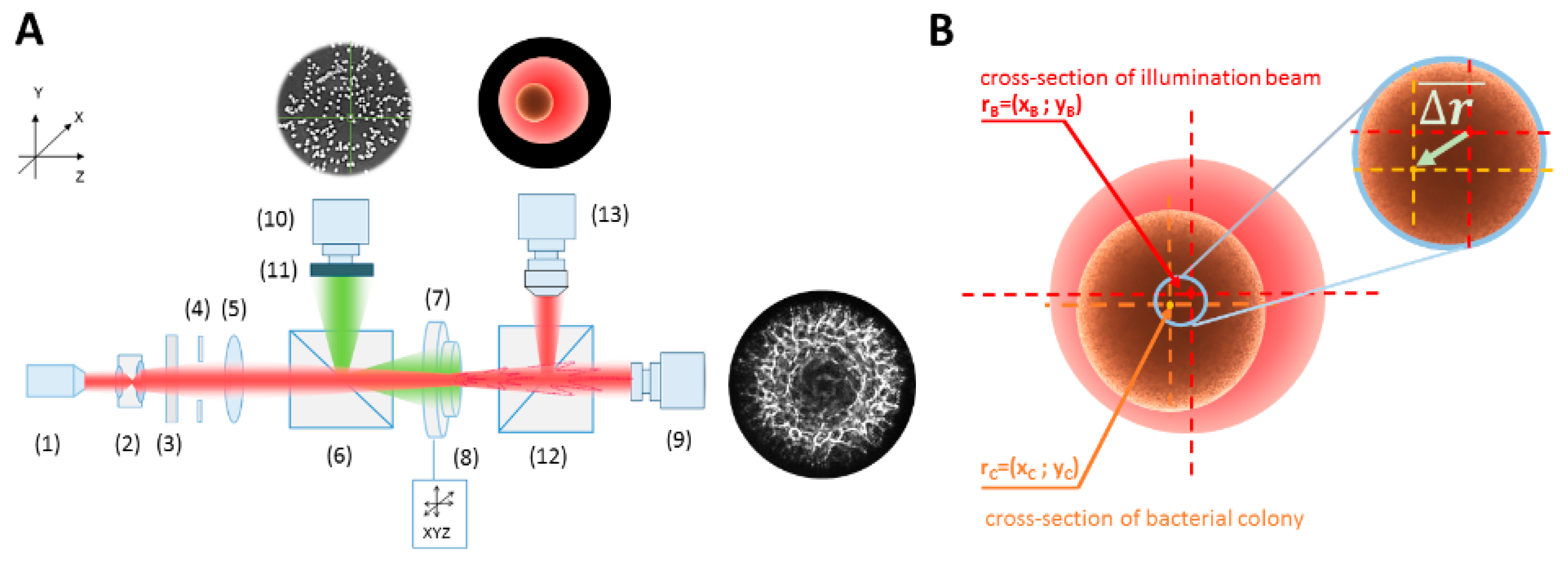
2.2. Optical System Configuration
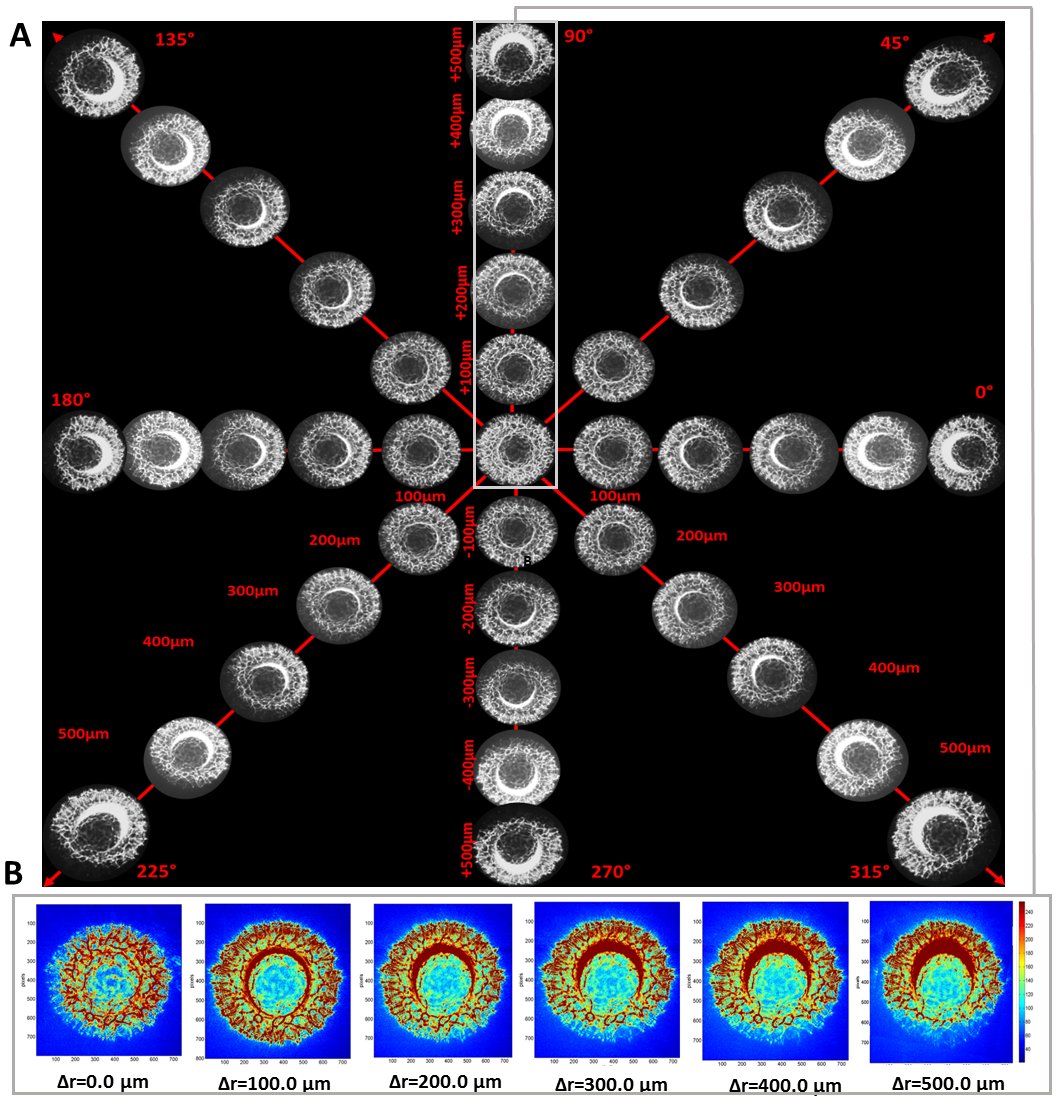
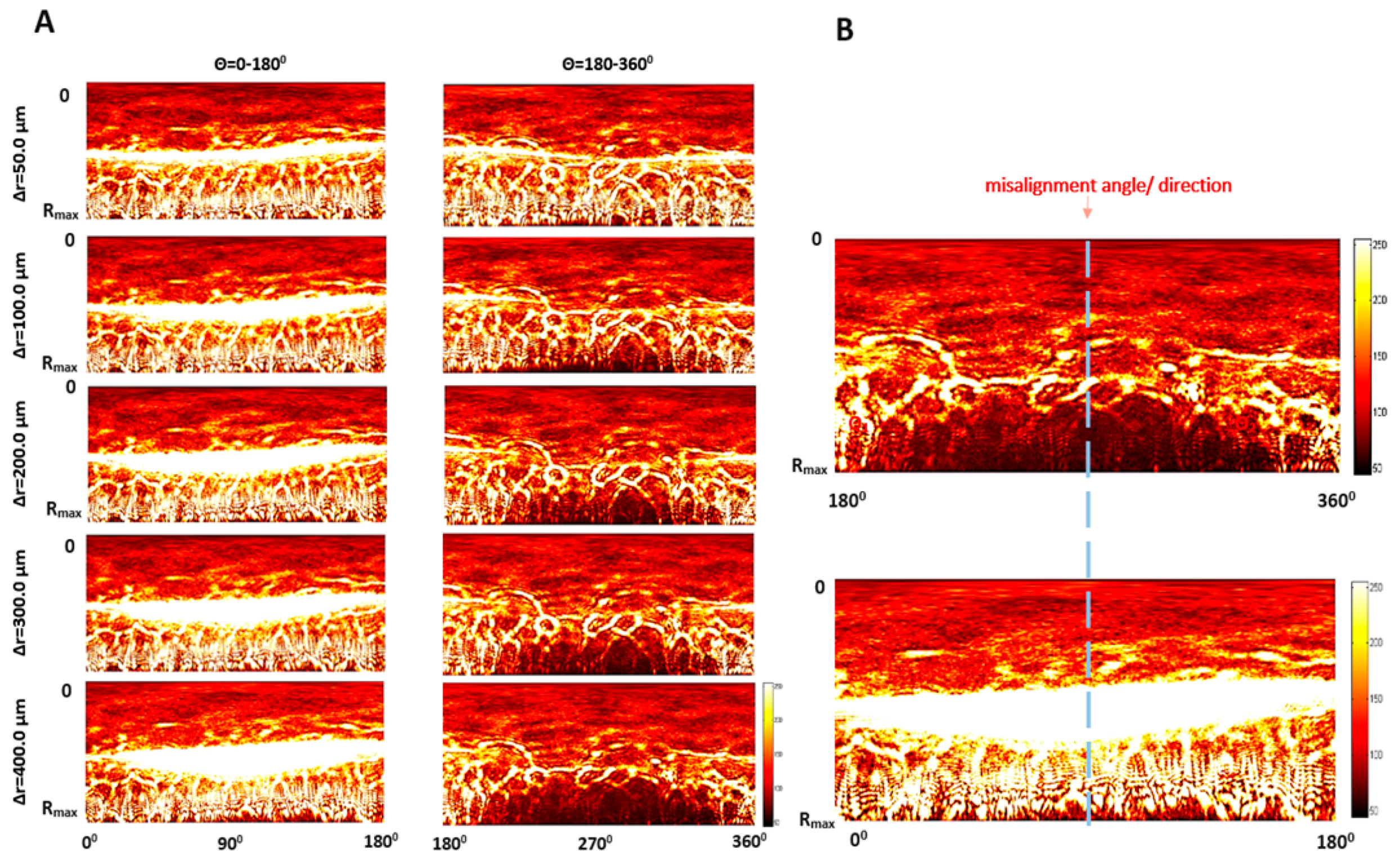
2.3. The Misalignment ∆r-Parameter Describing the Relative Dislocation of the Bacterial Colony and the Illuminating Beam’s Centers
