DNA/RNA Electrochemical Biosensing Devices a Future Replacement of PCR Methods for a Fast Epidemic Containment
Abstract
1. Introduction
2. Signal Transduction
3. Signal Amplification Approaches for the DNA/RNA Electrochemical Sensor
3.1. Enzyme Mediated Signal Amplification
3.2. Nanomaterial Enhanced Signal
3.3. Nucleic Acid Amplification and Processing Based Approaches
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Wittwer, C.T.; Makrigiorgos, G.M. Nucleic acid techniques. In Principles and Applications of Molecular Diagnostics; Elsevier: Amsterdam, The Netherlands, 2018; pp. 47–86. ISBN 978-0-12-816061-9. [Google Scholar]
- Cheng, V.C.C.; Wong, S.C.; Chen, J.H.K.; Yip, C.C.Y.; Chuang, V.W.M.; Tsang, O.T.Y.; Sridhar, S.; Chan, J.F.W.; Ho, P.L.; Yuen, K.Y. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect. Control Hosp. Epidemiol. 2020, 41, 493–498. [Google Scholar] [CrossRef]
- Fritsch, A.; Schweiger, B.; Biere, B. Influenza c virus in pre-school children with respiratory infections: Retrospective analysis of data from the national influenza surveillance system in germany, 2012 to 2014. Eurosurveillance 2019, 24. [Google Scholar] [CrossRef]
- Sonawane, A.A.; Shastri, J.; Bavdekar, S.B. Respiratory pathogens in infants diagnosed with acute lower respiratory tract infection in a tertiary care hospital of western India using multiplex Real Time PCR. Indian J. Pediatr. 2019, 86, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Deghmane, A.E.; Hong, E.; Taha, M.K. Diagnosis of meningococcal infection using internally controlled multiplex real-time PCR. In Neisseria Meningitidis: Methods and Protocols; Humana Press Inc.: New York, NY, USA, 2019; Volume 1969, pp. 17–31. ISBN 978-1-4939-9202-7. [Google Scholar]
- Wlassow, M.; Poiteau, L.; Roudot-Thoraval, F.; Rosa, I.; Soulier, A.; Hézode, C.; Ortonne, V.; Pawlotsky, J.M.; Chevaliez, S. The new Xpert HCV viral load real-time PCR assay accurately quantifies hepatitis C virus RNA in serum and whole-blood specimens. J. Clin. Virol. 2019, 117, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zheng, X.; Zhao, C.; Li, Y.; Chen, S.; Liu, G.; Wang, C.; Lv, Q.; Liu, P.; Zheng, Y.; et al. Detection of pathogenic microorganisms from bloodstream infection specimens using TaqMan array card technology. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Loonen, A.J.M.; Bos, M.P.; van Meerbergen, B.; Neerken, S.; Catsburg, A.; Dobbelaer, I.; Penterman, R.; Maertens, G.; van de Wiel, P.; Savelkoul, P.; et al. Comparison of pathogen DNA isolation methods from large volumes of whole blood to improve molecular diagnosis of bloodstream infections. PLoS ONE 2013, 8, e72349. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef] [PubMed]
- Myers, F.B.; Lee, L.P. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab. Chip 2008, 8, 2015–2031. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Jiang, B.; Xie, J.; Xiang, Y.; Yuan, R.; Chai, Y. Sensitive point-of-care monitoring of HIV related DNA sequences with a personal glucometer. Chem. Commun. 2012, 48, 10733–10735. [Google Scholar] [CrossRef]
