Estimation Method of Soluble Solid Content in Peach Based on Deep Features of Hyperspectral Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Data Collection
2.2.1. Hyperspectral Image Acquisition
2.2.2. Peach Soluble Solids Content Collection
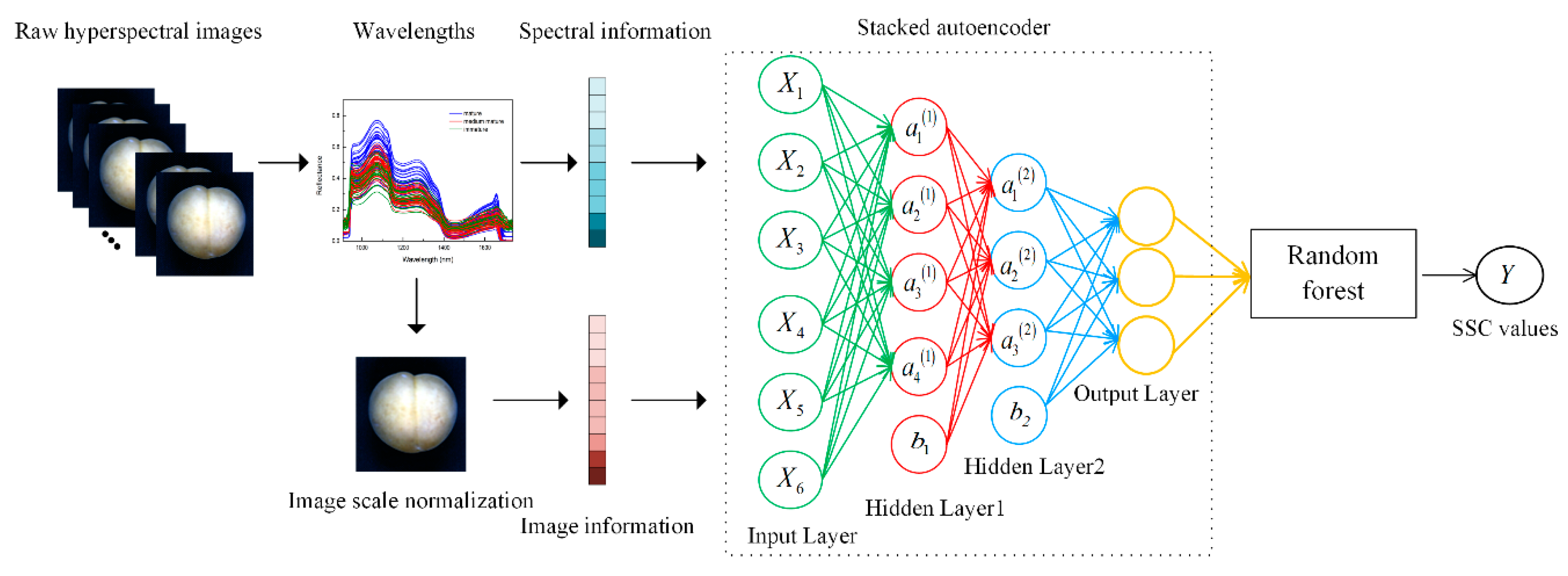
2.3. Methodology
2.3.1. Stacked Autoencoder
2.3.2. Information Extraction from Hyperspectral Images
2.3.3. Stacked Autoencoder–Random Forest
3. Results and Analysis
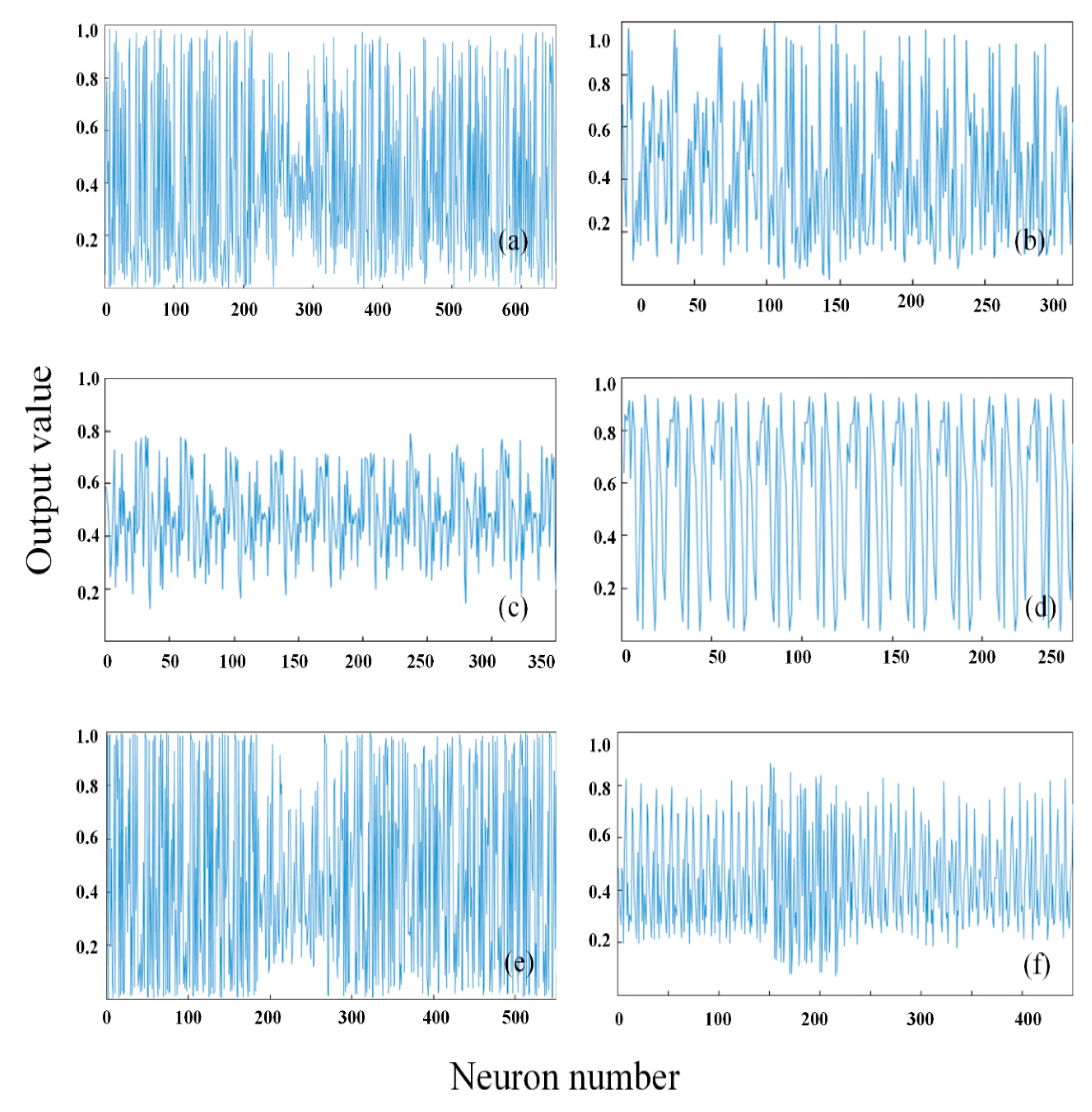
3.1. Deep Feature Extraction Results of Hyperspectral Images
3.2. Different Structures of SAE for Peach to Estimate Soluble Solids Content
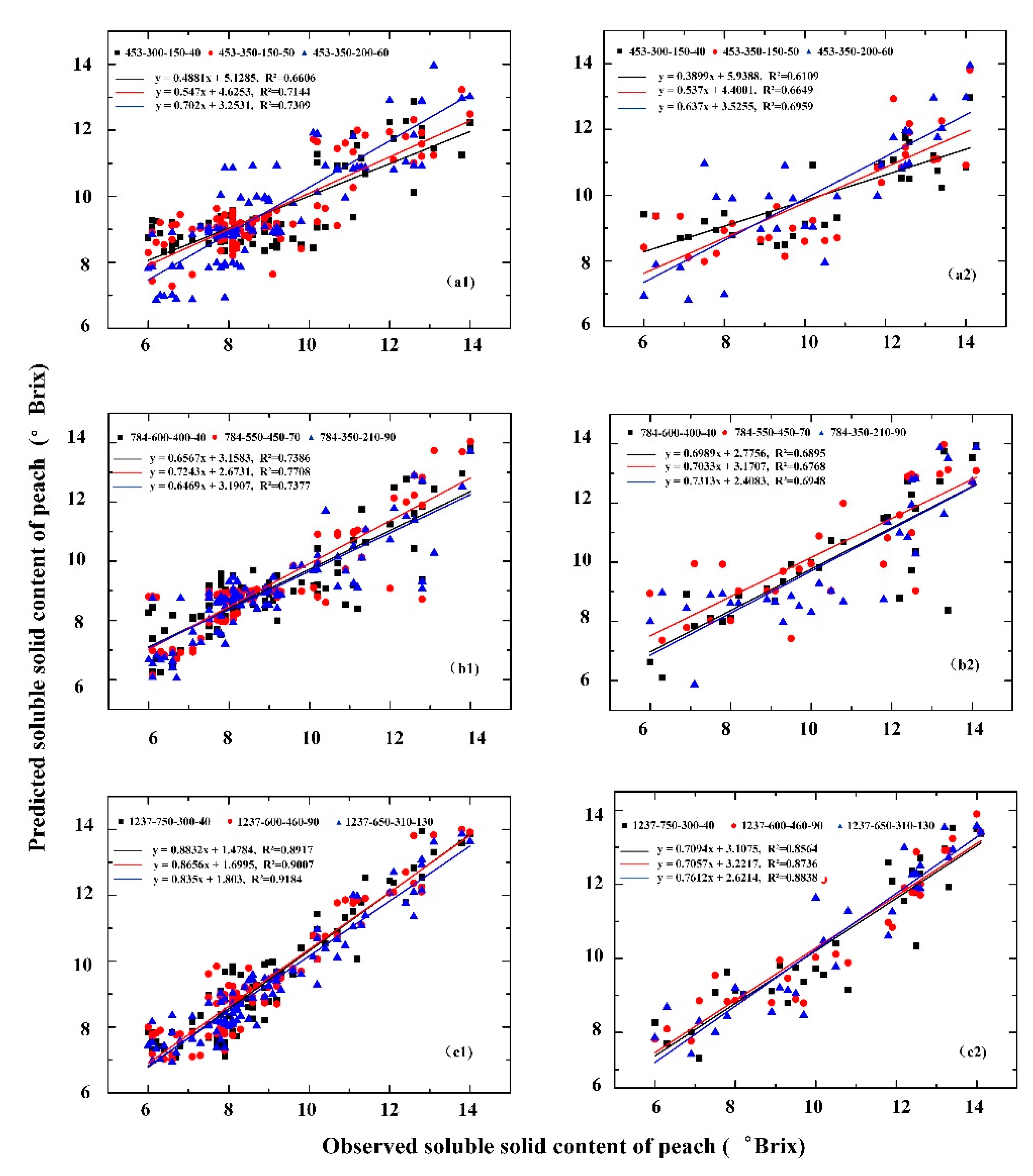
3.3. Comparison of Estimation Models of Peach SSC Based on Different Features
4. Discussion
4.1. Parameter Selection and Experimental Results
4.2. Different Structures and Different Features
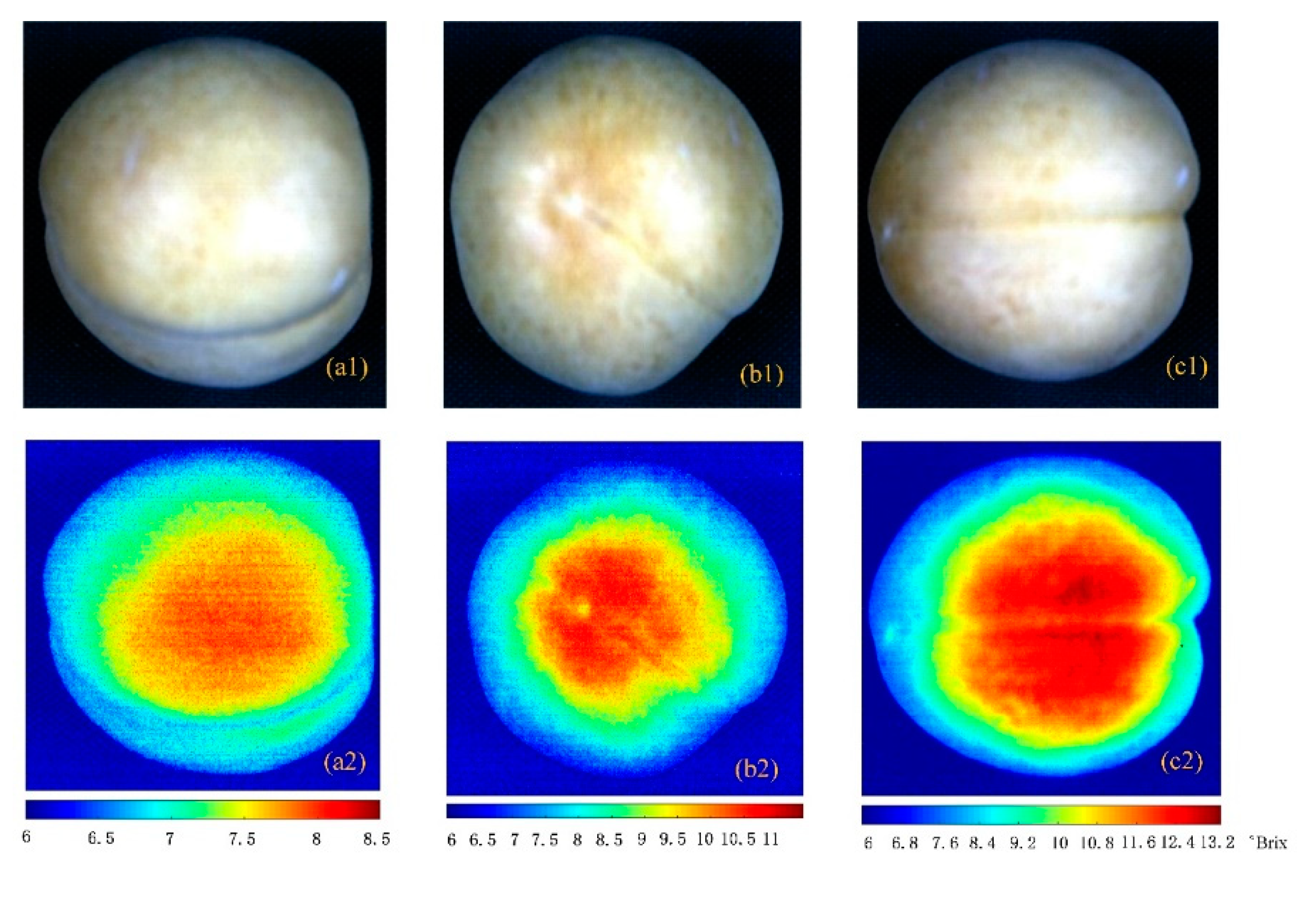
4.3. Visualized Results of Soluble Solid Content
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shah, S.T.; Sajid, M. Influence of calcium sources and concentrations on the quality and storage performance of peach. Sarhad J. Agric. 2017, 33, 532–539. [Google Scholar] [CrossRef]
- Pinto, C.D.; Reginato, G.; Mesa, K.; Shinya, P.; Diaz, M.; Infante, R. Monitoring the flesh softening and the ripening of peach during the last phase of growth on-tree. Hortscience 2016, 51, 995–1000. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Fu, Q.; Fu, Z.; Li, X.; Matetic, M.; Bakaric, M.B.; Jemric, T. A comprehensive peach fruit quality evaluation method for grading and consumption. Appl Sci. 2020, 10, 1348. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Lu, R. Prediction of apple fruit firmness and soluble solids content using characteristics of multispectral scattering images. J. Food Eng. 2007, 82, 142–152. [Google Scholar] [CrossRef]