2.4. Classification Features Extracted from Diffraction Patterns
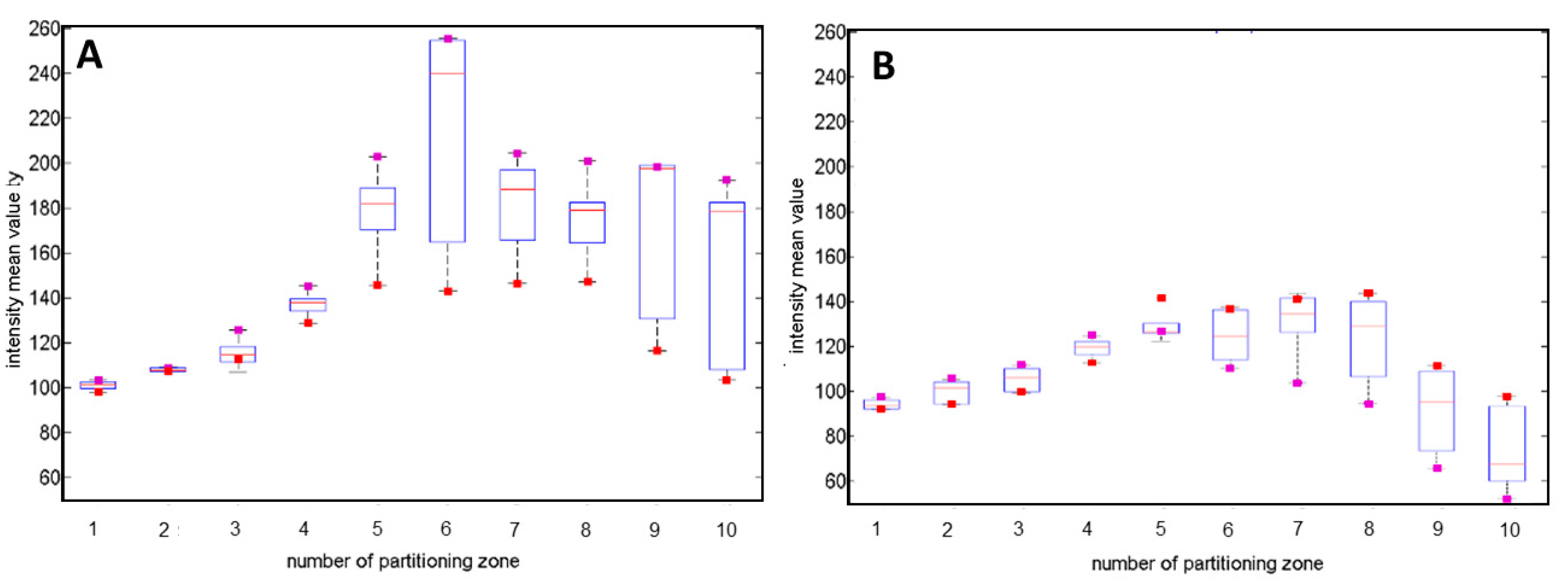
2.5. The Impact of the Misalignment ∆r-Parameter on the Variation of the Classification Features
2.6. The Sensitivity of Features Extracted form Diffraction Patterns on Misalignment ∆r-Parameter
2.7. The Concept of an Algorithm for Automatic Detection and Correction of the Colony–Illuminating Beam Misalignment
- Step 1:
- Register the experimental diffraction pattern of a bacterial colony.
