Research Progress on the Early Monitoring of Pine Wilt Disease Using Hyperspectral Techniques
Abstract
1. Introduction
2. PWD Symptoms
3. Traditional PWD Monitoring Technology
3.1. PWD Pine Tree Monitoring Methods
3.2. Pine Wood PWD Monitoring Technology
4. Principles of Monitoring PWD with Hyperspectral Remote Sensing Technology
5. Hyperspectral Technology in Monitoring of PWD in Forests
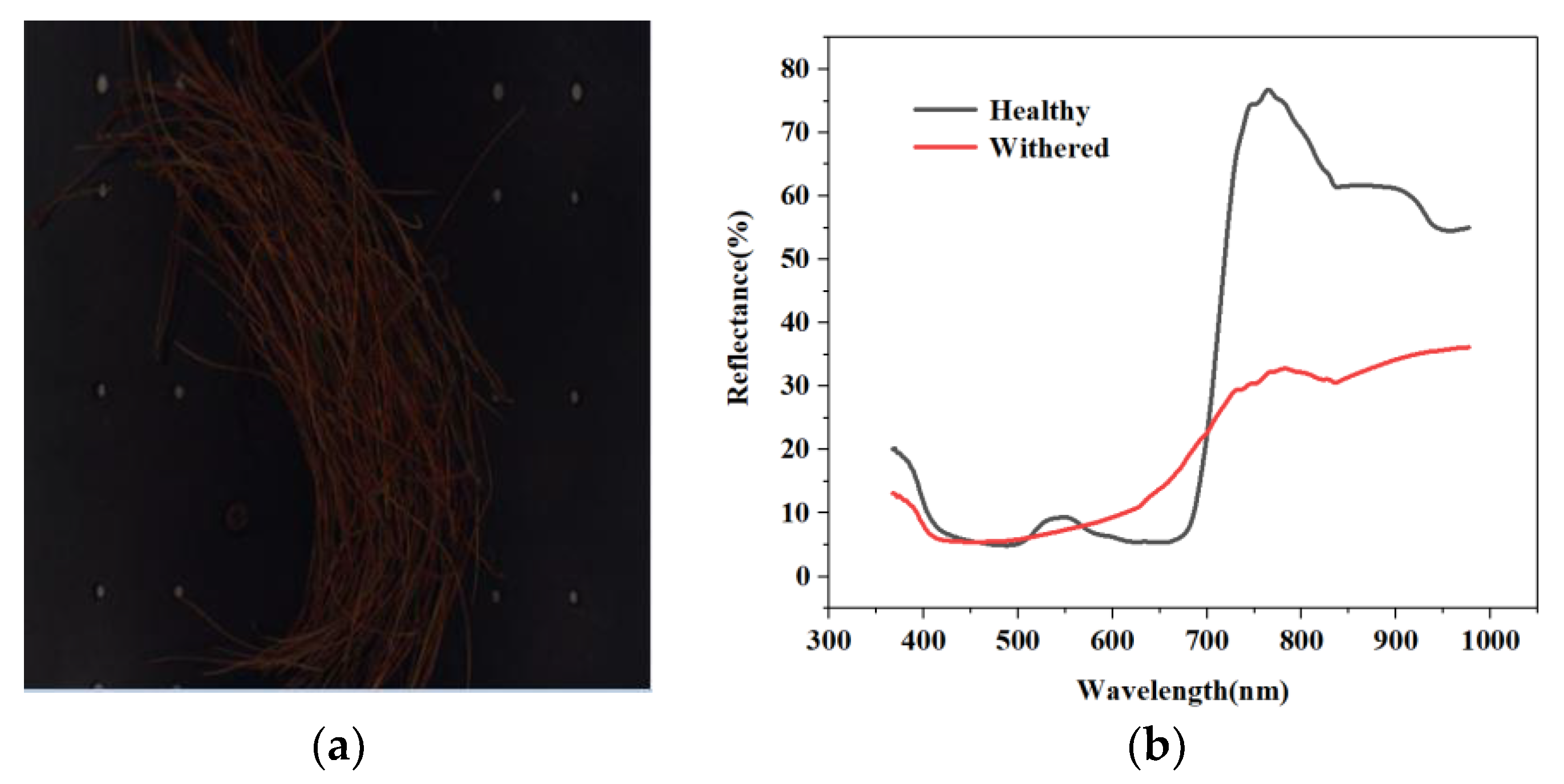
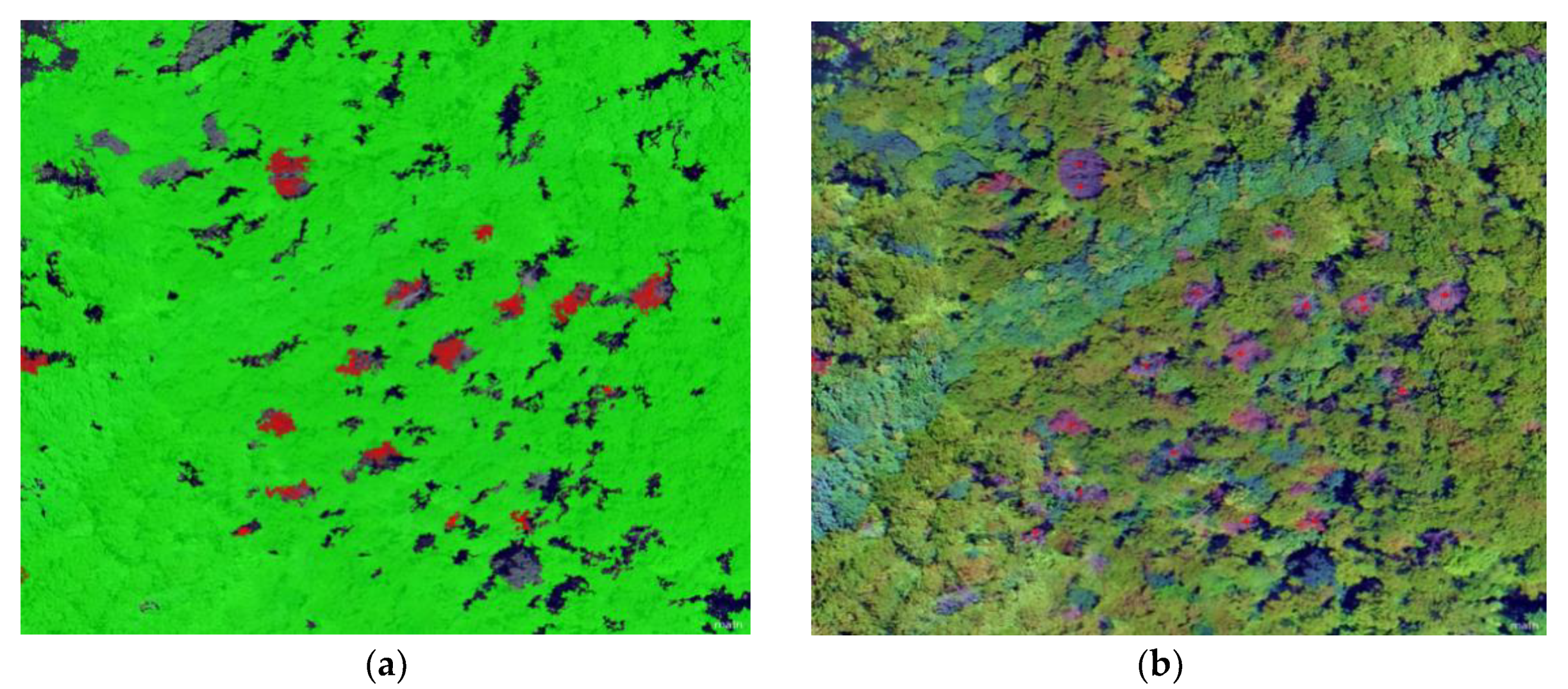
5.1. Forest Surveillance of Dead Pine Wilt Disease
lik(θ) = fD(x, x2, …, xn∣θ)
5.2. Forest Monitoring for Early PWD Detection
6. Drones Equipped with Hyperspectral Sensors to Monitor Forest PWD
7. Prospects of PWD Monitoring Using Hyperspectral Technology
7.1. Hyperspectral Technology in PWD Forest Monitoring
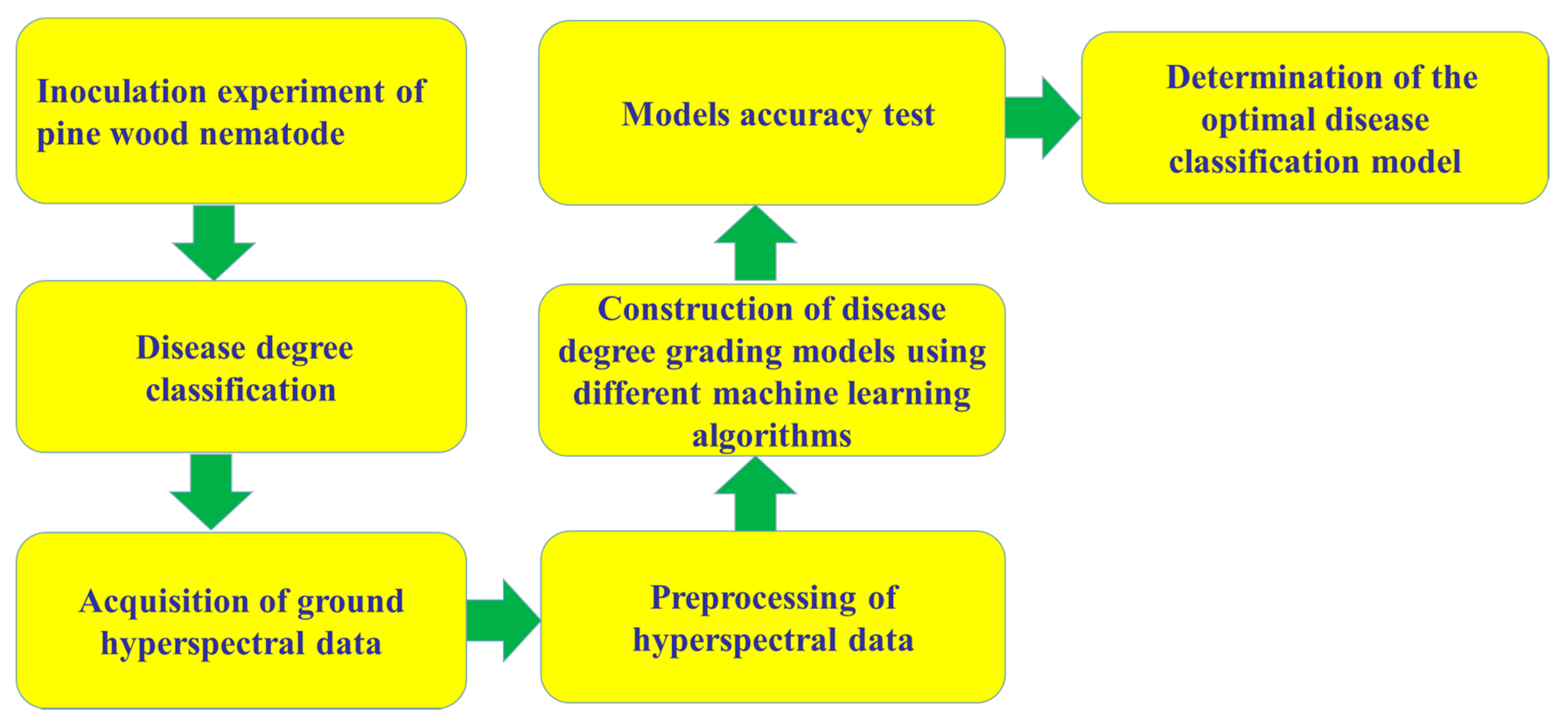
7.2. Hyperspectral Data Acquisition
7.3. Hyperspectral Image Acquisition Environment
7.4. Hyperspectral Data Analysis
7.4.1. Data Processing
7.4.2. Machine Learning Methods Used in PWN Research
7.5. Challenges and Countermeasures in PWD Monitoring with Spectral Technology