- Trotter, M.; Borst, N.; Thewes, R.; von Stetten, F. Review: Electrochemical DNA sensing–Principles, commercial systems, and applications. Biosens. Bioelectron. 2020, 154, 112069. [Google Scholar] [CrossRef]
- Liu, G.; Wan, Y.; Gau, V.; Zhang, J.; Wang, L.; Song, S.; Fan, C. An enzyme-based E-DNA sensor for sequence-specific detection of femtomolar DNA targets. J. Am. Chem. Soc. 2008, 130, 6820–6825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Chen, S.X.; Yin, P. Optimizing the specificity of nucleic acid hybridization. Nat. Chem. 2012, 4, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Nanomaterial-Based amplified transduction of biomolecular interactions. Small 2005, 1, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Blair, E.O.; Corrigan, D.K. A review of microfabricated electrochemical biosensors for DNA detection. Biosens. Bioelectron. 2019, 134, 57–67. [Google Scholar] [CrossRef]
- Hoilett, O.S.; Walker, J.F.; Balash, B.M.; Jaras, N.J.; Boppana, S.; Linnes, J.C. KickStat: A coin-sized potentiostat for high-resolution electrochemical analysis. Sensors 2020, 20, 2407. [Google Scholar] [CrossRef]
- Chapman, J.; Power, A.; Kiran, K.; Chandra, S. Review—New twists in the plot: Recent advances in electrochemical genosensors for disease screening. J. Electrochem. Soc. 2017, 164, B665. [Google Scholar] [CrossRef]
- Ozer, T.; Geiss, B.J.; Henry, C.S. Review—Chemical and biological sensors for viral detection. J. Electrochem. Soc. 2019, 167, 037523. [Google Scholar] [CrossRef]
- Yáñez-Sedeño, P.; Campuzano, S.; Pingarrón, J.M. Pushing the limits of electrochemistry toward challenging applications in clinical diagnosis, prognosis, and therapeutic action. Chem. Commun. 2019, 55, 2563–2592. [Google Scholar] [CrossRef]
- Smith, S.J.; Nemr, C.R.; Kelley, S.O. Chemistry-Driven approaches for ultrasensitive nucleic acid detection. J. Am. Chem. Soc. 2017, 139, 1020–1028. [Google Scholar] [CrossRef]
- Bonanni, A.; del Valle, M. Use of nanomaterials for impedimetric DNA sensors: A review. Anal. Chim. Acta 2010, 678, 7–17. [Google Scholar] [CrossRef]
- Kupis-Rozmysłowicz, J.; Antonucci, A.; Boghossian, A.A. Review—Engineering the selectivity of the DNA-SWCNT sensor. ECS J. Solid State Sci. Technol. 2016, 5, M3067. [Google Scholar] [CrossRef]
- Rasheed, P.A.; Sandhyarani, N. Electrochemical DNA sensors based on the use of gold nanoparticles: A review on recent developments. Microchim. Acta 2017, 184, 981–1000. [Google Scholar] [CrossRef]
- Vikrant, K.; Bhardwaj, N.; Bhardwaj, S.K.; Kim, K.-H.; Deep, A. Nanomaterials as efficient platforms for sensing DNA. Biomaterials 2019, 214, 119215. [Google Scholar] [CrossRef] [PubMed]
- Pellitero, M.A.; Shaver, A.; Arroyo-Currás, N. Critical review—Approaches for the electrochemical interrogation of DNA-based sensors: A critical review. J. Electrochem. Soc. 2020, 167, 037529. [Google Scholar] [CrossRef]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef]
- Leslie, A. Pray Discovery of DNA structure and function: Watson and Crick. Nat. Educ. 2008, 1, 100. [Google Scholar]
