Using the Redundant Convolutional Encoder–Decoder to Denoise QRS Complexes in ECG Signals Recorded with an Armband Wearable Device
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Extraction of Armband Sequences for Adding Colored Noises
2.3. Extraction of Armband Sequences with Motion Noise Artifacts
2.4. Extraction of Sequences from the MIT-BIH Arrhythmia Database
2.5. Denoising Algorithm
3. Results
3.1. Armband Sequences with Added Colored Noises
3.2. Armband Sequences with Motion Noise Artifacts
3.3. Sequences from MIT-BIH Arrhythmia Database
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hossain, M.B.; Bashar, S.K.; Walkey, A.J.; McManus, D.D.; Chon, K.H. An accurate QRS complex and P wave detection in ECG signals using complete ensemble empirical mode decomposition with adaptive noise approach. IEEE Access 2019, 7, 128869–128880. [Google Scholar] [CrossRef]
- Reyes, B.A.; Posada-Quintero, H.F.; Bales, J.R.; Clement, A.L.; Pins, G.D.; Swiston, A.; Riistama, J.; Florian, J.P.; Shykoff, B.; Qin, M.; et al. Novel electrodes for underwater ECG monitoring. IEEE Trans. Biomed. Eng. 2014, 61, 1863–1876. [Google Scholar] [CrossRef] [PubMed]
- Lobodzinski, S.S. ECG patch monitors for assessment of cardiac rhythm abnormalities. Prog. Cardiocasc. Dis. 2013, 56, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Lazaro, J.; Reljin, N.; Hossain, M.B.; Noh, Y.; Laguna, P.; Chon, K.H. Wearable armband device for daily life electrocardiogram monitoring. IEEE Trans. Biomed. Eng. 2020. [Google Scholar] [CrossRef] [PubMed]
- Guler, I.; Ubeyli, E.D. ECG beat classifier designed by combined neural network model. Pattern Recognit. 2005, 38, 199–208. [Google Scholar] [CrossRef]
- Marque, C.; Bisch, C.; Dantas, R.; Elayoubi, S.; Brosse, V.; Perot, C. Adaptive filtering for ECG rejection from surface EMG recordings. J. Electromyofr. Kinesiol. 2005, 15, 310–315. [Google Scholar] [CrossRef]
- Lu, G.; Brittain, J.S.; Holland, P.; Yianni, J.; Green, A.L.; Stein, J.F.; Aziz, T.Z.; Wang, S. Removing ECG noise from surface EMG signals using adaptive filtering. Neurosci. Lett. 2009, 462, 14–19. [Google Scholar] [CrossRef]