7.5.1. Challenges
7.5.2. Conclusions and Suggestions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, N.; Jeon, H.W.; Mannaa, M.; Jeong, S.; Kim, J.; Lee, C.; Park, A.R.; Kim, J.; Seo, Y. Induction of resistance against pine wilt disease caused by Bursaphelenchus xylophilus using selected pine endophytic bacteria. Plant Pathol. 2019, 68, 434–444. [Google Scholar] [CrossRef]
- Mota, M.M.; Vieira, P. Pine Wilt Disease: A Worldwide Threat to Forest Ecosystem; Springer: Heidelberg, Germany, 2008; pp. 1–10. [Google Scholar]
- Abelleira, A.; Picoaga, A.; Mansilla, J.P.; Aguin, O. Detection of Bursaphelenchus xylophilus, causal agent of pine wilt disease on Pinus pinaster in Northwest Spain. Plant Dis. 2011, 95, 776. [Google Scholar] [CrossRef] [PubMed]
- Futai, K. Pine Wood Nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 2013, 51, 61–83. [Google Scholar] [CrossRef] [PubMed]
- He, L.X.; Ji, J.; Qiu, X.W. Occurrence and control measures of pine wood nematode disease in the world. J. For. Eng. 2014, 28, 8–13. [Google Scholar]