- Inouye, M.; Ikeda, R.; Takase, M.; Tsuri, T.; Chiba, J. Single-nucleotide polymorphism detection with “wire-like” DNA probes that display quasi “on-off” digital action. Proc. Natl. Acad. Sci. USA 2005, 102, 11606–11610. [Google Scholar] [CrossRef]
- Gong, P.; Levicky, R. DNA surface hybridization regimes. Proc. Natl. Acad. Sci. USA 2008, 105, 5301–5306. [Google Scholar] [CrossRef]
- Levicky, R.; Horgan, A. Physicochemical perspectives on DNA microarray and biosensor technologies. Trends Biotechnol. 2005, 23, 143–149. [Google Scholar] [CrossRef]
- Ferapontova, E.E. DNA Electrochemistry and Electrochemical Sensors for Nucleic Acids. Annu. Rev. Anal. Chem. 2018. [Google Scholar] [CrossRef]
- Paleček, E.; Bartošík, M. Electrochemistry of nucleic acids. Chem. Rev. 2012, 112, 3427–3481. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E.; Shipovskov, S.V. Electrochemically induced oxidative damage to double stranded calf thymus DNA adsorbed on gold electrodes. Biochem. Mosc. 2003, 68, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhou, Y.; Ma, Q.; Ai, S.; Ju, P.; Zhu, L.; Lu, L. Electrochemical oxidation behavior of guanine and adenine on graphene-Nafion composite film modified glassy carbon electrode and the simultaneous determination. Process Biochem. 2010, 45, 1707–1712. [Google Scholar] [CrossRef]
- Zhou, M.; Zhai, Y.; Dong, S. Electrochemical sensing and biosensing platform based on chemically reduced graphene oxide. Anal. Chem. 2009, 81, 5603–5613. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.C.B.; Oliveira-Brett, A.M. In situ DNA oxidative damage by electrochemically generated hydroxyl free radicals on a boron-doped diamond electrode. Langmuir 2012, 28, 4896–4901. [Google Scholar] [CrossRef] [PubMed]
- Özcan, A.; Şahin, Y.; Özsöz, M.; Turan, S. Electrochemical oxidation of ds-DNA on polypyrrole nanofiber modified pencil graphite electrode. Electroanalysis 2007, 19, 2208–2216. [Google Scholar] [CrossRef]
- Pogacean, F.; Biris, A.R.; Coros, M.; Watanabe, F.; Biris, A.S.; Clichici, S.; Filip, A.; Pruneanu, S. Electrochemical oxidation of adenine using platinum electrodes modified with carbon nanotubes. Phys. E Low Dimens. Syst. Nanostruct. 2014, 59, 181–185. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, J.; Luo, J.; Lin, Z. Oxidation and adsorption of deoxyribonucleic acid at highly ordered pyrolytic graphite electrode. Electrochim. Acta 2000, 45, 2923–2927. [Google Scholar] [CrossRef]
- Wang, L.; Veselinovic, M.; Yang, L.; Geiss, B.J.; Dandy, D.S.; Chen, T. A sensitive DNA capacitive biosensor using interdigitated electrodes. Biosens. Bioelectron. 2017, 87, 646–653. [Google Scholar] [CrossRef]
- Manzano, M.; Viezzi, S.; Mazerat, S.; Marks, R.S.; Vidic, J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018, 100, 89–95. [Google Scholar] [CrossRef]
- Subak, H.; Ozkan-Ariksoysal, D. Label-free electrochemical biosensor for the detection of Influenza genes and the solution of guanine-based displaying problem of DNA hybridization. Sens. Actuators B Chem. 2018, 263, 196–207. [Google Scholar] [CrossRef]