- Lu, R. Multispectral imaging for predicting firmness and soluble solids content of apple fruit. Postharvest Biol. Technol. 2004, 31, 147–157. [Google Scholar] [CrossRef]
- Li, J.; Xue, L.; Liu, M.H.; Wang, X.; Luo, C.S. Study of fluorescence spectrum for measurement of soluble solids content in navel orange. Adv. Mater. Res. 2011, 186, 126–130. [Google Scholar] [CrossRef]
- Gao, F.; Dong, Y.; Xiao, W.; Yin, B.; Yan, C.; He, S. LED-induced fluorescence spectroscopy technique for apple freshness and quality detection. Postharvest Biol. Technol. 2016, 119, 27–32. [Google Scholar] [CrossRef]
- Moigne, M.L.; Dufour, E.; Bertrand, D.; Maury, C.; Seraphin, D.; Jourjon, F. Front face fluorescence spectroscopy and visible spectroscopy coupled with chemometrics have the potential to characterise ripening of Cabernet Franc grapes. Anal. Chim. Acta 2008, 621, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Penchaiya, P.; Bobelyn, E.; Verlinden, B.; Nicolai, B.; Saeys, W. Non-destructive measurement of firmness and soluble solids content in bell pepper using NIR spectroscopy. J. Food Eng. 2009, 94, 267–273. [Google Scholar] [CrossRef]
- Xie, L.; Ying, Y.; Lin, H.; Zhou, Y.; Niu, X. Nondestructive determination of soluble solids content and pH in tomato juice using NIR transmittance spectroscopy. Sens. Instrum. Food Qual. Saf. 2008, 2, 111–115. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, B.; Li, J.; Huang, W.; Wang, C. Effect of spectrum measurement position variation on the robustness of NIR spectroscopy models for soluble solids content of apple. Biosyst. Eng. 2016, 143, 9–19. [Google Scholar] [CrossRef]