- Tracey, B.H.; Miller, E.L. Nonlocal means denoising of ECG signals. IEEE Trans. Biomed. Eng. 2012, 59, 2383–2386. [Google Scholar] [CrossRef]
- Selesnick, I. Sparsity-assisted signal smoothing (revisited). In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), New Orleans, LA, USA, 5–9 March 2017. [Google Scholar]
- He, T.; Clifford, G.; Tarassenko, L. Application of independent component analysis in removing artefacts from electrocardiogram. Neural Comput. Appl. 2006, 15, 105–116. [Google Scholar] [CrossRef]
- Sameni, R.; Shamsollahi, M.B.; Jutten, C.; Clifford, G.D. A nonlinear Bayesian filtering framework for ECG denoising. IEEE Trans. Biomed. Eng. 2007, 54, 2172–2185. [Google Scholar] [CrossRef] [Green Version]
- Sayadi, O.; Shamsollahi, M.B. ECG denoising and compression using a modified extended Kalman filter structure. IEEE Trans. Biomed. Eng. 2008, 55, 2240–2248. [Google Scholar] [CrossRef] [PubMed]
- Hesar, H.D.; Mohebbi, M. An adaptive particle weighting strategy for ECG denoising using marginalized particle extended Kalman filter: An evaluation in arrhythmia context. IEEE J. Biomed. Health Inform. 2017, 21, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Tiwari, A.K. Optimal selection of wavelet basis function applied to ECG signal denoisisng. Digit. Signal Process. 2006, 16, 275–287. [Google Scholar] [CrossRef]
- Awal, M.A.; Mostafa, S.S.; Ahmad, M.; Rashid, M.A. An adaptive level dependent wavelet thresholding for ECG denoising. Biocybern. Biomed. Eng. 2014, 34, 238–249. [Google Scholar] [CrossRef]
- Jenkal, W.; Latif, R.; Toumanari, A.; Dliou, A.; B’charri, O.E.; Maoulainine, F.M.R. An efficient algorithm of ECG signal denoising using the adaptive dual threshold filter and the discrete wavelet transform. Biocybern. Biomed. Eng. 2016, 36, 499–508. [Google Scholar] [CrossRef]