- Step 2:
- Limit the diffraction pattern by the shape function and determine its center.
- Step 3:
- Divide the pattern into 10 partitioning zones of equal widths. The number of limiting zones should be limited only to the peripheral ones. Based on the experimental measurement performed on colonies with a radius in the range 0.6–0.8 mm, this condition applies to the 6th to 10th zone.
- Step 4:
- Measure the mean intensity values of the 6th partitioning zone (mean.6) and determine the two parts of the diffraction patterns, with the highest and the lowest values of the mean.6. The most reliable indicator of the smallest misalignment is the mean.10 value. The Fresnel pattern is then divided into two parts with maximal (+) and minimal (−) mean intensity.
- Step 5:
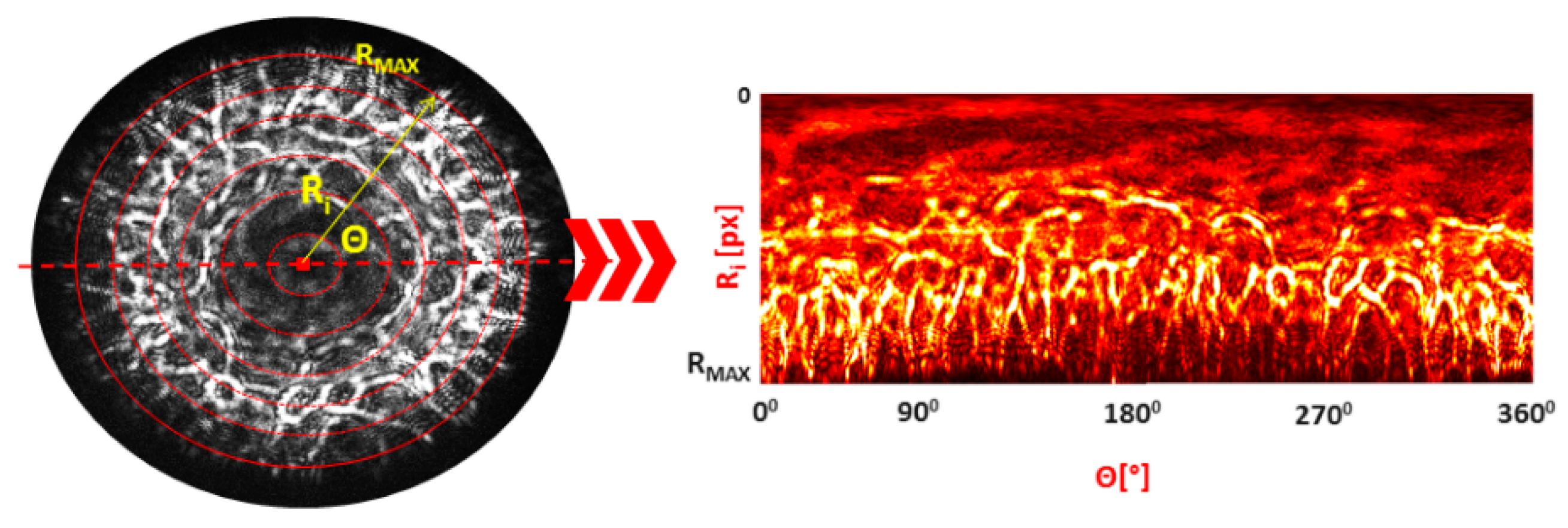
- Perform the inverse polar transformation of (−) part of the pattern with the 1° angular resolution and divide into radial zones of the width 0.1 RMAX, where RMAX is the radius of the circle limiting the diffraction pattern (determined in Step 2).