- Moller, S.M.; Travers, S.; Bertram, H.C.; Bertelsen, M.G. Prediction of postharvest dry matter, soluble solids content, firmness and acidity in apples (cv. Elshof) using NMR and NIR spectroscopy: A comparative study. Eur. Food Res. Technol. 2013, 237, 1021–1024. [Google Scholar] [CrossRef]
- Zhang, W.; Pan, L.; Zhao, X.; Tu, K. A Study on soluble solids content assessment using electronic nose: Persimmon fruit picked on different dates. Int. J. Food Prop. 2016, 19, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, J.; Ye, S. Prediction of soluble solids content, firmness and pH of pear by signals of electronic nose sensors. Anal. Chim. Acta 2008, 606, 112–118. [Google Scholar] [CrossRef]
- Xu, S.; Lu, H.; Ference, C.; Zhang, Q. Visible/near infrared reflection spectrometer and electronic nose data fusion as an accuracy improvement method for portable total soluble solid content detection of orange. Appl. Sci. 2019, 9, 3761. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Guo, W. Nondestructive determination of soluble solids content of persimmons by using dielectric spectroscopy. Int. J. Food Prop. 2018, 20, S2596–S2611. [Google Scholar] [CrossRef]
- Guo, W.; Fang, L.; Liu, D.; Wang, Z. Determination of soluble solids content and firmness of pears during ripening by using dielectric spectroscopy. Comput. Electron. Agric. 2015, 117, 226–233. [Google Scholar] [CrossRef]
- Li, J.; Peng, Y.K.; Chen, L.P.; Huang, W.Q. Near-infrared hyperspectral imaging combined with cars algorithm to quantitatively determine soluble solids content in “Ya” pear. Spectrosc. Spectr. Anal. 2014, 34, 1264–1269. [Google Scholar]