- B’charri, O.E.; Latif, R.; Elmansouri, K.; Abenaou, A.; Jenkal, W. ECG signal performance de-noising assessment based on threshold tuning of dual-tree wavelet transform. Biomed. Eng. Online 2017, 16, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhu, J.; Yan, T.; Yang, L. A new modified wavelet-based ECG denoising. Comput. Assist. Surg. 2019, 24 (Suppl. 1), 174–183. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Velasco, M.; Weng, B.; Barner, K.E. ECG signal denoising and baseline wander correction based on the empirical mode decomposition. Comput. Biol. Med. 2008, 38, 1–13. [Google Scholar] [CrossRef]
- Kabir, M.A.; Shahnaz, C. Denoising of ECG signals based on noise reduction algorithms in EMD and wavelet domains. Biomed. Signal Process. Control 2012, 7, 481–489. [Google Scholar] [CrossRef]
- Nguyen, P.; Kim, J.-M. Adaptive ECG denoising using genetic algorithm-based thresholding and ensemble empirical mode decomposition. Inf. Sci. 2016, 373, 499–511. [Google Scholar] [CrossRef]
- Rakshit, M.; Das, S. An efficient ECG denoising methodology using empirical mode decomposition and adaptive switching mean filter. Biomed. Signal Process. Control 2018, 40, 140–148. [Google Scholar] [CrossRef]
- Akhbari, M.; Shamsollahi, M.B.; Jutten, C.; Armoundas, A.A.; Sayadi, O. ECG denoising and fiducial point extraction using an extended Kalman filtering framework with linear and nonlinear phase observations. Phys. Meas. 2016, 37, 203–226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Hu, X.; Tang, Z.; Ahn, A.C. Sparse representation-based ECG signal enhancement and QRS detection. Phys. Meas. 2016, 37, 2093–2110. [Google Scholar] [CrossRef] [PubMed]
- Xiong, P.; Wang, H.; Liu, M.; Lin, F.; Hou, Z.; Liu, X. A stacked contractive denoising auto-encoder for ECG signal denoising. Phys. Meas. 2016, 37, 2214–2230. [Google Scholar] [CrossRef]
- Chiang, H.-T.; Hsieh, Y.-Y.; Fu, S.-W.; Hung, K.-H.; Tsao, Y.; Chien, S.-Y. Noise reduction in ECG signals using fully convolutional denoising autoencoders. IEEE Access 2019, 7, 60806–60813. [Google Scholar] [CrossRef]
- Park, S.R.; Lee, J.W. A fully convolutional neural network for speech enhancement. In Proceedings of the Interspeech Conference, Stockholm, Sweden, 20–24 August 2017. [Google Scholar]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH Arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- Dash, S.; Chon, K.H.; Lu, S.; Raeder, E.A. Automatic real time detection of atrial fibrillation. Ann. BME 2009, 37, 1701–1709. [Google Scholar] [CrossRef]