- Step 6:
- Transform the processed pattern including the peripheral (6–10) zones into a binary mask and determine the number of the dark pixels at each angle Θ (from 10th to 7th).
- Step 7:
- Derive the histogram representing the number of dark pixels at each Θ. Using the fitted dependence of the dark pixels’ number at specific angles, determine the misalignment angle based on the symmetry of the dependence.
- Step 8:
- Adjust positioning of the colony at the lowest possible resolution of the translation stage. Aligning conditions are considered with respect to the mean value of the 10th partitioning zone. For each dislocation the comparability condition relative to the values (mean.10(+) and mean.10(−)) of the mean.10 feature extracted from the peripheral zone of the (+) and (−) parts of decentered patterns is estimated. If mean.10(+) = mean.10(−), the colony and beam alignment is successfully achieved. If mean.10(+) ≠ mean.10(−), the dislocation should be continued until the intensity distribution of the diffraction pattern is symmetrical (mean.10(+) = mean.10(+)). When this condition is met, the colony and illuminating beam are aligned and the centered diffraction pattern can be registered.
3. Results and Discussions
3.1. The Dependence of Bacterial Colonies’ Diffraction Patterns on Misalignment
3.2. The Quantitative Analysis of the Influence of the ∆r-Parameter on the Classification Features
3.3. The Algorithm for Automatic Detection of the Colony–Illuminating Beam Misalignment and Correction of Diffraction Pattern Deformation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Banada, P.P.; Guo, S.; Bayraktar, B.; Bae, E.; Rajwa, B.; Robinson, J.P.; Hirleman, E.D.; Bhunia, A.K. Optical forward-scattering for detection of Listeria monocytogenes and other Listeria species. Biosens. Bioelectron. 2007, 22, 1664–1671. [Google Scholar] [CrossRef] [PubMed]
- Alsulami, T.S.; Zhu, X.; Abdelhaseib, M.U.; Singh, A.K.; Bhunia, A.K. Rapid detection and differentiation of Staphylococcus colonies using an optical scattering technology. Anal. Bioanal. Chem. 2018, 410. [Google Scholar] [CrossRef] [PubMed]
- Suchwalko, A.; Buzalewicz, I.; Wieliczko, A.; Podbielska, H. Bacteria species identification by the statistical analysis of bacterial colonies Fresnel patterns. Opt. Express 2013, 21. [Google Scholar] [CrossRef]
- Suchwalko, A.; Buzalewicz, I.; Podbielska, H. Computer-based classification of bacteria species by analysis of their colonies fresnel diffraction patterns. In Proceedings of the Progress in Biomedical Optics and Imaging—Proceedings of SPIE, San Fancisco, CA, USA, 31 January 2012; Volume 8212. [Google Scholar]
- Buzalewicz, I.; Suchwałko, A.; Trzciński, P.; Sas-Paszt, L.; Sumorok, B.; Kowal, K.; Kozera, R.; Wieliczko, A.; Podbielska, H. Integrated multi-channel optical system for bacteria characterization and its potential use for monitoring of environmental bacteria. Biomed. Opt. Express 2019, 10, 1165. [Google Scholar] [CrossRef] [PubMed]
- Marcoux, P.R.; Dupoy, M.; Cuer, A.; Kodja, J.-L.; Lefebvre, A.; Licari, F.; Louvet, R.; Narassiguin, A.; Mallard, F. Optical forward-scattering for identification of bacteria within microcolonies. Appl. Microbiol. Biotechnol. 2014, 98, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Minoni, U.; Signoroni, A.; Nassini, G. On the application of optical forward-scattering to bacterial identification in an automated clinical analysis perspective. Biosens. Bioelectron. 2015, 68, 536–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kok, J.; Thomas, L.C.; Olma, T.; Chen, S.C.A.; Iredell, J.R. Identification of bacteria in blood culture broths using matrix-assisted laser desorption-ionization SepsityperTM and time of flight mass spectrometry. PLoS ONE 2011, 6, e23285. [Google Scholar] [CrossRef] [PubMed]