- Esuer, M.; Arias, M.; Bello, A. Occurrence of the genus Bursaphelenchus Fuchs, 1937 (Nematoda: Aphelenchida) in the Spanish conifer forests. Nematology 2004, 6, 155–156. [Google Scholar]
- Futai, K. Role of asymptomatic carrier trees in epidemic spread of pine wilt disease. J. For. Res. 2003, 8, 253–260. [Google Scholar] [CrossRef]
- Jones, J.T.; Moens, M.; Mota, M. Bursaphelenchus xylophilus: Opportunities in comparative genomics and molecular host-parasite interactions. Mol. Plant Pathol. 2008, 9, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Proenca, D.; Romeu, F.; Santos, C.V.; Andre, L.; Luis, F.; Abrantes, L.M.; Morais, P.V. Diversity of bacteria associated with Bursaphelenchus xylophilus and other nematodes isolated from Pinus pinaster trees with pine wilt disease. PLoS ONE 2010, 5, e15191. [Google Scholar] [CrossRef]
- Fonseca, L.; Cardoso, J.; Lopes, A. The pinewood nematode, Bursaphelenchus xylophsilus, in Madeira Island. Helminthologia 2012, 49, 96–103. [Google Scholar] [CrossRef]
- Erevková, A.; Mota, M.; Vieira, P. Bursaphelenchus xylophilus (Steiner & Bührer, 1934) Nickle 1970—Pinewood nematode: A threat to European forests. For. J. 2014, 60, 125–129. [Google Scholar]
- Diogo, N.P.; Gregor, G.; Paula, V.M. Understanding pine wilt disease: Roles of the pine endophytic bacteria and of the bacteria carried by the disease—Causing pinewood nematode. Microbiol. Open 2017, 6, e415. [Google Scholar]
- Kishi, Y. The Pine Wood Nematode and the Japanese Pine Sawyer; Thomas Company Limited: Tokyo, Japan, 1995; p. 302. [Google Scholar]
- Shin, H.; Lee, H.; Woo, K.S.; Noh, E.W.; Koo, Y.B.; Lee, K.J. Identification of genes up regulated by pine wood nematode inoculation in Japanese red pine. Tree Physiol. 2009, 29, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, X. Analysis on the trend of invasion and expansion of Bursaphelenchus xylophilus. Pest Dis. 2018, 37, 5–8. [Google Scholar]
- Announcement of the State Forestry and Grassland Administration (2019 No. 4) (Pinewood Nematode Epidemic Area in 2019). Available online: http://www.forestry.gov.cn/ (accessed on 1 February 2019).
