Deep Recurrent Neural Networks for Automatic Detection of Sleep Apnea from Single Channel Respiration Signals
Abstract
:1. Introduction
2. Background & Problem Statement
2.1. Standards for Scoring Sleep Apnea
2.2. Algorithms for Automated Apnea Detection with PSG Respiration Signals
2.3. Problem Statement
3. Materials and Methods
3.1. Data Set
3.2. Data Preprocessing
3.3. Recurrent Neural Network (RNN)
3.3.1. Long Short-Term Memory (LSTM)
3.3.2. Bidirectional LSTM (BiLSTM)
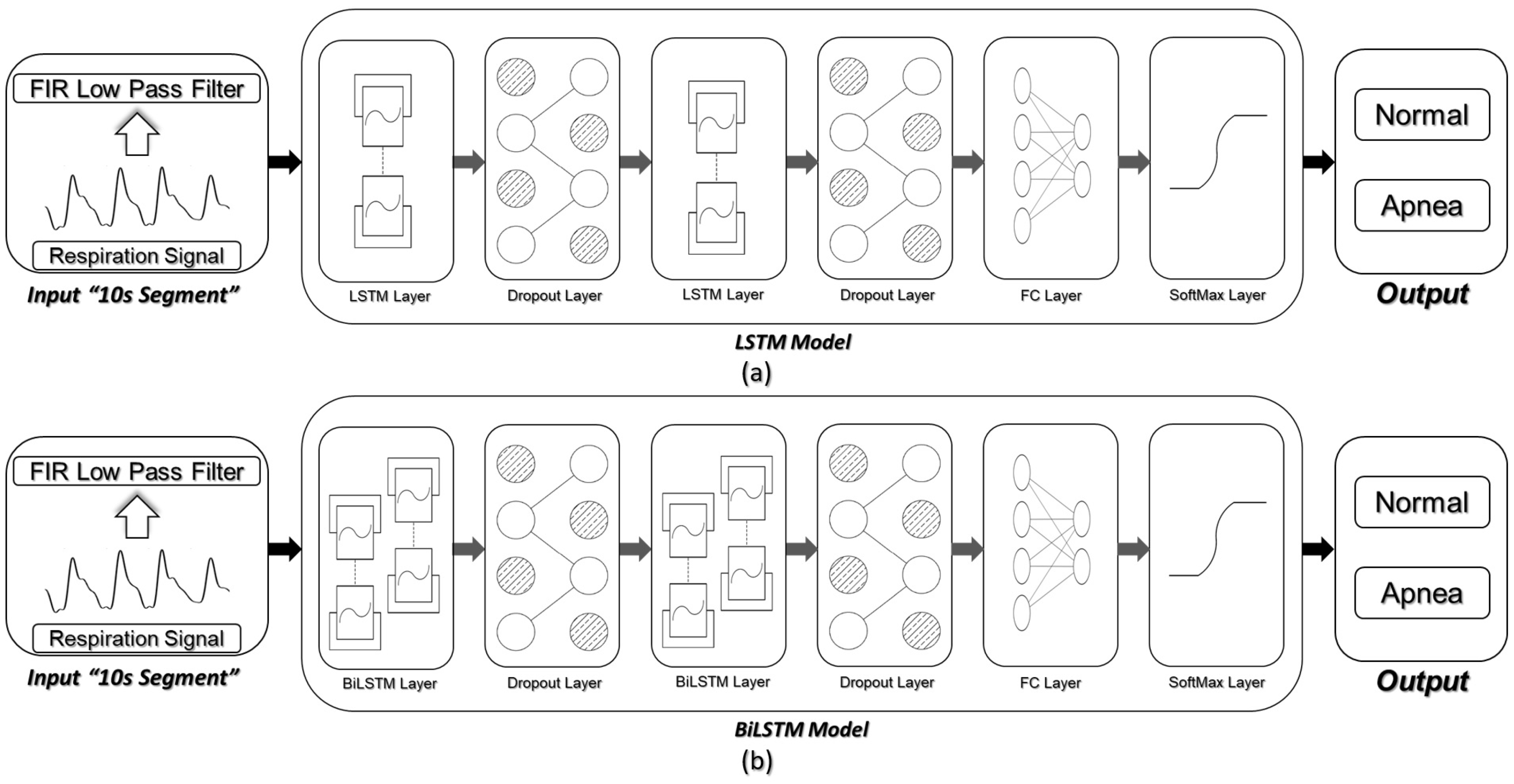
3.4. Network Architecture and Detection Scenarios
3.5. Evaluation of Detection Results
3.5.1. Classification Performance over Detection Windows
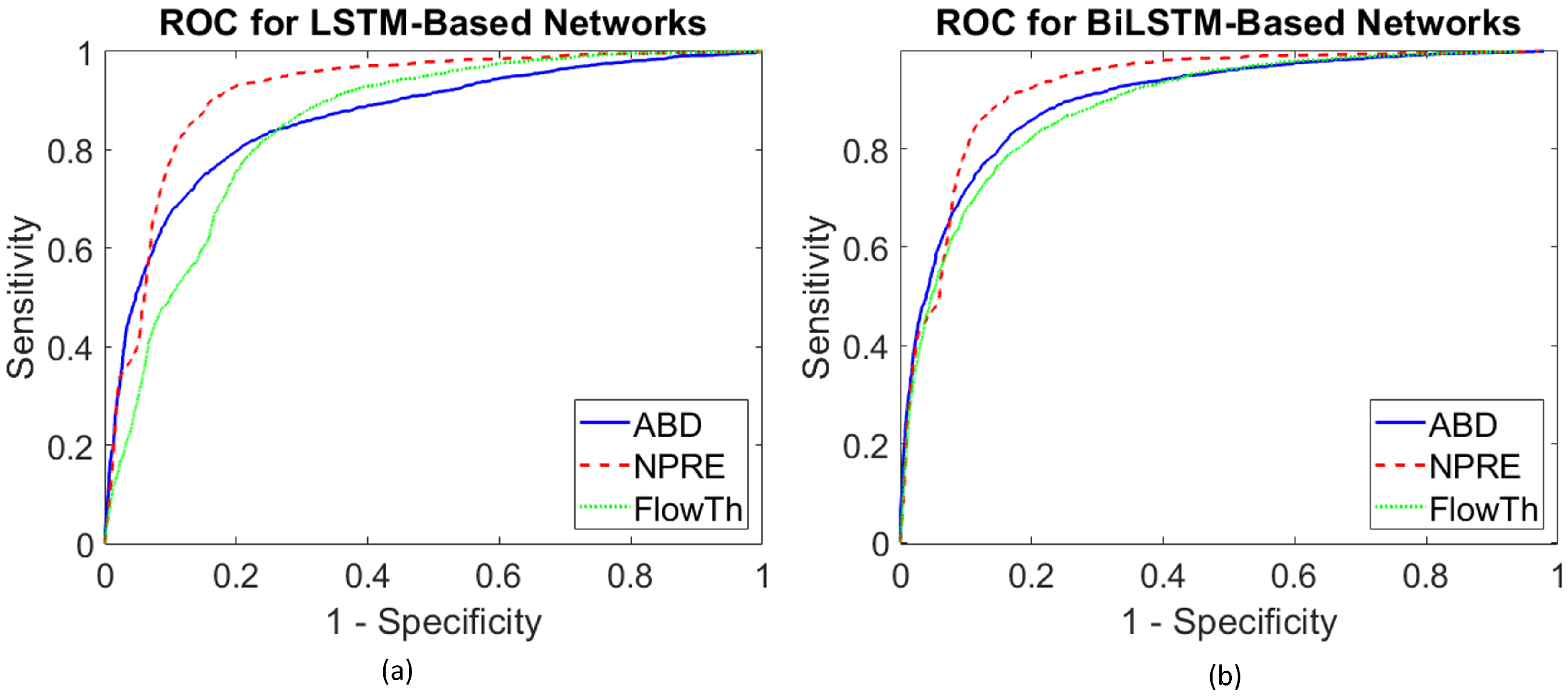
3.5.2. Receiver Operating Characteristics () Curve
4. Results
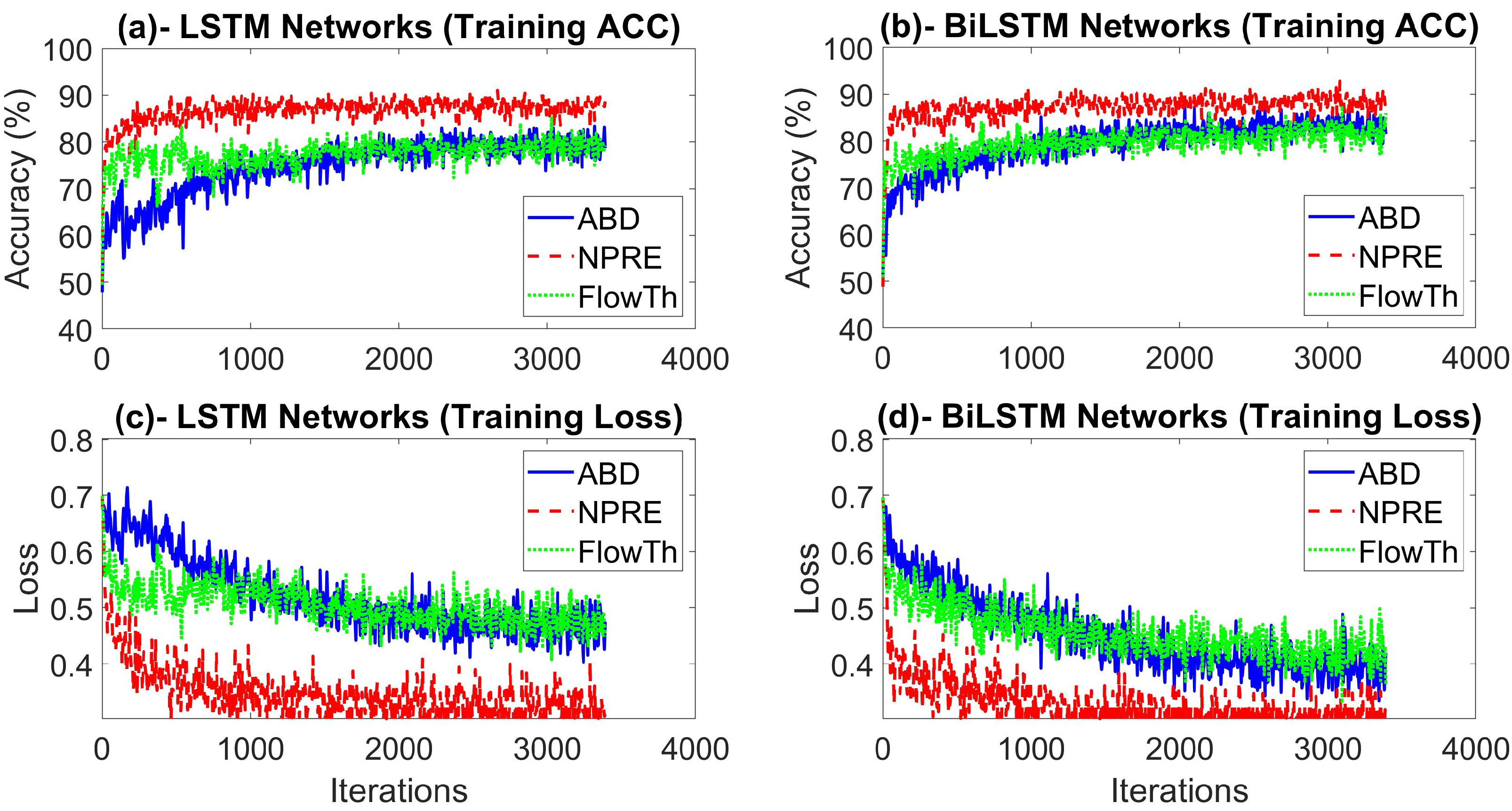
4.1. Experimental Setting and Network Optimization
4.2. Overall Performance over Different Respiration Signals
4.3. Individualized Patient Based Performance for the Best Detection Scenarios
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Berry, R.B.; Brooks, R.; Gamaldo, C.E.; Harding, S.M.; Lloyd, R.M.; Marcus, C.L.; Vaughn, B.V. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications; Version 2.6; American Academy of Sleep Medicine: Darien, IL, USA, 2020. [Google Scholar]
- Quan, S.; Gillin, J.C.; Littner, M.; Shepard, J. Sleep-related breathing disorders in adults: Recommendations for syndrome definition and measurement techniques in clinical research. editorials. Sleep 1999, 22, 662–689. [Google Scholar] [CrossRef] [Green Version]
- De Chazal, P.; Penzel, T.; Heneghan, C. Automated detection of obstructive sleep apnoea at different time scales using the electrocardiogram. Physiol. Meas. 2004, 25, 967. [Google Scholar] [CrossRef] [Green Version]
- Somers, V.K.; White, D.P.; Amin, R.; Abraham, W.T.; Costa, F.; Culebras, A.; Daniels, S.; Floras, J.S.; Hunt, C.E.; Olson, L.J.; et al. Sleep apnea and cardiovascular disease: An American heart association/American college of cardiology foundation scientific statement from the American heart association council for high blood pressure research professional education committee, council on clinical cardiology, stroke council, and council on cardiovascular nursing in collaboration with the national heart, lung, and blood institute national center on sleep disorders research (National Institutes of Health). J. Am. Coll. Cardiol. 2008, 52, 686–717. [Google Scholar] [PubMed] [Green Version]
- Botros, N.; Concato, J.; Mohsenin, V.; Selim, B.; Doctor, K.; Yaggi, H.K. Obstructive sleep apnea as a risk factor for type 2 diabetes. Am. J. Med. 2009, 122, 1122–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, T.; Palta, M.; Dempsey, J.; Skatrud, J.; Weber, S.; Badr, S. The occurrence of sleep-disordered breathing among middle-aged adults. N. Engl. J. Med. 1993, 328, 1230–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, Q.; Wang, Y.; Qiu, C. A real-time auto-adjustable smart pillow system for sleep apnea detection and treatment. In Proceedings of the 2013 ACM/IEEE International Conference on Information Processing in Sensor Networks (IPSN), Philadelphia, PA, USA, 8–11 April 2013; pp. 179–190. [Google Scholar]