- Deshmukh, R.; Prusty, A.K.; Roy, U.; Bhand, S. A capacitive DNA sensor for sensitive detection of: Escherichia coli O157:H7 in potable water based on the z3276 genetic marker: Fabrication and analytical performance. Analyst 2020, 145, 2267–2278. [Google Scholar] [CrossRef] [PubMed]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors—Sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, I.; Malecka, K.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecka, H.; Radecki, J. Single electrode genosensor for simultaneous determination of sequences encoding hemagglutinin and neuraminidase of avian influenza virus type H5N1. Anal. Chem. 2013, 85, 10167–10173. [Google Scholar] [CrossRef]
- Malecka, K.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecka, H.; Radecki, J. Electrochemical genosensor based on disc and screen printed gold electrodes for detection of specific DNA and RNA sequences derived from Avian Influenza Virus H5N1. Sens. Actuators B Chem. 2016, 224, 290–297. [Google Scholar] [CrossRef]
- Ilkhani, H.; Farhad, S. A novel electrochemical DNA biosensor for Ebola virus detection. Anal. Biochem. 2018, 557, 151–155. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, R.; Zhu, J.; Lu, X.; Li, Y.; Qiu, S.; Jia, L.; Jiao, X.; Song, S.; Fan, C.; et al. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of avian influenza A (H7N9) virus. ACS Appl. Mater. Interfaces 2015, 7, 8834–8842. [Google Scholar] [CrossRef]
- Wang, Q.; Wen, Y.; Li, Y.; Liang, W.; Li, W.; Li, Y.; Wu, J.; Zhu, H.; Zhao, K.; Zhang, J.; et al. Ultrasensitive electrochemical biosensor of bacterial 16S rRNA gene based on polyA DNA probes. Anal. Chem. 2019, 91, 9277–9283. [Google Scholar] [CrossRef]
- Alzate, D.; Cajigas, S.; Robledo, S.; Muskus, C.; Orozco, J. Genosensors for differential detection of Zika virus. Talanta 2020, 210, 120648. [Google Scholar] [CrossRef]
- Azek, F.; Grossiord, C.; Joannes, M.; Limoges, B.; Brossier, P. Hybridization assay at a disposable electrochemical biosensor for the attomole detection of amplified human cytomegalovirus DNA. Anal. Biochem. 2000, 284, 107–113. [Google Scholar] [CrossRef]
- Walter, A.; Wu, J.; Flechsig, G.-U.; Haake, D.A.; Wang, J. Redox cycling amplified electrochemical detection of DNA hybridization: Application to pathogen E. coli bacterial RNA. Anal. Chim. Acta 2011, 689, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Shipovskov, S.; Saunders, A.M.; Nielsen, J.S.; Hansen, M.H.; Gothelf, K.V.; Ferapontova, E.E. Electrochemical sandwich assay for attomole analysis of DNA and RNA from beer spoilage bacteria Lactobacillus brevis. Biosens. Bioelectron. 2012, 37, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Chumbimuni-Torres, K.Y.; Wu, J.; Clawson, C.; Galik, M.; Walter, A.; Flechsig, G.-U.; Bakker, E.; Zhang, L.; Wang, J. Amplified potentiometric transduction of DNA hybridization using ion-loaded liposomes. Analyst 2010, 135, 1618–1623. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Ganganboina, A.B.; Nasrin, F.; Takemura, K.; Doong, R.A.; Utomo, D.I.S.; Lee, J.; Khoris, I.M.; Park, E.Y. Femtomolar detection of Dengue virus DNA with serotype identification ability. Anal. Chem. 2018, 90, 12464–12474. [Google Scholar] [CrossRef]