- Clifford, G.D. ECG statistics, noise, artifacts, and missing data. In Advanced Methods and Tools for ECG Data Analysis; Clifford, G.D., Azuaje, F., McSharry, P., Eds.; Artech House Publishing: Boston, MA, USA; London, UK, 2006; pp. 55–99. [Google Scholar]
- Marouf, M. High Frequency Noise Approximation and Adaptive Reduction in the ECG Signals. Ph.D. Thesis, School of Electrical Engineering, University of Belgrade, Belgrade, Serbia, December 2017. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Liu, D.; Smaragdis, P.; Kim, M. Experiments on deep learning for speech denoising. In Proceedings of the Interspeech Conference, Singapore, 14–18 September 2014. [Google Scholar]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, BME-32, 230–236. [Google Scholar] [CrossRef]
- Clifford, G.D.; Lopez, D.; Li, Q.; Rezek, I. Signal quality indices and data fusion for determining acceptability of electrocardiograms collected in noisy ambulatory environments. In Proceedings of the Computing in Cardiology Conference, Hangzhou, China, 18–21 September 2011. [Google Scholar]
- Hossain, M.B.; Lazaro, J.; Noh, Y.; Chon, K. Denoising wearable armband ECG data using the variable frequency complex demodulation technique. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020. [Google Scholar]
- Torres, M.E.; Colominas, M.A.; Schlotthauer, G.; Flandrin, P. A complete ensemble empirical mode decomposition with adaptive noise. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Prague, Czech Republic, 22–27 May 2011. [Google Scholar]

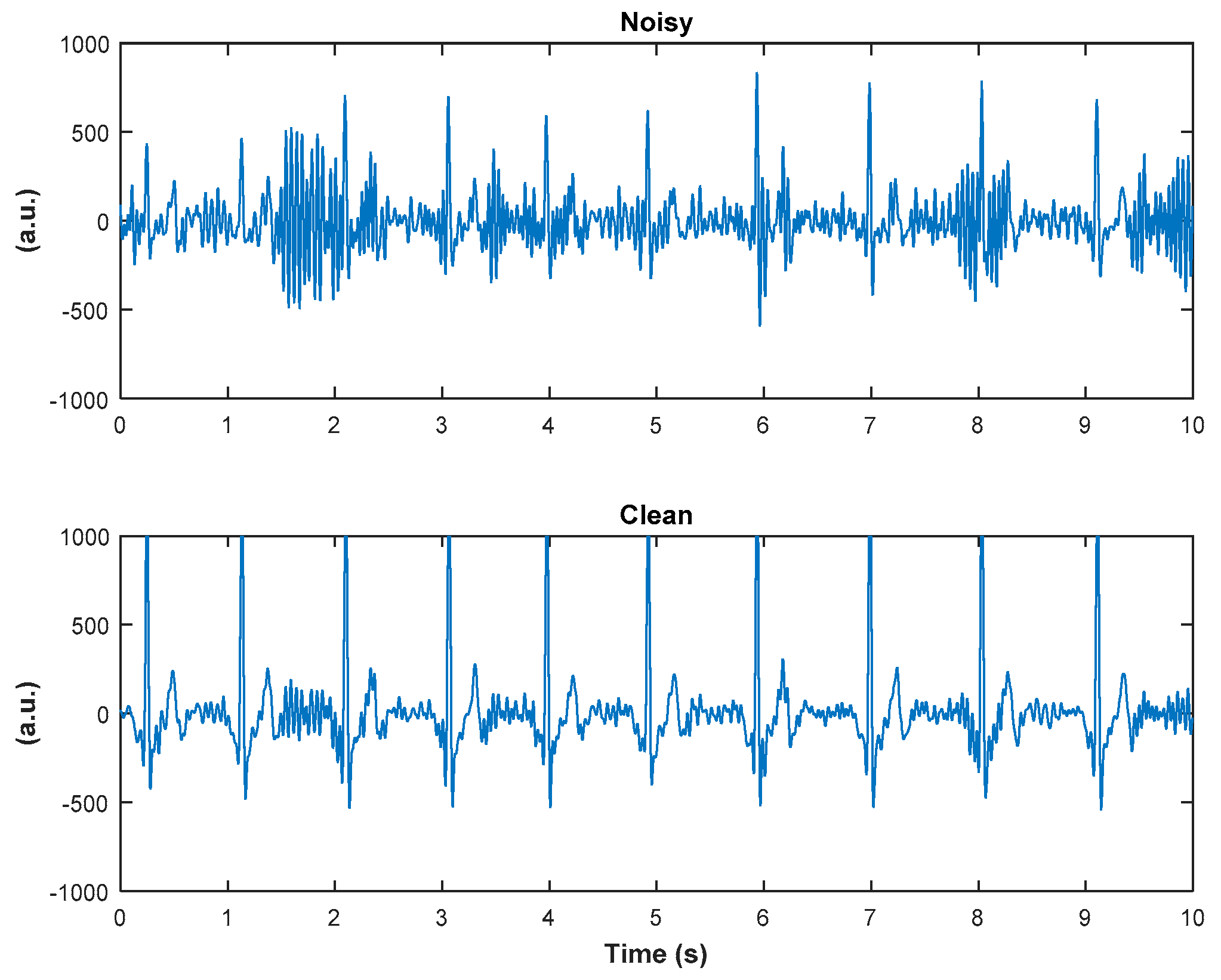


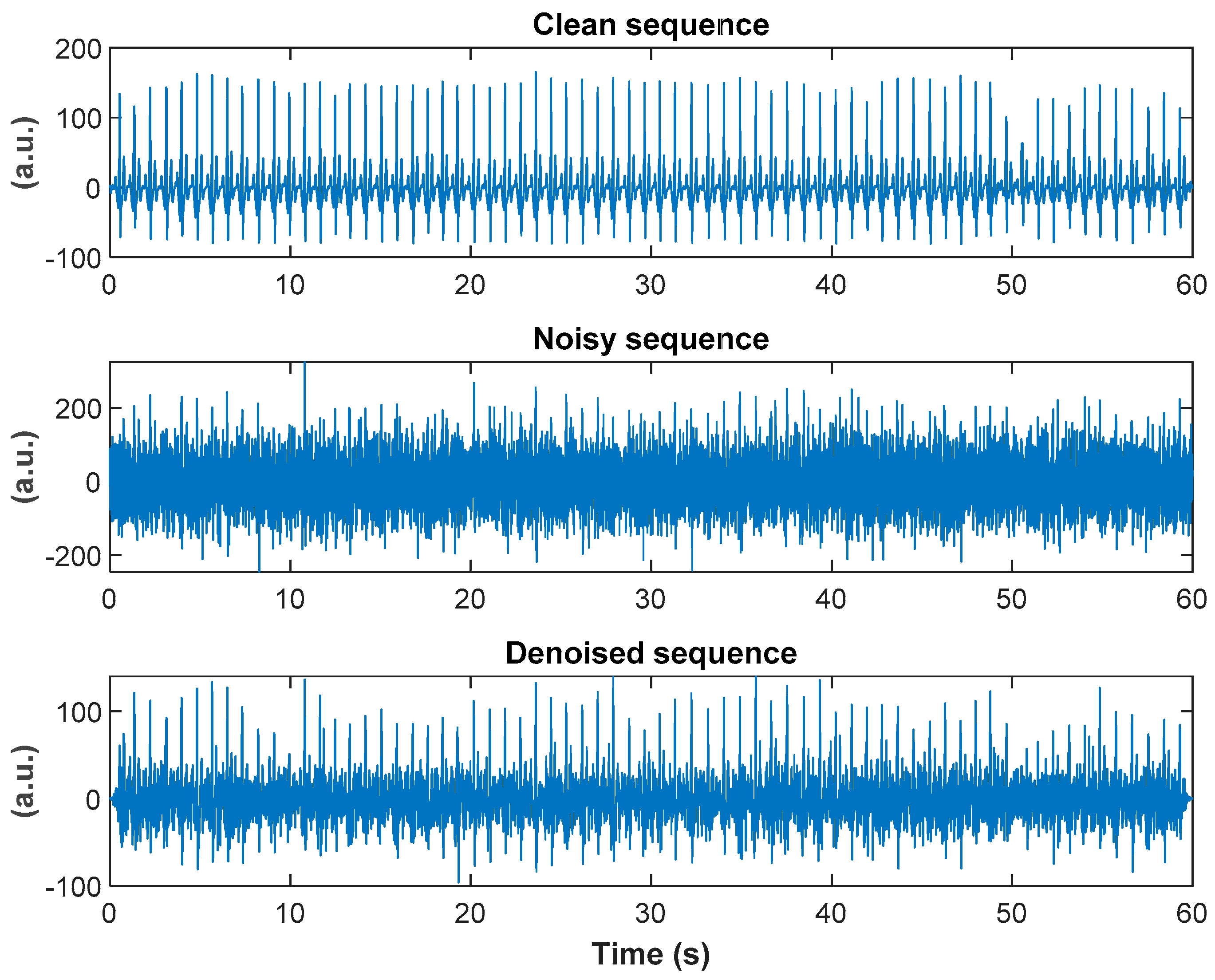
| Clean (Reference) | Noisy | Denoised | |
|---|---|---|---|
| White noise, SNR −5 dB | 100% | 80% | 95.92% |
| White noise, SNR −7 dB | 100% | 83.87% | 86.54% |
| Blue noise, SNR −5 dB | 100% | 97.78% | 100% |
| Blue noise, SNR −7 dB | 100% | 95.65% | 100% |
| Pink noise, SNR −5 dB | 100% | 62.5% | 93.75% |
| Pink noise, SNR −7 dB | 100% | 66.67% | 70.49% |