- Patil, S.P.; Schneider, H.; Schwartz, A.R.; Smith, P.L. Adult obstructive sleep apnea: Pathophysiology and diagnosis. Chest 2007, 132, 325–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitney, C.W.; Gottlieb, D.J.; Redline, S.; Norman, R.G.; Dodge, R.R.; Shahar, E.; Surovec, S.; Nieto, F.J. Reliability of scoring respiratory disturbance indices and sleep staging. Sleep 1998, 21, 749–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, J.; Kinnear, W. Sleep on the cheap: The role of overnight oximetry in the diagnosis of sleep apnoea hypopnoea syndrome. Thorax 1999, 54, 958–959. [Google Scholar] [CrossRef] [Green Version]
- de Almeida, F.R.; Ayas, N.T.; Otsuka, R.; Ueda, H.; Hamilton, P.; Ryan, F.C.; Lowe, A.A. Nasal pressure recordings to detect obstructive sleep apnea. Sleep Breath. 2006, 10, 62–69. [Google Scholar] [CrossRef]
- Magalang, U.J.; Dmochowski, J.; Veeramachaneni, S.; Draw, A.; Mador, M.J.; El-Solh, A.; Grant, B.J. Prediction of the apnea-hypopnea index from overnight pulse oximetry. Chest 2003, 124, 1694–1701. [Google Scholar] [CrossRef] [Green Version]
- Nandakumar, R.; Gollakota, S.; Watson, N. Contactless sleep apnea detection on smartphones. In Proceedings of the 13th Annual International Conference on Mobile Systems, Applications, and Services, Florence, Italy, 18–22 May 2015; pp. 45–57. [Google Scholar]
- Mendez, M.O.; Bianchi, A.M.; Matteucci, M.; Cerutti, S.; Penzel, T. Sleep apnea screening by autoregressive models from a single ECG lead. Biomed. Eng. IEEE Trans. 2009, 56, 2838–2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khandoker, A.H.; Gubbi, J.; Palaniswami, M. Automated scoring of obstructive sleep apnea and hypopnea events using short-term electrocardiogram recordings. Inf. Technol. Biomed. IEEE Trans. 2009, 13, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Penzel, T.; McNames, J.; De Chazal, P.; Raymond, B.; Murray, A.; Moody, G. Systematic comparison of different algorithms for apnoea detection based on electrocardiogram recordings. Med. Biol. Eng. Comput. 2002, 40, 402–407. [Google Scholar] [CrossRef]
- Nigro, C.A.; Dibur, E.; Aimaretti, S.; González, S.; Rhodius, E. Comparison of the automatic analysis versus the manual scoring from ApneaLink™ device for the diagnosis of obstructive sleep apnoea syndrome. Sleep Breath. 2011, 15, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Chaudhuri, S.; Munshi, S. Obstructive sleep apnoea detection using convolutional neural network based deep learning framework. Biomed. Eng. Lett. 2018, 8, 95–100. [Google Scholar] [CrossRef]
- Urtnasan, E.; Park, J.U.; Joo, E.Y.; Lee, K.J. Automated detection of obstructive sleep apnea events from a single-lead electrocardiogram using a convolutional neural network. J. Med. Syst. 2018, 42, 104. [Google Scholar] [CrossRef]
- Urtnasan, E.; Park, J.U.; Lee, K.J. Automatic detection of sleep-disordered breathing events using recurrent neural networks from an electrocardiogram signal. Neural Comput. Appl. 2020, 32, 4733–4742. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, X.; Ho, J.K.; Chow, T.W. Object-level video advertising: An optimization framework. IEEE Trans. Ind. Inform. 2016, 13, 520–531. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Zhang, H.; Ji, Y.; Huang, W.; Liu, L. Sitcom-star-based clothing retrieval for video advertising: A deep learning framework. Neural Comput. Appl. 2019, 31, 7361–7380. [Google Scholar] [CrossRef]
- Long Short-Term Memory Based Recurrent Neural Network Architectures for Large Vocabulary Speech Recognition. Available online: https://arxiv.org/abs/1402.1128 (accessed on 10 July 2020).