- Khater, M.; de la Escosura-Muñiz, A.; Quesada-González, D.; Merkoçi, A. Electrochemical detection of plant virus using gold nanoparticle-modified electrodes. Anal. Chim. Acta 2019, 1046, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Singhal, C.; Dubey, A.; Mathur, A.; Pundir, C.S.; Narang, J. Paper based DNA biosensor for detection of chikungunya virus using gold shells coated magnetic nanocubes. Process Biochem. 2018, 74, 35–42. [Google Scholar] [CrossRef]
- Shariati, M.; Ghorbani, M.; Sasanpour, P.; Karimizefreh, A. An ultrasensitive label free human papilloma virus DNA biosensor using gold nanotubes based on nanoporous polycarbonate in electrical alignment. Anal. Chim. Acta 2019, 1048, 31–41. [Google Scholar] [CrossRef]
- Lee, J.; Morita, M.; Takemura, K.; Park, E.Y. A multi-functional gold/iron-oxide nanoparticle-CNT hybrid nanomaterial as virus DNA sensing platform. Biosens. Bioelectron. 2018, 102, 425–431. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, H.; Jiang, B.; Chai, Y.; Yuan, R. Quantum dot layer-by-layer assemblies as signal amplification labels for ultrasensitive electronic detection of uropathogens. Anal. Chem. 2011, 83, 4302–4306. [Google Scholar] [CrossRef]
- Xu, J.; Jiang, B.; Su, J.; Xiang, Y.; Yuan, R.; Chai, Y. Background current reduction and biobarcode amplification for label-free, highly sensitive electrochemical detection of pathogenic DNA. Chem. Commun. 2012, 48, 3309–3311. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, S.; Zhao, M.; Zhang, P.; Xin, Z.; Tao, J.; Bai, L. Amperometric DNA biosensor for Mycobacterium tuberculosis detection using flower-like carbon nanotubes-polyaniline nanohybrid and enzyme-assisted signal amplification strategy. Biosens. Bioelectron. 2018, 119, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Tang, H.; Cheng, W.; Yan, L.; Zhang, D.; Ju, H.; Ding, S. A sensitive electrochemical DNA biosensor for specific detection of Enterobacteriaceae bacteria by Exonuclease III-assisted signal amplification. Biosens. Bioelectron. 2013, 48, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Feng, M.; Li, J.; Liu, Y.; Xiao, Q. Voltammetric determination of attomolar levels of a sequence derived from the genom of hepatitis B virus by using molecular beacon mediated circular strand displacement and rolling circle amplification. Microchim. Acta 2018, 185, 206. [Google Scholar] [CrossRef]
- Yan, Y.; Ding, S.; Zhao, D.; Yuan, R.; Zhang, Y.; Cheng, W. Direct ultrasensitive electrochemical biosensing of pathogenic DNA using homogeneous target-initiated transcription amplification. Sci. Rep. 2016, 6, 18810. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, Z.; Jian, W.; Sun, D.; Zhang, B.; Li, X.; Yao, M. Ultrasensitive electrochemical detection of avian influenza A (H7N9) virus DNA based on isothermal exponential amplification coupled with hybridization chain reaction of DNAzyme nanowires. Biosens. Bioelectron. 2015, 64, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, S.; Cánovas, R.; Neumann, F.; Paulraj, T.; Nilsson, M.; Crespo, G.A.; Madaboosi, N. The sweet detection of rolling circle amplification: Glucose-based electrochemical genosensor for the detection of viral nucleic acid. Biosens. Bioelectron. 2020, 151, 112002. [Google Scholar] [CrossRef]