- Croxatto, A.; Prod’hom, G.; Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. Fems Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef]
- Ziegler, D.; Mariotti, A.; Pflüger, V.; Saad, M.; Vogel, G.; Tonolla, M.; Perret, X. In situ identification of plant-invasive bacteria with MALDI-TOF mass spectrometry. PLoS ONE 2012, 7, e37189. [Google Scholar] [CrossRef]
- Ho, C.-S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef]
- Pahlow, S.; Meisel, S.; Cialla-May, D.; Weber, K.; Rösch, P.; Popp, J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv. Drug Deliv. Rev. 2015, 89, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Strola, S.A.; Baritaux, J.-C.; Schultz, E.; Simon, A.C.; Allier, C.; Espagnon, I.; Jary, D.; Dinten, J.-M. Single bacteria identification by Raman spectroscopy. J. Biomed. Opt. 2014, 19, 111610. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.; Yang, S.-M.; Lim, B.; Park, S.H.; Rackerby, B.; Kim, H.-Y. Design of PCR assays to specifically detect and identify 37 Lactobacillus species in a single 96 well plate. BMC Microbiol. 2020, 20, 96. [Google Scholar] [CrossRef] [Green Version]
- Podbielska, H.; Buzalewicz, I.; Suchwalko, A.; Wieliczko, A. Bacteria Classification by Means of the Statistical Analysis of Fresnel Diffraction Patterns of Bacteria Colonies. In Proceedings of the Biomedical Optics and 3-D Imaging; OSA, Miami, FL, USA, 28 April–2 May 2012; OSA: Washington, DC, USA, 2012; p. BSu5A.5. [Google Scholar]
- Kim, H.; Rajwa, B.; Bhunia, A.K.; Robinson, J.P.; Bae, E. Development of a multispectral light-scatter sensor for bacterial colonies. J. Biophotonics 2017, 10, 634–644. [Google Scholar] [CrossRef]
- Suchwalko, A.; Buzalewicz, I.; Podbielska, H. Statistical identification of bacteria species. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2013; pp. 711–721. ISBN 978-84-939843-9-7. [Google Scholar]
- Tang, Y.; Kim, H.; Singh, A.K.; Aroonnual, A.; Bae, E.; Rajwa, B.; Fratamico, P.M.; Bhunia, A.K. Light scattering sensor for direct identification of colonies of Escherichia coli serogroups O26, O45, O103, O111, O121, O145 and O157. PLoS ONE 2014, 9, e105272. [Google Scholar] [CrossRef]
- Suchwałko, A.; Buzalewicz, I.; Podbielska, H. Bacteria identification in an optical system with optimized diffraction pattern registration condition supported by enhanced statistical analysis. Opt. Express 2014, 22. [Google Scholar] [CrossRef]
- Buzalewicz, I.; Wieliczko, A.; Podbielska, H. Influence of various growth conditions on Fresnel diffraction patterns of bacteria colonies examined in the optical system with converging spherical wave illumination. Opt. Express 2011, 19, 21768. [Google Scholar] [CrossRef]
- Mialon, M.; Tang, Y.; Singh, A.K.; Bae, E.; Bhunia, A.K. Effects of Preparation and Storage of Agar Media on the Sensitivity of Bacterial Forward Scattering Patterns. Open J. Appl. Biosens. 2012, 1, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Drolia, R.; Bai, X.; Bhunia, A.K. Streptomycin induced stress response in salmonella enterica serovar typhimurium shows distinct colony scatter signature. PLoS ONE 2015, 10, e135035. [Google Scholar] [CrossRef] [Green Version]
- Suchwalko, A.; Buzalewicz, I.; Podbielska, H. Identification of bacteria species by using morphological and textural properties of bacterial colonies diffraction patterns. In Proceedings of the SPIE, Videometrics, Range Imaging and Applications XII; and Automated Visual Inspection, Munich, Germany, 24 May 2013; pp. 87911M--1–87911M--7. [Google Scholar]