- Berry, R.B.; Budhiraja, R.; Gottlieb, D.J.; Gozal, D.; Iber, C.; Kapur, V.K.; Marcus, C.L.; Mehra, R.; Parthasarathy, S.; Quan, S.F.; et al. Rules for scoring respiratory events in sleep: Update of the 2007 AASM Manual for the Scoring of Sleep and Associated Events. J. Clin. Sleep Med. 2012, 8, 597–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemons, W.W.; Littner, M.R.; Rowley, J.A.; Gay, P.; Anderson, W.M.; Hudgel, D.W.; McEvoy, R.D.; Loube, D.I. Home diagnosis of sleep apnea: A systematic review of the literature: An evidence review cosponsored by the American Academy of Sleep Medicine, the American College of Chest Physicians, and the American Thoracic Society. Chest 2003, 124, 1543–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farre, R.; Montserrat, J.; Navajas, D. Noninvasive monitoring of respiratory mechanics during sleep. Eur. Respir. J. 2004, 24, 1052–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornton, A.T.; Singh, P.; Ruehland, W.R.; Rochford, P.D. AASM criteria for scoring respiratory events: Interaction between apnea sensor and hypopnea definition. Sleep 2012, 35, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Hernández, L.; Ballester, E.; Farré, R.; Badia, J.R.; Lobelo, R.; Navajas, D.; Montserrat, J.M. Performance of nasal prongs in sleep studies: Spectrum of flow-related events. Chest 2001, 119, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Masa, J.; Corral, J.; Martin, M.; Riesco, J.; Sojo, A.; Hernández, M.; Douglas, N. Assessment of thoracoabdominal bands to detect respiratory effort-related arousal. Eur. Respir. J. 2003, 22, 661–667. [Google Scholar] [CrossRef] [Green Version]
- Koo, B.B.; Drummond, C.; Surovec, S.; Johnson, N.; Marvin, S.A.; Redline, S. Validation of a polyvinylidene fluoride impedance sensor for respiratory event classification during polysomnography. J. Clin. Sleep Med. 2011, 7. [Google Scholar] [CrossRef] [Green Version]
- Tobin, M.J.; Cohn, M.A.; Sackner, M.A. Breathing abnormalities during sleep. Arch. Intern. Med. 1983, 143, 1221–1228. [Google Scholar] [CrossRef]
- Boudewyns, A.; Willemen, M.; Wagemans, M.; De Cock, W.; Van de Heyning, P.; De Backer, W. Assessment of respiratory effort by means of strain gauges and esophageal pressure swings: A comparative study. Sleep 1997, 20, 168–170. [Google Scholar] [CrossRef]
- Fontenla-Romero, O.; Guijarro-Berdiñas, B.; Alonso-Betanzos, A.; Moret-Bonillo, V. A new method for sleep apnea classification using wavelets and feedforward neural networks. Artif. Intell. Med. 2005, 34, 65–76. [Google Scholar] [CrossRef]
- Nakano, H.; Tanigawa, T.; Furukawa, T.; Nishima, S. Automatic detection of sleep-disordered breathing from a single-channel airflow record. Eur. Respir. J. 2007, 29, 728–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Shin, H.B.; Jeong, D.U.; Park, K.S. Detection of apneic events from single channel nasal airflow using 2nd derivative method. Comput. Methods Programs Biomed. 2008, 91, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, N.; Narasimhan, R. Detection of sleep apnea on a per-second basis using respiratory signals. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 2124–2127. [Google Scholar]