- Barreda-García, S.; González-Álvarez, M.J.; de-los-Santos-Álvarez, N.; Palacios-Gutiérrez, J.J.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Attomolar quantitation of Mycobacterium tuberculosis by asymmetric helicase-dependent isothermal DNA-amplification and electrochemical detection. Biosens. Bioelectron. 2015, 68, 122–128. [Google Scholar] [CrossRef]
- Li, L.; Wang, L.; Xu, Q.; Xu, L.; Liang, W.; Li, Y.; Ding, M.; Aldalbahi, A.; Ge, Z.; Wang, L.; et al. Bacterial analysis using an electrochemical DNA biosensor with poly-adenine-mediated DNA self-assembly. ACS Appl. Mater. Interfaces 2018, 10, 6895–6903. [Google Scholar] [CrossRef]
- Pei, H.; Lu, N.; Wen, Y.; Song, S.; Liu, Y.; Yan, H.; Fan, C. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv. Mater. 2010, 22, 4754–4758. [Google Scholar] [CrossRef]
- Pei, H.; Li, F.; Wan, Y.; Wei, M.; Liu, H.; Su, Y.; Chen, N.; Huang, Q.; Fan, C. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA–gold nanoparticle nanoconjugates. J. Am. Chem. Soc. 2012, 134, 11876–11879. [Google Scholar] [CrossRef]
- Xu, L.; Liang, W.; Wen, Y.; Wang, L.; Yang, X.; Ren, S.; Jia, N.; Zuo, X.; Liu, G. An ultrasensitive electrochemical biosensor for the detection of mecA gene in methicillin-resistant Staphylococcus aureus. Biosens. Bioelectron. 2018, 99, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E.; Hansen, M.N.; Saunders, A.M.; Shipovskov, S.; Sutherland, D.S.; Gothelf, K.V. Electrochemical DNA sandwich assay with a lipase label for attomole detection of DNA. Chem. Commun. 2010, 46, 1836–1838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Huarng, M.C.; Alocilja, E.C. A multiplex nanoparticle-based bio-barcoded DNA sensor for the simultaneous detection of multiple pathogens. Biosens. Bioelectron. 2010, 26, 1736–1742. [Google Scholar] [CrossRef]
- Asiello, P.J.; Baeumner, A.J. Miniaturized isothermal nucleic acid amplification, a review. Lab. Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Zhang, W.; Yan, Y.; Shen, B.; Zhu, D.; Lei, P.; Ding, S. A novel electrochemical biosensor for ultrasensitive and specific detection of DNA based on molecular beacon mediated circular strand displacement and rolling circle amplification. Biosens. Bioelectron. 2014, 62, 274–279. [Google Scholar] [CrossRef]
- Huang, Y.L.; Gao, Z.F.; Luo, H.Q.; Li, N.B. Sensitive detection of HIV gene by coupling exonuclease III-assisted target recycling and guanine nanowire amplification. Sens. Actuators B Chem. 2017, 238, 1017–1023. [Google Scholar] [CrossRef]
- Carinelli, S.; Kühnemund, M.; Nilsson, M.; Pividori, M.I. Yoctomole electrochemical genosensing of Ebola virus cDNA by rolling circle and circle to circle amplification. Biosens. Bioelectron. 2017, 93, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Barreda-García, S.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Helicase-dependent isothermal amplification: A novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal. Bioanal. Chem. 2018, 410, 679–693. [Google Scholar] [CrossRef]