- Pu, Y.; Sun, D.; Riccioli, C.; Buccheri, M.; Grassi, M.; Cattaneo, T.M.; Gowen, A. Calibration transfer from micro nir spectrometer to hyperspectral imaging: A case study on predicting soluble solids content of bananito fruit (Musa acuminata). Food Anal. Meth. 2018, 11, 1021–1033. [Google Scholar] [CrossRef]
- Dong, J.; Guo, W.; Wang, Z.; Liu, D.; Zhao, F. Nondestructive determination of soluble solids content of ‘fuji’ apples produced in different areas and bagged with different materials during ripening. Food Anal. Meth. 2016, 9, 1087–1095. [Google Scholar] [CrossRef]
- Leiva-Valenzuela, G.A.; Lu, R.; Aguilera, J.M. Prediction of firmness and soluble solids content of blueberries using hyperspectral reflectance imaging. J. Food Eng. 2013, 115, 91–98. [Google Scholar] [CrossRef]
- Guo, W.; Zhao, F.; Dong, J. Nondestructive measurement of soluble solids content of kiwifruits using near-infrared hyperspectral imaging. Food Anal. Meth. 2016, 9, 38–47. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, L.; Guo, E. Hyperspectral laser-induced fluorescence imaging for nondestructive assessing soluble solids content of orange. In Proceedings of the International Conference on Computer and Computing Technologies in Agriculture, Wuyishan, China, 18–20 August 2007; pp. 51–59. [Google Scholar]
- Baiano, A.; Terracone, C.; Peri, G.; Romaniello, R. Application of hyperspectral imaging for prediction of physico-chemical and sensory characteristics of table grapes. Comput. Electron. Agric. 2012, 87, 142–151. [Google Scholar] [CrossRef]
- Ma, T.; Li, X.; Inagaki, T.; Yang, H.; Tsuchikawa, S. Noncontact evaluation of soluble solids content in apples by near-infrared hyperspectral imaging. J. Food Eng. 2017, 224, 53–61. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, B.; Li, J.; Liu, C.; Huang, W.; Tian, X. Prediction of soluble solids content of apple using the combination of spectra and textural features of hyperspectral reflectance imaging data. Postharvest Biol. Technol. 2016, 121, 51–61. [Google Scholar] [CrossRef]