- Ciołek, M.; Niedźwiecki, M.; Sieklicki, S.; Drozdowski, J.; Siebert, J. Automated detection of sleep apnea and hypopnea events based on robust airflow envelope tracking in the presence of breathing artifacts. IEEE J. Biomed. Health Inform. 2015, 19, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Koley, B.L.; Dey, D. Automatic detection of sleep apnea and hypopnea events from single channel measurement of respiration signal employing ensemble binary SVM classifiers. Measurement 2013, 46, 2082–2092. [Google Scholar] [CrossRef]
- Koley, B.L.; Dey, D. Real-time adaptive apnea and hypopnea event detection methodology for portable sleep apnea monitoring devices. IEEE Trans. Biomed. Eng. 2013, 60, 3354–3363. [Google Scholar] [CrossRef]
- Várady, P.; Micsik, T.; Benedek, S.; Benyó, Z. A novel method for the detection of apnea and hypopnea events in respiration signals. Biomed. Eng. IEEE Trans. 2002, 49, 936–942. [Google Scholar] [CrossRef]
- Tian, J.; Liu, J. Apnea detection based on time delay neural network. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 2571–2574. [Google Scholar]
- Norman, R.G.; Rapoport, D.M.; Ayappa, I. Detection of flow limitation in obstructive sleep apnea with an artificial neural network. Physiol. Meas. 2007, 28, 1089. [Google Scholar] [CrossRef]
- Gutiérrez-Tobal, G.C.; Álvarez, D.; Marcos, J.V.; Del Campo, F.; Hornero, R. Pattern recognition in airflow recordings to assist in the sleep apnoea–hypopnoea syndrome diagnosis. Med. Biol. Eng. Comput. 2013, 51, 1367–1380. [Google Scholar] [CrossRef]
- Gutiérrez-Tobal, G.C.; Álvarez, D.; del Campo, F.; Hornero, R. Utility of adaboost to detect sleep apnea-hypopnea syndrome from single-channel airflow. IEEE Trans. Biomed. Eng. 2016, 63, 636–646. [Google Scholar] [CrossRef] [Green Version]
- Mostafa, S.S.; Mendonça, F.; G Ravelo-García, A.; Morgado-Dias, F. A Systematic Review of Detecting Sleep Apnea Using Deep Learning. Sensors 2019, 19, 4934. [Google Scholar] [CrossRef] [Green Version]
- Pathinarupothi, R.K.; Rangan, E.S.; Gopalakrishnan, E.; Vinaykumar, R.; Soman, K. Single sensor techniques for sleep apnea diagnosis using deep learning. In Proceedings of the 2017 IEEE International Conference on Healthcare Informatics (ICHI), Park City, UT, USA, 23–26 August 2017; pp. 524–529. [Google Scholar]
- Pathinarupothi, R.K.; Vinaykumar, R.; Rangan, E.; Gopalakrishnan, E.; Soman, K. Instantaneous heart rate as a robust feature for sleep apnea severity detection using deep learning. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Orlando, FL, USA, 16–19 February 2017; pp. 293–296. [Google Scholar]
- Cheng, M.; Sori, W.J.; Jiang, F.; Khan, A.; Liu, S. Recurrent neural network based classification of ECG signal features for obstruction of sleep apnea detection. In Proceedings of the 2017 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC), Guangzhou, China, 21–24 July 2017; pp. 199–202. [Google Scholar]
- De Falco, I.; De Pietro, G.; Sannino, G.; Scafuri, U.; Tarantino, E.; Della Cioppa, A.; Trunfio, G.A. Deep neural network hyper-parameter setting for classification of obstructive sleep apnea episodes. In Proceedings of the 2018 IEEE Symposium on Computers and Communications (ISCC), Natal, Brazil, 25–28 June 2018; pp. 01187–01192. [Google Scholar]
- Banluesombatkul, N.; Rakthanmanon, T.; Wilaiprasitporn, T. Single channel ECG for obstructive sleep apnea severity detection using a deep learning approach. In Proceedings of the TENCON 2018—2018 IEEE Region 10 Conference, Jeju, Korea, 28–31 October 2018; pp. 2011–2016. [Google Scholar]