- Barreda-García, S.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Sequence-specific electrochemical detection of enzymatic amplification products of Salmonella genome on ITO electrodes improves pathogen detection to the single copy level. Sens. Actuators B Chem. 2018, 268, 438–445. [Google Scholar] [CrossRef]
- Barreda-García, S.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Solid-phase helicase dependent amplification and electrochemical detection of Salmonella on highly stable oligonucleotide-modified ITO electrodes. Chem. Commun. 2017, 53, 9721–9724. [Google Scholar] [CrossRef] [PubMed]
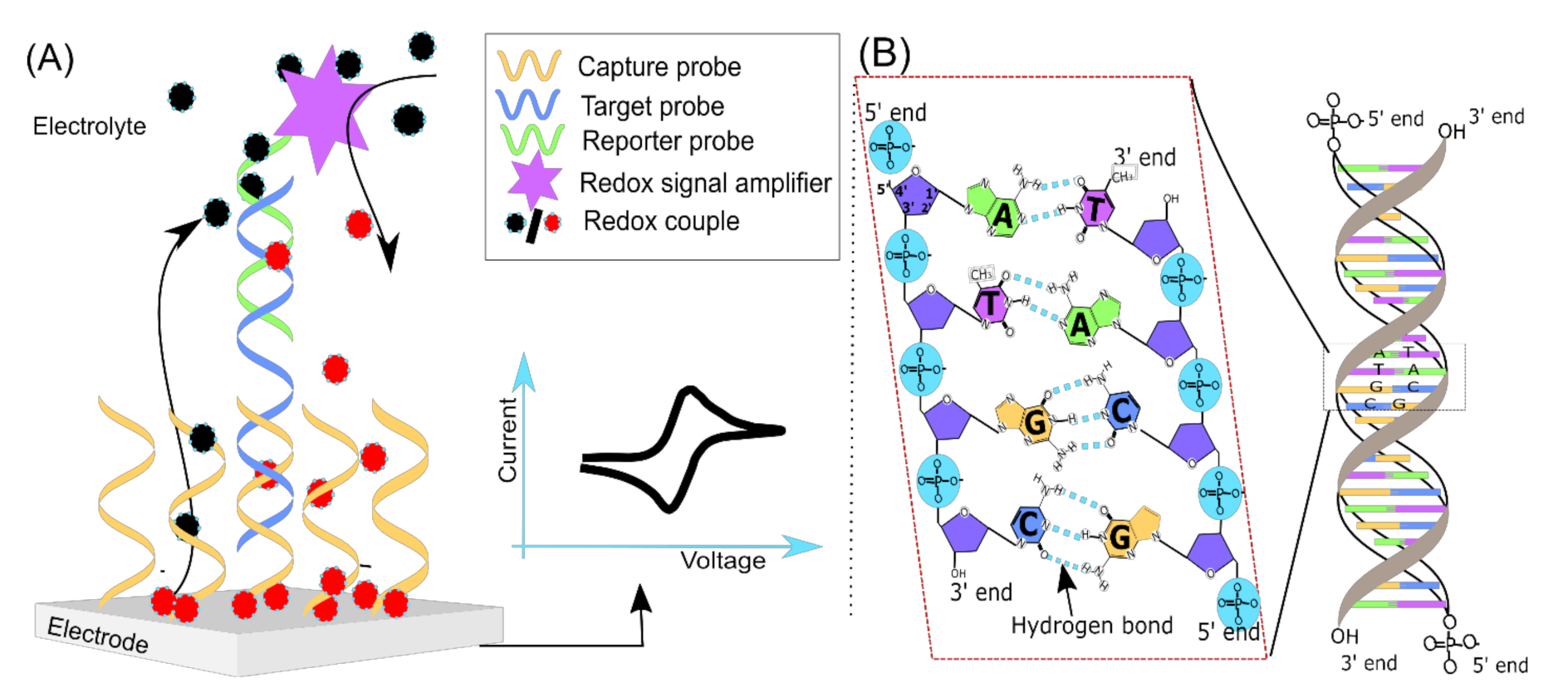
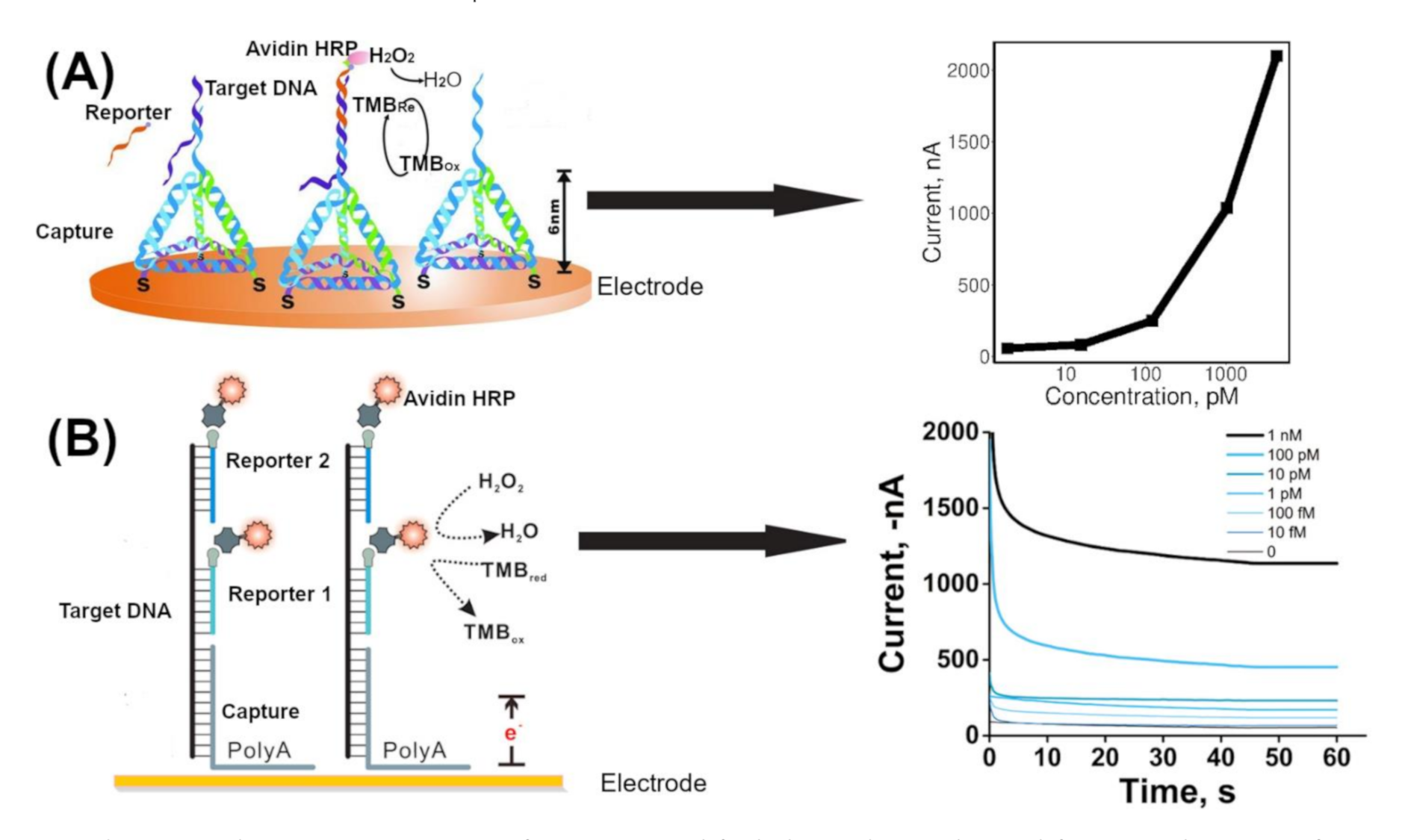
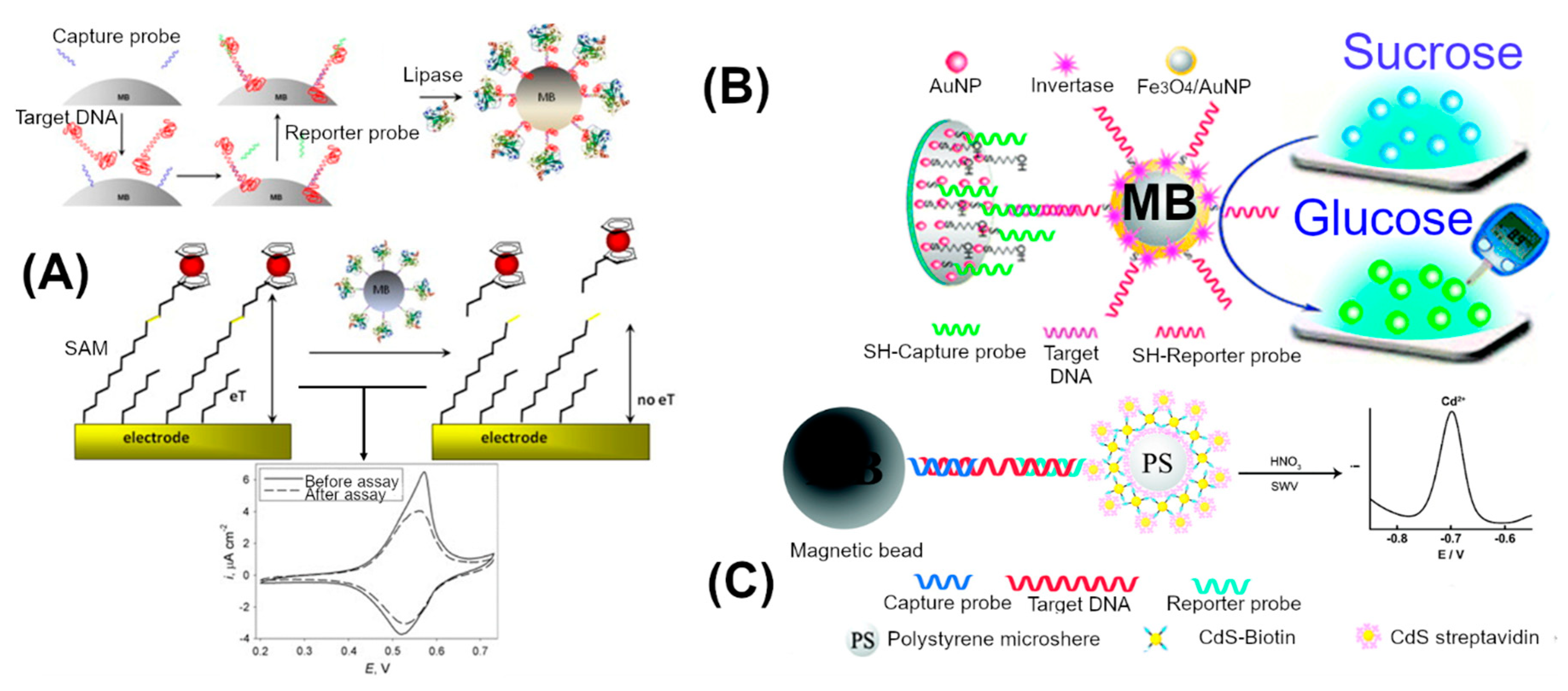
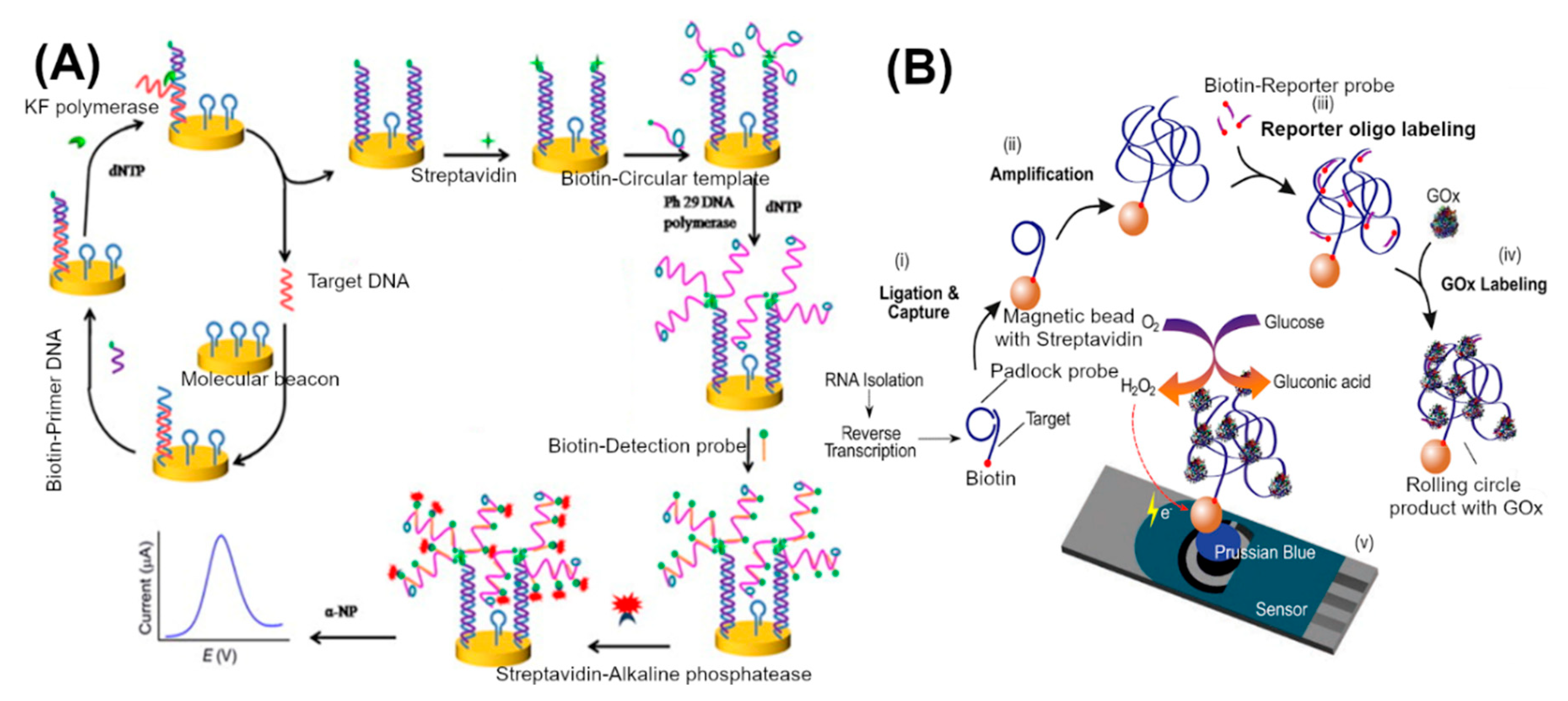
| Electrode | Reference Electrode | Electrolyte | Guanine Oxidation Peak (Ep) (V) | Reference |
|---|---|---|---|---|
| Gold | Ag/AgCl | PBS, pH 7.4 | +0.7/+0.8 | [34] |
| Nafion/Graphene | SCE | 0.1 M PBS (pH 4.4) | +0.8 | [35] |
| Glassy carbon electrode | Ag/AgCl | 0.1 M PBS (pH 7.0) | +0.6 | [36] |
| Boron doped diamond | Ag/AgCl | 0.1 M acetate buffer (pH 4.5) | +0.9 | [37] |
| Pencil graphite | Ag/AgCl | 0.5 M acetate buffer and 20 mM LiCIO4 | +0.76 | [38] |
| DWNTs, and MWNTs | Ag/AgCl | PBS (pH 6) | +1 | [39] |
| HOPGE | Ag/AgCl | 0.1 M sodium acetate buffer (pH 7.6) | 0.9 | [40] |
| Pathogen | Target | Capture Probe | Reporter Probe | Electrode Modification | Amplification Strategy | Redox Signal | Limit of Detection (LOD) * | Analytical Technique | References |
|---|---|---|---|---|---|---|---|---|---|
| Ebolavirus | Biotin-ssDNA | HS-ssDNA | NA | Au | Strep-alkaline phosphatase | 4-aminophenol | 4.7 nM | DPV | [48] |
| Avian influenza A (H7N9) virus | ssDNA | SH-tetrahedral DNA | Biotin-ssDNA | Au | Strep-HRP | TMP | 0.75 pM | Amperometric | [49] |
| Bacteria 16s RNA gene | ssDNA and genomic DNA | ssDNA (polydA SAM) | Biotin-ssDNA | Au | Strep-HRP | TMB | 10 fM | Amperometric | [50] |
| Zika virus | ssDNA | Biotin-ssDNA (Strept-magentic beads) | Digoxigenin -ssDNA | Au | Anti- Digoxigenin coupled HRP | TMB | 0.7 pM | Chronoamperometry | [51] |
| HIV DNA | ssDNA | SH-ssDNA | SH-ssDNA | Glucose meter | Invertase-Fe3O4-Au | Glucose | 0.5 pM | Amperometry | [11] |
| Human cytomegalovirus | ssDNA(PCR product) | NA | Biotin-ssDNA | Carbon | Strep-HRP | Ophenyldimine/2,2′-diaminoazobenene | 3.6 × 105 copies/mL | DPV | [52] |
| E. coli | gDNA | SH-ssDNA | Biotin-ssDNA | Au | Strep-HRP and redox cycling | p-aminophenyl phosphate | 0.5416667 | Chronoamperometric | [53] |
| Lactobacillus brevis | gDNA, RNA | Biotin-ssDNA | Biotin-ssDNA | Au | Strep-Lipase | Ferrocene | 16 amole | CV | [54] |
| E. coli | ssDNA | HS-ssDNA | Biotin-ssDNA | Au | Liposome loaded with Ca2+ | Ca2+ ion-selective electrode (No redox reaction) | 0.2 nM | Potentiometric method using | [55] |