| Purple noise, SNR −5 dB | 100% | 93.62% | 100% |
| Purple noise, SNR −7 dB | 100% | 95.65% | 100% |
| Brown noise, SNR −15 dB | 100% | 43.56% | 80.85% |
| Brown noise, SNR −17 dB | 100% | 34.33% | 79.17% |
| SNRimp (dB) Mean ± sd | |
|---|---|
| White noise, SNR −5 dB | 7.32 ± 0.2 |
| White noise, SNR −7 dB | 8.43 ± 0.17 |
| Blue noise, SNR −5 dB | 8.76 ± 0.24 |
| Blue noise, SNR −7 dB | 9.9 ± 0.49 |
| Pink noise, SNR −5 dB | 6.45 ± 0.26 |
| Pink noise, SNR −7 dB | 7.64 ± 0.42 |
| Purple noise, SNR −5 dB | 8.81 ± 0.26 |
| Purple noise, SNR −7 dB | 9.77 ± 0.33 |
| Brown noise, SNR −15 dB | 17.39 ± 1.11 |
| Brown noise, SNR −17 dB | 18.79 ± 1.73 |
| Clean (Reference) | Noisy (p-Values) | Denoised (p-Values) | |
|---|---|---|---|
| White noise, SNR −5 dB | 0.73 ± 0.02 | 0.26 ± 0.01 *& (<3 × 10−8) | 0.63 ± 0.01 * (<2 × 10−5) |
| White noise, SNR −7 dB | 0.22 ± 0.01 *& (<2 × 10−7) | 0.61 ± 0.01 * (<6 × 10−5) | |
| Blue noise, SNR −5 dB | 0.20 ± 0.01 *& (<2 × 10−8) | 0.60 ± 0.02 * (<2 × 10−5) | |
| Blue noise, SNR −7 dB | 0.15 ± 0.01 *& (<5 × 10−6) | 0.55 ± 0.04 * (<2 × 10−4) | |
| Pink noise, SNR −5 dB | 0.32 ± 0.02 *& (<7 × 10−6) | 0.64 ± 0.02 * (<2 × 10−4) | |
| Pink noise, SNR −7 dB | 0.28 ± 0.02 *& (<1 × 10−6) | 0.62 ± 0.01 * (<2 × 10−4) | |
| Purple noise, SNR −5 dB | 0.17 ± 0.00 *& (<3 × 10−7) | 0.53 ± 0.02 * (<2 × 10−6) | |
| Purple noise, SNR −7 dB | 0.13 ± 0.01 *& (<3 × 10−6) | 0.47 ± 0.03 * (<2 × 10−5) | |
| Brown noise, SNR −15 dB | 0.03 ± 0.02 *& (<4 × 10−7) | 0.63 ± 0.02 * (<4 × 10−5) | |
| Brown noise, SNR −17 dB | 0.02 ± 0.01 *& (<2 × 10−7) | 0.59 ± 0.02 * (<2 × 10−4) |
| Noisy Sequences (p-Values) | Denoised Sequences | |
|---|---|---|
| White noise, SNR −5 dB | 0.49 ± 0.01 * (4 × 10−12) | 0.71 ± 0.01 |
| White noise, SNR −7 dB | 0.41 ± 0.01 * (4 × 10−11) | 0.64 ± 0.01 |
| Blue noise, SNR −5 dB | 0.48 ± 0.01 * (9 × 10−13) | 0.79 ± 0.01 |
| Blue noise, SNR −7 dB | 0.41 ± 0.01 * (1 × 10−10) | 0.74 ± 0.03 |
| Pink noise, SNR −5 dB | 0.50 ± 0.02 * (1 × 10−7) | 0.64 ± 0.02 |
| Pink noise, SNR −7 dB | 0.43 ± 0.03 * (6 × 10−5) | 0.57 ± 0.04 |
| Purple noise, SNR −5 dB | 0.48 ± 0.01 * (2 × 10−13) | 0.79 ± 0.01 |