- Li, K.; Pan, W.; Li, Y.; Jiang, Q.; Liu, G. A method to detect sleep apnea based on deep neural network and hidden markov model using single-lead ECG signal. Neurocomputing 2018, 294, 94–101. [Google Scholar] [CrossRef]
- Erdenebayar, U.; Kim, Y.J.; Park, J.U.; Joo, E.Y.; Lee, K.J. Deep learning approaches for automatic detection of sleep apnea events from an electrocardiogram. Comput. Methods Programs Biomed. 2019, 180, 105001. [Google Scholar] [CrossRef] [PubMed]
- Haidar, R.; McCloskey, S.; Koprinska, I.; Jeffries, B. Convolutional neural networks on multiple respiratory channels to detect hypopnea and obstructive apnea events. In Proceedings of the 2018 International Joint Conference on Neural Networks (IJCNN), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–7. [Google Scholar]
- Cen, L.; Yu, Z.L.; Kluge, T.; Ser, W. Automatic system for obstructive sleep apnea events detection using convolutional neural network. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 3975–3978. [Google Scholar]
- Biswal, S.; Sun, H.; Goparaju, B.; Westover, M.B.; Sun, J.; Bianchi, M.T. Expert-level sleep scoring with deep neural networks. J. Am. Med. Inform. Assoc. 2018, 25, 1643–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Steenkiste, T.; Groenendaal, W.; Deschrijver, D.; Dhaene, T. Automated sleep apnea detection in raw respiratory signals using long short-term memory neural networks. IEEE J. Biomed. Health Inform. 2018, 23, 2354–2364. [Google Scholar] [CrossRef] [Green Version]
- Haidar, R.; Koprinska, I.; Jeffries, B. Sleep apnea event detection from nasal airflow using convolutional neural networks. In Proceedings of the International Conference on Neural Information Processing, Guangzhou, China, 14–18 November 2017; Springer: Cham, Switzerland, 2017; pp. 819–827. [Google Scholar]
- McCloskey, S.; Haidar, R.; Koprinska, I.; Jeffries, B. Detecting hypopnea and obstructive apnea events using convolutional neural networks on wavelet spectrograms of nasal airflow. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Melbourne, VIC, Australia, 3–6 June 2018; Springer: Cham, Switzerland, 2018; pp. 361–372. [Google Scholar]
- Lakhan, P.; Ditthapron, A.; Banluesombatkul, N.; Wilaiprasitporn, T. Deep neural networks with weighted averaged overnight airflow features for sleep apnea-hypopnea severity classification. In Proceedings of the TENCON 2018—2018 IEEE Region 10 Conference, Jeju, Korea, 28–31 October 2018; pp. 0441–0445. [Google Scholar]
- Choi, S.H.; Yoon, H.; Kim, H.S.; Kim, H.B.; Kwon, H.B.; Oh, S.M.; Lee, Y.J.; Park, K.S. Real-time apnea-hypopnea event detection during sleep by convolutional neural networks. Comput. Biol. Med. 2018, 100, 123–131. [Google Scholar] [CrossRef]
- Werbos, P.J. Backpropagation through time: What it does and how to do it. Proc. IEEE 1990, 78, 1550–1560. [Google Scholar] [CrossRef] [Green Version]
- Bengio, Y.; Simard, P.; Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 1994, 5, 157–166. [Google Scholar] [CrossRef]
- Rajendran, S.; Meert, W.; Giustiniano, D.; Lenders, V.; Pollin, S. Deep learning models for wireless signal classification with distributed low-cost spectrum sensors. IEEE Trans. Cogn. Commun. Netw. 2018, 4, 433–445. [Google Scholar] [CrossRef] [Green Version]