- Wang, X.R.; Kong, X.C.; Jia, W.H.; Zhu, X.W.; Ren, L.L.; Moto, M.M. A rapid staining-assisted wood sampling method for PCR-based detection of pine wood nematode Bursaphelenchus xylophilus in Pinus massoniana wood tissue. For. Pathol. 2010, 40, 510–520. [Google Scholar] [CrossRef]
- Wang, X.R.; Zhu, X.W.; Kong, X.C.; Moto, M.M. A rapid detection of the pinewood nematode, Bursaphelenchus xylophilus in stored Monochamus alternatus by rDNA amplification. J. Appl. Entomol. 2011, 135, 156–159. [Google Scholar] [CrossRef]
- Abedin, M.N.; Refaat, T.F.; Bhat, I.B. Progress of Multicolor Single Detector to Detector Array Development for Remote Sensing. Proc. Spie Int. Soc. Opt. Eng. 2004, 5543, 239–247. [Google Scholar]
- Krezhova, D.; DiKova, B.; Maneva, S. Ground based hyperspectral remote sensing for disease detection of tobacco plants. Bulg. J. Agric. Sci. 2014, 20, 1142–1150. [Google Scholar]
- Vanegas, F.; Bratanov, D.; Powell, K.; Weiss, J.; Gonzalez, F. A Novel Methodology for Improving Plant Pest Surveillance in Vineyards and Crops Using UAV-Based Hyperspectral and Spatial Data. Sensors 2018, 18, 260. [Google Scholar] [CrossRef]
- Mewes, T.; Franke, J.; Menz, G. Spectral requirements on airborne hyperspectral remote sensing data for wheat disease detection. Springer Sci. Bus. Media 2011, 12, 795–812. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis., Y.; Kakarla, S.C.; Roberts, P. Detection of target spot and bacterial spot diseases in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precis. Agric. 2019. [Google Scholar] [CrossRef]
- Shadrin, D.; Pukalchik, M.; Uryasheva, A.; Tsykunov, E.; Yashin, G.; Rodichenko, N.; Tsetserukou, D. Hyper-spectral NIR and MIR data and optimal wavebands for detection of apple tree diseases. arXiv 2004, arXiv:2004.02325. [Google Scholar]
- Gu, Q.; Sheng, L.; Zhang, T.; Lu, Y.; Zhang, Z.; Zheng, K.; Hu, H.; Zhou, H. Early detection of tomato spotted wilt virus infection in tobacco using the hyperspectral imaging technique and machine learning algorithms. Comput. Electron. Agric. 2019, 167, 105066. [Google Scholar] [CrossRef]
- Smith, M.L.; Ollinger, S.V.; Martin, M.E.; Aber, J.D.; Hallett, R.A.; Goodale, C.L. Direct estimation of aboveground forest productivity through hyperspectral remote sensing of canopy nitrogen. Ecol. Appl. 2002, 12, 5. [Google Scholar] [CrossRef]
- Nigam, R.; Kot, R.; Sandhu, S.S.; Bhattacharya1, B.K.; Chandi, R.S.; Singh, M.; Singh, J.; Manjunath, K.R. Ground Based Hyperspectral Remote Sensing to Discriminate Biotic Stress in Cotton Crop. Proc. Spie 2016, 9880, 98800H-10. [Google Scholar]
- Santos, C.S.S.D.; Vasconcelos, M.W.D. Identification of genes differentially expressed in Pinus pinaster and Pinus pinea after infection with the pine wood nematode. Eur. J. Plant. Pathol. 2012, 132, 407–418. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Koolstra, L. Using Hyperspectral Remote Sensing Data for Retrieving Canopy Chlorophyll and Nitrogen Content. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 574–583. [Google Scholar] [CrossRef]
- Koksal, E.S.; Üstun, H.; Özcan, H.; Gunturk, A. Estimating water stressed dwarf green bean pigment concentration through hyperspectral indices. Pak. J. Bot. 2010, 42, 1895–1901. [Google Scholar]
- Kim, S.R.; Kim, E.S.; Nam, Y.; Choi, W.I.; Kim, C.W. Distribution characteristics analysis of pine wilt disease using time series hyperspectral aerial imagery. Korean J. Remote Sens. 2015, 31, 385–394. [Google Scholar] [CrossRef]
- Moens, M.; Subbotin, S.; Maafi, Z.T. Molecular identification of cyst-forming nematodes (Heteroderidae) from Iran and a phylogeny based on ITS-rDNA sequences. Nematology 2003, 5, 99–111. [Google Scholar]
- Akintayo, A.; Tylka, G.L.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. A deep learning framework to discern and count microscopic nematode eggs. Sci. Rep. 2018, 8, 9145. [Google Scholar] [CrossRef]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.K.; Ganapathysubramanian, B. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 2017, 13, 23. [Google Scholar] [CrossRef]
- Zhang, J.; Naik, H.S.; Assefa, T.; Sarkar, S.; Chowda Reddy, R.V.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci. Rep. 2017, 7, 44048. [Google Scholar] [CrossRef] [PubMed]