| Dengue virus | PCR amplified target with poly (dT) | HS-ssPoly(dA) | Fluro-ssDNA | Au-Polyaniline/N,S-GQDs@AuNP-dA | Nanomaterial as carrier | Methylene blue-intercalation | 9.4 fM | DPV | [56] |
| Citrus tristeza virus | ssDNA | HS-ssDNA | NA | Au/AuNPs | Nanoparticle as carrier | [Fe(CN)63−/4−] | 100 nM | Impedance | [57] |
| Chikungunya virus | ssDNA | ssDNA | NA | Carbon/Fe3O4@Au (+ and − charge interaction to accumulate the DNA) | Nanoparticle as carrier | Methylene blue | 0.1 nM | DPV and CV | [58] |
| Human papilloma virus | ssDNA | HS-ssDNA | NA | Nanoporous polycarbonate-AuNTs | Nanoparticles as carrier | [Fe(CN)63−/4−] | 1 fM | Impedance | [59] |
| Influenza and Norovirus | ssDNA | SH-ssDNA | NA | Pt-Au/Iron Oxide-CNT | Nanoparticles as carrier | NA | 8.8 pM | Conductivity (the resistance change) | [60] |
| E. coli uropathogens | ssDNA | Biotin-ssDNA | Biotin-ssDNA | Glassy carbon | CdS quantum dots as reporter | Cd2+ | 0.22 fM | SWV | [61] |
| E. coli | ssDNA | SH-ssDNA | ssDNA | AuNP-deposited on glassy carbon electrode | Nanoparticle as high amount reporter probe carrier | [Ru(NH3)6]3+ | 1 fM | DPV | [62] |
| Mycobacterium tuberculosis | PCR product | SH-ssDNA | SH-ssDNA loaded AuNPs@ CNT-PANI | Au | Endonuclease | Polyaniline | 0.33 fM | DPV | [63] |
| Enterobacteriaceae | ssDNA and HAV cDNA | HS-ssDNA | biotin-ssDNA | Au | Exonuclease III and Strep-alkaline phosphatase | α-naphthyl phosphate | 8.7 fM | DPV | [64] |
| Hepatitis B virus (HBV) | ssDNA | HS-ssDNA | ssDNA as primer | Au | CSD and RCA | Methylene blue | 2.6 aM | DPV | [65] |
| Salmonella | gDNA | SH-ssDNA | Biotin-ssDNA | Au | DNA polymerase, T4 RNA polymerase and Strep-alkaline phosphatase | α-naphthyl phosphate | 0.97 fM | DPV | [66] |
| Avian influenza A (H7N9) virus | ssDNA | SH-ssDNA | Molecular beacons | Au | EXPAR-HCR and G-quadruplex–hemin-(HRP like catalysis) | TMB | 9.4 fM | DPV | [67] |
| Ebolavirus | RNA | cDNA synthesized from target RNA | Biotin-ssDNA | Carbon | RCA and Strep-glucose oxidase | H2O2 | 1 pM | Chronoamperometry | [68] |
| Mycobacterium tuberculosis | gDNA | Biotin-PCR product from target | Fluorescein-ssDNA | Carbon | HDA and Antifluorescein-POD Fab | TMP | 0.5 aM | Chronoamperometry | [69] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santhanam, M.; Algov, I.; Alfonta, L. DNA/RNA Electrochemical Biosensing Devices a Future Replacement of PCR Methods for a Fast Epidemic Containment. Sensors 2020, 20, 4648. https://doi.org/10.3390/s20164648
Santhanam M, Algov I, Alfonta L. DNA/RNA Electrochemical Biosensing Devices a Future Replacement of PCR Methods for a Fast Epidemic Containment. Sensors. 2020; 20(16):4648. https://doi.org/10.3390/s20164648
Chicago/Turabian StyleSanthanam, Manikandan, Itay Algov, and Lital Alfonta. 2020. "DNA/RNA Electrochemical Biosensing Devices a Future Replacement of PCR Methods for a Fast Epidemic Containment" Sensors 20, no. 16: 4648. https://doi.org/10.3390/s20164648
APA StyleSanthanam, M., Algov, I., & Alfonta, L. (2020). DNA/RNA Electrochemical Biosensing Devices a Future Replacement of PCR Methods for a Fast Epidemic Containment. Sensors, 20(16), 4648. https://doi.org/10.3390/s20164648