- Gal, Y.; Ghahramani, Z. A theoretically grounded application of dropout in recurrent neural networks. In Proceedings of the Advances in Neural Information Processing Systems 29 (NIPS 2016), Barcelona, Spain, 5–10 December 2016; pp. 1019–1027. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Zweig, M.H.; Campbell, G. Receiver-Operating Characteristic (ROC) Plots: A Fundamental Evaluation Tool in Clinical Medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar] [CrossRef]
- Kim, J.; ElMoaqet, H.; Tilbury, D.M.; Ramachandran, S.K.; Penzel, T. Time domain characterization for sleep apnea in oronasal airflow signal: A dynamic threshold classification approach. Physiol. Meas. 2019, 40, 054007. [Google Scholar] [CrossRef] [PubMed]
- Adam: A Method for Stochastic Optimization. Available online: https://arxiv.org/abs/1412.6980 (accessed on 15 July 2020).
- Berg, S.; Haight, J.S.; Yap, V.; Hoffstein, V.; Cole, P. Comparison of direct and indirect measurements of respiratory airflow: Implications for hypopneas. Sleep 1997, 20, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Farré, R.; Montserrat, J.; Rotger, M.; Ballester, E.; Navajas, D. Accuracy of thermistors and thermocouples as flow-measuring devices for detecting hypopnoeas. Eur. Respir. J. 1998, 11, 179–182. [Google Scholar] [CrossRef] [Green Version]
- Thurnheer, R.; Xie, X.; Bloch, K.E. Accuracy of nasal cannula pressure recordings for assessment of ventilation during sleep. Am. J. Respir. Crit. Care Med. 2001, 164, 1914–1919. [Google Scholar] [CrossRef] [PubMed]
- Azimi, H.; Gilakjani, S.S.; Bouchard, M.; Goubran, R.A.; Knoefel, F. Automatic apnea-hypopnea events detection using an alternative sensor. In Proceedings of the 2018 IEEE Sensors Applications Symposium (SAS), Seoul, Korea, 12–14 March 2018; pp. 1–5. [Google Scholar]
- Koley, B.; Dey, D. Adaptive classification system for real-time detection of apnea and hypopnea events. In Proceedings of the 2013 IEEE Point-of-Care Healthcare Technologies (PHT), Bangalore, India, 16–18 January 2013; pp. 42–45. [Google Scholar]
- Koley, B.; Dey, D. Automated detection of apnea and hypopnea events. In Proceedings of the 2012 Third International Conference on Emerging Applications of Information Technology, Kolkata, India, 30 November–1 December 2012; pp. 85–88. [Google Scholar]
- Gutiérrez-Tobal, G.; Hornero, R.; Álvarez, D.; Marcos, J.; Del Campo, F. Linear and nonlinear analysis of airflow recordings to help in sleep apnoea–hypopnoea syndrome diagnosis. Physiol. Meas. 2012, 33, 1261. [Google Scholar] [CrossRef]
- Wong, K.K.; Jankelson, D.; Reid, A.; Unger, G.; Dungan, G.; Hedner, J.A.; Grunstein, R.R. Diagnostic test evaluation of a nasal flow monitor for obstructive sleep apnea detection in sleep apnea research. Behav. Res. Methods 2008, 40, 360–366. [Google Scholar] [CrossRef] [Green Version]
- BaHammam, A.; Sharif, M.; Gacuan, D.E.; George, S. Evaluation of the accuracy of manual and automatic scoring of a single airflow channel in patients with a high probability of obstructive sleep apnea. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2011, 17, MT13. [Google Scholar] [CrossRef] [Green Version]
- Rathnayake, S.I.; Wood, I.A.; Abeyratne, U.R.; Hukins, C. Nonlinear features for single-channel diagnosis of sleep-disordered breathing diseases. IEEE Trans. Biomed. Eng. 2010, 57, 1973–1981. [Google Scholar] [CrossRef]