| Purple noise, SNR −7 dB | 0.41 ± 0.01 * (8 × 10−12) | 0.74 ± 0.02 |
| Brown noise, SNR −15 dB | 0.23 ± 0.04 * (2 × 10−7) | 0.71 ± 0.08 |
| Brown noise, SNR −17 dB | 0.27 ± 0.18 * (0.002) | 0.65 ± 0.13 |
| Clean (Reference) | Noisy | Denoised | |
|---|---|---|---|
| Correctly detected peaks | 88.60% | 61.16% | 91.86% |
| Missed peaks | 11.40% | 38.84% | 8.14% |
| Method | Cross-Correlation | Ratio of Power | Correctly Detected Peaks |
|---|---|---|---|
| DWT-based [14] | 0.74 ± 0.08 | 0.63 ± 0.06 | 76.28% |
| EMD-DWT-based [20] | 0.77 ± 0.07 | 0.70 ± 0.04 | 87.27% |
| EMD-ASMF-based [22] | 0.79 ± 0.06 | 0.70 ± 0.03 | 92.83% |
| SASS-based [9] | 0.79 ± 0.07 | 0.69 ± 0.05 | 84.98% |
| VFCDM-based [37] | 0.81 ± 0.06 | 0.71 ± 0.03 | 92.83% |
| Proposed method | 0.77 ± 0.06 | 0.71 ± 0.03 | 91.86% |
| Test 1 | Test 2 | |
|---|---|---|
| Mean ± sd (dB) | 7.08 ± 0.25 | 7.43 ± 0.81 |
| Max (dB) | 7.55 | 9.03 |
| Min (dB) | 6.78 | 6.32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reljin, N.; Lazaro, J.; Hossain, M.B.; Noh, Y.S.; Cho, C.H.; Chon, K.H. Using the Redundant Convolutional Encoder–Decoder to Denoise QRS Complexes in ECG Signals Recorded with an Armband Wearable Device. Sensors 2020, 20, 4611. https://doi.org/10.3390/s20164611
Reljin N, Lazaro J, Hossain MB, Noh YS, Cho CH, Chon KH. Using the Redundant Convolutional Encoder–Decoder to Denoise QRS Complexes in ECG Signals Recorded with an Armband Wearable Device. Sensors. 2020; 20(16):4611. https://doi.org/10.3390/s20164611
Chicago/Turabian StyleReljin, Natasa, Jesus Lazaro, Md Billal Hossain, Yeon Sik Noh, Chae Ho Cho, and Ki H. Chon. 2020. "Using the Redundant Convolutional Encoder–Decoder to Denoise QRS Complexes in ECG Signals Recorded with an Armband Wearable Device" Sensors 20, no. 16: 4611. https://doi.org/10.3390/s20164611
APA StyleReljin, N., Lazaro, J., Hossain, M. B., Noh, Y. S., Cho, C. H., & Chon, K. H. (2020). Using the Redundant Convolutional Encoder–Decoder to Denoise QRS Complexes in ECG Signals Recorded with an Armband Wearable Device. Sensors, 20(16), 4611. https://doi.org/10.3390/s20164611