- Li, J.; Chen, L. Comparative analysis of models for robust and accurate evaluation of soluble solids content in ‘Pinggu’ peaches by hyperspectral imaging. Comput. Electron. Agric. 2017, 142, 524–535. [Google Scholar] [CrossRef]
- Yang, R.; Kan, J. Classification of tree species at the leaf level based on hyperspectral imaging technology. J. Appl. Spectrosc. 2020, 87, 184–193. [Google Scholar] [CrossRef]
- Fernandez, D.; Gonzalez, C.; Mozos, D.; Lopez, S. Fpga implementation of the principal component analysis algorithm for dimensionality reduction of hyperspectral images. J. Real-Time Image Process. 2019, 16, 1395–1406. [Google Scholar] [CrossRef]
- Dye, M.; Mutanga, O.; Ismail, R. Examining the utility of random forest and AISA Eagle hyperspectral image data to predict Pinus patula age in KwaZulu-Natal, South Africa. Geocarto Int. 2011, 26, 275–289. [Google Scholar] [CrossRef]
- Li, S.; Yu, B.; Wu, W.; Su, S.; Ji, R. Feature learning based on sae-pca network for human gesture recognition in rgbd images. Neurocomputing 2015, 151, 565–573. [Google Scholar] [CrossRef]
- Han, X.; Zhong, Y.; Zhang, L. Spatial-spectral unsupervised convolutional sparse auto-encoder classifier for hyperspectral imagery. Photogramm. Eng. Remote Sens. 2017, 83, 195–206. [Google Scholar] [CrossRef]
- Yu, X.; Lu, H.; Wu, D. Development of deep learning method for predicting firmness and soluble solid content of postharvest korla fragrant pear using Vis/Nir hyperspectral reflectance imaging. Postharvest Biol. Technol. 2018, 141, 39–49. [Google Scholar] [CrossRef]