- Rofail, L.M.; Wong, K.K.; Unger, G.; Marks, G.B.; Grunstein, R.R. The role of single-channel nasal airflow pressure transducer in the diagnosis of OSA in the sleep laboratory. J. Clin. Sleep Med. 2010, 6, 349–356. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, A.C.T.; Martinez, D.; Vasconcelos, L.F.T.; Gonçalves, S.C.; do Carmo Lenz, M.; Fuchs, S.C.; Gus, M.; de Abreu-Silva, E.O.; Moreira, L.B.; Fuchs, F.D. Diagnosis of obstructive sleep apnea syndrome and its outcomes with home portable monitoring. Chest 2009, 135, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.G.; Yang, M.C.; Chung, C.Y.; Chen, Y.T.; Chang, E.T. Respiratory-inductive-plethysmography-derived flow can be a useful clinical tool to detect patients with obstructive sleep apnea syndrome. J. Formos. Med. Assoc. 2011, 110, 642–645. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.Y.; Wu, H.T.; Hsu, C.A.; Huang, P.C.; Huang, Y.H.; Lo, Y.L. Sleep apnea detection based on thoracic and abdominal movement signals of wearable piezoelectric bands. IEEE J. Biomed. Health Inform. 2016, 21, 1533–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaughn, C.M.; Clemmons, P. Piezoelectric belts as a method for measuring chest and abdominal movement for obstructive sleep apnea diagnosis. Neurodiagn. J. 2012, 52, 275–280. [Google Scholar]
- Avcı, C.; Akbaş, A. Sleep apnea classification based on respiration signals by using ensemble methods. Bio-Med. Mater. Eng. 2015, 26, S1703–S1710. [Google Scholar]
- Redline, S.; Budhiraja, R.; Kapur, V.; Marcus, C.L.; Mateika, J.H.; Mehra, R.; Parthasarthy, S.; Somers, V.K.; Strohl, K.P.; Gozal, D.; et al. The scoring of respiratory events in sleep: Reliability and validity. J. Clin. Sleep Med. 2007, 3, 169–200. [Google Scholar] [CrossRef] [PubMed]
- Norman, R.G.; Ahmed, M.M.; Walsleben, J.A.; Rapoport, D.M. Detection of respiratory events during NPSG: Nasal cannula/pressure sensor versus thermistor. Sleep 1997, 20, 1175–1184. [Google Scholar]
- Series, F.; Marc, I. Nasal pressure recording in the diagnosis of sleep apnoea hypopnoea syndrome. Thorax 1999, 54, 506–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabil, A.; Glos, M.; Günther, A.; Schöbel, C.; Veauthier, C.; Fietze, I.; Penzel, T. Comparison of apnea detection using oronasal thermal airflow sensor, nasal pressure transducer, respiratory inductance plethysmography and tracheal sound sensor. J. Clin. Sleep Med. 2019, 15, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Data Summary | |||
|---|---|---|---|
| Labels/Segments | Training Set | Testing Set | Total |
| Normal | 29,078 | 7270 | 36,348 |
| Apnea | 7493 | 1873 | 9366 |
| Total | 36,571 | 9143 | 45,714 |
| Ref = +1 | Ref = 0 | |
|---|---|---|
| Det. = +1 | ||
| Det. = 0 |
| LSTM-Based Network—Overall Test Performance over 20% Hold Out of Data | |||||||
|---|---|---|---|---|---|---|---|
| Input Signal | |||||||
| NPRE | 90.0 (0.6) | 83.8 (0.5) | 85.1 (0.3) | 58.9 (0.6) | 97.0 (0.2) | 91.7 (0.2) | 71.2 (0.3) |
| ABD | 77.0 (1.1) | 82.7 (1.5) | 81.5 (1.1) | 53.4 (2.0) | 93.3 (0.3) | 86.5 (0.8) | 63.1 (1.5) |
| FlowTh | 85.1 (1.7) | 72.9 (2.1) | 75.4 (1.4) | 44.7 (1.4) | 95.0 (0.4) | 85.1 (1.8) | 58.6 (0.9) |
| BiLSTM-Based Network: Overall Performance over 20% Hold Out of Data | |||||||
|---|---|---|---|---|---|---|---|
| Input Signal | |||||||
| NPRE | 90.3 (0.5) | 83.7 (0.6) | 85.0 (0.4) | 58.8 (0.9) | 97.1 (0.1) | 92.4 (0.3) | 71.2 (0.5) |
| ABD | 78.5 (2.7) | 85.9 (0.7) | 84.4 (1.1) | 59.0 (2.0) | 94.0 (0.8) | 90.1 (2.1) | 67.4 (2.3) |
| FlowTh | 80.5 (1.7) | 81.6 (2.3) | 81.4 (1.5) | 53.0 (2.3) | 94.2 (0.3) | 89.0 (0.3) | 63.9 (1.3) |