- Singh, A.K.; Bhunia, A.K. Optical scatter patterns facilitate rapid differentiation of Enterobacteriaceae on CHROMagarTM Orientation medium. Microb. Biotechnol. 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Liao, S.X.; Pawlak, M. On the accuracy of Zernike moments for image analysis. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 1358–1364. [Google Scholar] [CrossRef] [Green Version]
- Mukundan, R.; Ramakrishnan, K.R. Fast computation of Legendre and Zernike moments. Pattern Recognit. 1995, 28, 1433–1442. [Google Scholar] [CrossRef]


| F-value | Δr [µm] | Number of Partitioning Zone | |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| 50 | 0.09 | 0.01 | 0.12 | 0.04 | 0.55 | 4.63 | 2.02 | 0.66 | 0.21 | 4.32 | |
| 100 | 0.72 | 0.01 | 0.18 | 0.21 | 1.42 | 4.42 | 1.82 | 2.06 | 4.51 | 11.18 | |
| 200 | 0.20 | 3.46 | 3.52 | 6.24 | 17.43 | 19.49 | 182.20 | 49.12 | 51.24 | 61.05 | |
| 300 | 1.42 | 0.01 | 1.03 | 4.45 | 19.55 | 24.90 | 193.11 | 52.39 | 60.21 | 72.22 | |
| 400 | 0.01 | 0.02 | 1.66 | 4.72 | 20.48 | 26.14 | 202.21 | 54.92 | 59.85 | 71.87 | |
| 500 | 0.43 | 4.43 | 4.52 | 4.95 | 14.55 | 24.90 | 193.11 | 52.39 | 7.34 | 4.91 | |
| F-critical = 4.30 | |||||||||||
| Set of All Classification Features | |||||
| Δr = 50 µm | Δr = 100 µm | Δr = 200 µm | Δr = 300 µm | Δr = 400 µm | Δr = 500 µm |
| 92.96% | 76.06% | 45.07% | 28.17% | 15.49% | 7.04% |
| Selected Subsets of the Classification Features | |||||
| Δr = 50 µm | Δr = 100 µm | Δr = 200.µm | Δr = 300 µm | Δr = 400 µm | Δr = 500 µm |
| 95.12% | 87.80% | 46.34% | 36.59% | 21.95% | 9.76% |
| Introduced Misalignment Angle Θ0 | Estimated Mean Angle ΘE | ΔΘ = |Θ0−ΘE| | |
|---|---|---|---|
| 280° | 278° | 2° | 0.71% |
| 120° | 122° | 5° | 4.16% |
| 45° | 40° | 2° | 4.44% |
| 85° | 89° | 4° | 4.71% |
| 28° | 32° | 2° | 7.14% |
| 180° | 185° | 5° | 2.78% |
| 220° | 119° | 1° | 0.45% |
| 140° | 144° | 4° | 2.86% |
| 90° | 86° | 4° | 4.44% |
| 340° | 335° | 5° | 1.47% |
| Δr [µm] | |
|---|---|
| 50 | 9.12% |
| 100 | 8.21% |
| 200 | 7.14% |
| 300 | 6.54% |
| 400 | 5.89% |
| 500 | 5.45% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buzalewicz, I.; Suchwałko, A.; Karwańska, M.; Wieliczko, A.; Podbielska, H. Development of the Correction Algorithm to Limit the Deformation of Bacterial Colonies Diffraction Patterns Caused by Misalignment and Its Impact on the Bacteria Identification in the Proposed Optical Biosensor. Sensors 2020, 20, 5797. https://doi.org/10.3390/s20205797
Buzalewicz I, Suchwałko A, Karwańska M, Wieliczko A, Podbielska H. Development of the Correction Algorithm to Limit the Deformation of Bacterial Colonies Diffraction Patterns Caused by Misalignment and Its Impact on the Bacteria Identification in the Proposed Optical Biosensor. Sensors. 2020; 20(20):5797. https://doi.org/10.3390/s20205797
Chicago/Turabian StyleBuzalewicz, Igor, Agnieszka Suchwałko, Magdalena Karwańska, Alina Wieliczko, and Halina Podbielska. 2020. "Development of the Correction Algorithm to Limit the Deformation of Bacterial Colonies Diffraction Patterns Caused by Misalignment and Its Impact on the Bacteria Identification in the Proposed Optical Biosensor" Sensors 20, no. 20: 5797. https://doi.org/10.3390/s20205797
APA StyleBuzalewicz, I., Suchwałko, A., Karwańska, M., Wieliczko, A., & Podbielska, H. (2020). Development of the Correction Algorithm to Limit the Deformation of Bacterial Colonies Diffraction Patterns Caused by Misalignment and Its Impact on the Bacteria Identification in the Proposed Optical Biosensor. Sensors, 20(20), 5797. https://doi.org/10.3390/s20205797