- Shen, L.X.; Wang, H.H.; Liu, Y.; Liu, Y.; Zhang, X.; Fei, Y.Q. Prediction of soluble solids content in green plum by using a sparse autoencoder. Appl. Sci. 2020, 10, 3769. [Google Scholar] [CrossRef]
- Yang, B.; Zhu, Y.; Wang, M.; Ning, J. A model for yellow tea polyphenols content estimation based on multi-feature fusion. IEEE Access 2019, 7, 180054–180063. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Feng, J.; Xiao, S.; Yau, W.; Zhou, J.T.; Yang, S. Structured autoencoders for subspace clustering. IEEE Trans. Image Process. 2018, 27, 5076–5086. [Google Scholar] [CrossRef]
- Blaschke, T.; Olivecrona, M.; Engkvist, O.; Bajorath, J.; Chen, H.M. Application of generative autoencoder in de novo molecular design. Mol. Inform. 2018, 37, 1700123. [Google Scholar] [CrossRef] [Green Version]
- Hassairi, S.; Ejbali, R.; Zaied, M. A deep stacked wavelet auto-encoders to supervised feature extraction to pattern classification. Multimed Tools Appl. 2018, 77, 5443–5459. [Google Scholar] [CrossRef]
- Yang, B.; Qi, L.; Wang, M.; Hussain, S.; Wang, H.; Wang, B.; Ning, J. Cross-category tea polyphenols evaluation model based on feature fusion of electronic nose and hyperspectral imagery. Sensors 2020, 20, 50. [Google Scholar] [CrossRef] [Green Version]
- Ding, H.; Xu, L.; Wu, Y.; Shi, W. Classification of hyperspectral images by deep learning of spectral-spatial features. Arab J. Geosci. 2020, 13, 464. [Google Scholar] [CrossRef]
- Seifi Majdar, R.; Ghassemian, H. A probabilistic SVM approach for hyperspectral image classification using spectral and texture features. Int. J. Remote Sens. 2017, 38, 4265–4284. [Google Scholar] [CrossRef]


| Dataset | Number | Content Range (%) | Mean (%) | SD (%) |
|---|---|---|---|---|
| Full | 120 | 6–14.1 | 9.29 | 2.17 |