| Leave One Out Test Results—LSTM-Based Detection Model with NPRE Signal | ||||||||
|---|---|---|---|---|---|---|---|---|
| # | Patient ID | |||||||
| 1 | 1 | 90.7 | 88.0 | 89.1 | 83.8 | 93.2 | 93.4 | 87.1 |
| 2 | 2 | 66.9 | 93.8 | 91.0 | 55.6 | 96.1 | 89.6 | 60.7 |
| 3 | 3 | 97.9 | 74.7 | 80.9 | 58.3 | 99.0 | 91.8 | 73.1 |
| 4 | 4 | 94.8 | 92.1 | 92.6 | 75.7 | 98.5 | 96.6 | 84.2 |
| 5 | 8 | 81.3 | 89.4 | 87.5 | 70.6 | 93.8 | 91.5 | 75.6 |
| 6 | 9 | 92.6 | 86.1 | 88.4 | 78.2 | 95.6 | 94.6 | 84.8 |
| 7 | 10 | 41.9 | 98.9 | 98.0 | 37.5 | 99.1 | 93.9 | 39.6 |
| 8 | 15 | 91.7 | 77.0 | 78.8 | 35.3 | 98.5 | 90.9 | 51.0 |
| 9 | 16 | 89.8 | 76.7 | 77.1 | 12.9 | 99.5 | 91.4 | 22.6 |
| 10 | 17 | 97.4 | 78.2 | 84.0 | 65.7 | 98.6 | 92.7 | 78.5 |
| 11 | 18 | 97.0 | 50.6 | 59.5 | 31.9 | 98.6 | 74.8 | 48.0 |
| 14 | 21 | 96.3 | 70.9 | 82.8 | 74.3 | 95.7 | 89.7 | 83.9 |
| 15 | 22 | 77.3 | 94.5 | 93.1 | 56.4 | 97.9 | 93.4 | 65.2 |
| 16 | 23 | 93.9 | 92.9 | 93.8 | 62.0 | 99.2 | 97.6 | 74.7 |
| 17 | 24 | 88.7 | 76.3 | 81.9 | 75.4 | 89.2 | 90.1 | 81.5 |
| Average | 86.7 | 83.0 | 85.4 | 57.4 | 96.9 | 91.7 | 69.0 | |
| Leave One Out Test Results—BiLSTM-Based Detection Model with NPRE Signal | ||||||||
|---|---|---|---|---|---|---|---|---|
| # | Patient ID | |||||||
| 1 | 1 | 91.7 | 89.5 | 90.4 | 85.7 | 94.0 | 95.8 | 88.6 |
| 2 | 2 | 63.3 | 94.5 | 91.2 | 56.9 | 95.7 | 88.7 | 59.9 |
| 3 | 3 | 97.9 | 72.6 | 79.3 | 56.3 | 99.0 | 92.0 | 71.5 |
| 4 | 4 | 97.1 | 87.4 | 89.4 | 66.7 | 99.1 | 97.0 | 79.1 |
| 5 | 8 | 74.8 | 92.0 | 87.9 | 74.5 | 92.1 | 92.4 | 74.6 |
| 6 | 9 | 94.7 | 84.3 | 87.9 | 76.5 | 96.7 | 95.2 | 84.6 |
| 7 | 10 | 62.8 | 98.3 | 97.7 | 37.0 | 99.4 | 96.9 | 46.6 |
| 8 | 15 | 92.0 | 78.7 | 80.3 | 37.2 | 98.6 | 91.6 | 53.0 |
| 9 | 16 | 88.0 | 80.1 | 80.4 | 14.6 | 99.4 | 92.2 | 25.0 |
| 10 | 17 | 96.5 | 81.0 | 85.6 | 68.5 | 98.2 | 95.9 | 80.1 |
| 11 | 18 | 96.6 | 51.9 | 60.5 | 32.4 | 98.5 | 73.2 | 48.5 |
| 12 | 19 | 78.0 | 95.9 | 94.6 | 58.7 | 98.3 | 96.7 | 67.0 |
| 13 | 20 | 90.2 | 74.7 | 77.6 | 44.8 | 97.1 | 89.8 | 59.9 |
| 14 | 21 | 92.9 | 75.6 | 83.7 | 76.8 | 92.5 | 90.9 | 84.1 |
| 15 | 22 | 72.5 | 94.5 | 92.7 | 54.8 | 97.4 | 92.4 | 62.4 |
| 16 | 23 | 93.5 | 93.3 | 93.3 | 60.2 | 99.3 | 97.7 | 73.2 |
| 17 | 24 | 79.0 | 85.6 | 82.6 | 81.7 | 83.2 | 90.0 | 80.3 |
| Average | 86.0 | 84.1 | 85.6 | 57.8 | 96.4 | 92.3 | 69.2 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
ElMoaqet, H.; Eid, M.; Glos, M.; Ryalat, M.; Penzel, T. Deep Recurrent Neural Networks for Automatic Detection of Sleep Apnea from Single Channel Respiration Signals. Sensors 2020, 20, 5037. https://doi.org/10.3390/s20185037
ElMoaqet H, Eid M, Glos M, Ryalat M, Penzel T. Deep Recurrent Neural Networks for Automatic Detection of Sleep Apnea from Single Channel Respiration Signals. Sensors. 2020; 20(18):5037. https://doi.org/10.3390/s20185037
Chicago/Turabian StyleElMoaqet, Hisham, Mohammad Eid, Martin Glos, Mutaz Ryalat, and Thomas Penzel. 2020. "Deep Recurrent Neural Networks for Automatic Detection of Sleep Apnea from Single Channel Respiration Signals" Sensors 20, no. 18: 5037. https://doi.org/10.3390/s20185037
APA StyleElMoaqet, H., Eid, M., Glos, M., Ryalat, M., & Penzel, T. (2020). Deep Recurrent Neural Networks for Automatic Detection of Sleep Apnea from Single Channel Respiration Signals. Sensors, 20(18), 5037. https://doi.org/10.3390/s20185037