| Calibration set | 90 | 6–14 | 8.91 | 1.94 |
| Validation | 30 | 6–14.1 | 10.41 | 2.41 |
| Features | SAE Optimal Scale | Calibration Set | Validation Set | ||
|---|---|---|---|---|---|
| R2 | RMSE | R2 | RMSE | ||
| Deep feature of spectral | 453-300-150-40 | 0.6606 | 1.3323 | 0.6109 | 1.699 |
| 453-350-150-50 | 0.7144 | 1.2551 | 0.6649 | 1.5033 | |
| 453-350-200-60 | 0.7309 | 1.1744 | 0.6959 | 0.2486 | |
| Deep feature of image | 784-600-400-40 | 0.7386 | 1.0163 | 0.6895 | 1.3886 |
| 784-550-450-70 | 0.7708 | 0.9613 | 0.6747 | 1.3733 | |
| 784-350-210-90 | 0.7377 | 1.018 | 0.6948 | 1.3825 | |
| Deep feature of fusion information | 1237-750-300-40 | 0.8917 | 0.7756 | 0.8564 | 0.9922 |
| 1237-600-460-90 | 0.9007 | 0.7956 | 0.8736 | 0.9715 | |
| 1237-650-310-130 | 0.9184 | 0.6693 | 0.8838 | 0.8887 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Gao, Y.; Yan, Q.; Qi, L.; Zhu, Y.; Wang, B. Estimation Method of Soluble Solid Content in Peach Based on Deep Features of Hyperspectral Imagery. Sensors 2020, 20, 5021. https://doi.org/10.3390/s20185021
Yang B, Gao Y, Yan Q, Qi L, Zhu Y, Wang B. Estimation Method of Soluble Solid Content in Peach Based on Deep Features of Hyperspectral Imagery. Sensors. 2020; 20(18):5021. https://doi.org/10.3390/s20185021
Chicago/Turabian StyleYang, Baohua, Yuan Gao, Qian Yan, Lin Qi, Yue Zhu, and Bing Wang. 2020. "Estimation Method of Soluble Solid Content in Peach Based on Deep Features of Hyperspectral Imagery" Sensors 20, no. 18: 5021. https://doi.org/10.3390/s20185021
APA StyleYang, B., Gao, Y., Yan, Q., Qi, L., Zhu, Y., & Wang, B. (2020). Estimation Method of Soluble Solid Content in Peach Based on Deep Features of Hyperspectral Imagery. Sensors, 20(18), 5021. https://doi.org/10.3390/s20185021