- Harmey, J.H.; Harmey, M.A. Detection and identification of Bursaphelenchus species with DNA fingerprinting and polymerase chain reaction. J. Nematol. 1993, 25, 406–415. [Google Scholar] [PubMed]
- Abad, P.; Mota, M.; Vieira, P. Satellite DNA used as a species specific probe for identification of the pine wood nematode Bursaphelenchus xylophilus. EPPO Bull. 2004, 30, 571–574. [Google Scholar] [CrossRef]
- Akeuchi, Y.; Kanzaki, N.; Futai, K. A nested PCR-based method for detecting the pine wood nematode, Bursaphelenchus xylophilus, from pine wood. Nematology 2005, 7, 775–782. [Google Scholar]
- Wang, X.; Zhu, X.; Hu, Y.; Huang, H.; Kong, X.; Jia, W. A PCR-Based Method for Detecting Bursaphelenchus xylophilus from Monochamus alternatus. Sci. Silvae Sin. 2009, 45, 70–75. [Google Scholar]
- Kenichi, Y.; Takuma, T.; Natsumi, K.; Komatsu, M.; Levia, D.F.; Kabeya, D.; Tobita, H.; Kitao, M.; Ishida, A. Pine wilt disease causes cavitation around the resin canals and irrecoverable xylem conduit dysfunction. J. Exp. Bot. 2018, 69, 589–602. [Google Scholar]
- Kuroda, K. Physiological incidences related to symptom development and wilting mechanism. In Pine Wilt Disease; Springer: Tokyo, Japan, 2008; pp. 204–222. [Google Scholar] [CrossRef]
- Goet, A.F.H.; Vane, G.; Solomon, J.E.; Rock, B.N. Imaging spectrometry for earth remote sensing. Science 1985, 228, 1147–1153. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral sensors and imaging technologies in phytopathology: State of the Art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Zarco-Tejada, P.J.; Núñez, L.C.; Sepulcre-Cantób, G.; Gonzálezc, M.R.; Martin, P. Grape quality assessment in vineyards affected by iron deficiency chlorosis using narrow-band physiological remote sensing indices. Remote Sens. Environ. 2010, 114, 1968–1986. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Digital image processing techniques for detecting, quantifying and classifying plant diseases. Springerplus 2013, 2, 660. [Google Scholar] [CrossRef] [PubMed]
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267. [Google Scholar] [CrossRef]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef]
- Kim, S.R.; Lee, W.K.; Lim, C.H.; Kim, M.; Kafatos, M.C.; Lee, S.H.; Lee, S.S. Hyperspectral analysis of pine wilt disease to determine an optimal detection index. Forests 2018, 9, 115. [Google Scholar] [CrossRef]
- Wendel, A.; Underwood, J. Illumination compensation in ground based hyperspectral imaging. ISPRS J. Photogramm. Remote Sens. 2017, 129, 162–178. [Google Scholar] [CrossRef]
- Shamsoddini, A.; Trinder, J.C.; Turner, R. Pine plantation structure mapping using worldview-2 multispectral image. Int. J. Remote Sens. 2013, 34, 3986–4007. [Google Scholar] [CrossRef]
- Lorente, D.; Aleixos, N.; Gómez-Sanchis, J.; Cubero, S.; García-Navarrete, O.L.; Blasco, J. Recent advances and applications of hyperspectral imaging for fruit and vegetable quality assessment. Neuroimage 2012, 5, 1121–1142. [Google Scholar] [CrossRef]
- Prieto, A.; Bellas, F.; Lopez-Pena, F.; Duro, R.J. Automatic preprocessing and classification system for high resolution ultra and hyperspectral images. In Computational Intelligence for Remote Sensing; Springer: Berlin, Heidelberg, 2008; pp. 313–340. [Google Scholar]
- Wu, J.; Ma, X.M.; Li, Z.Q.; Gao, S. An evaluation of airborne videography for detecting and monitorIng forest insect and disease. Rorest Res. 1994, 7, 579–584. [Google Scholar]
- Kim, J.B.; Jo, M.H.; Oh, J.S.; Lee, K.J.; Park, S.J. Extraction method of dam-aged area by pine wilt disease (bursaphelenchus xylophilus) using remotely sensed data and gIS. In Proceedings of the ACRS 2001—22nd Asian Conference on Remote Sensing, Singapore, 5–9 November 2001. [Google Scholar]
- Zhou, X.H.; Bian, G.B.; Xie, X.L.; Hou, Z.G.; Li, R.Q.; Zhou, Y.J. Qualitative and quantitative assessment of technical skills in percutaneous coronary intervention: In vivo porcine studies. IEEE Trans. Biomed. Eng. 2019, 99, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Eilertson, K.E.; Fricks, J.; Ferrari, M.J. Estimation and prediction for a mechanistic model of measles transmission using particle filtering and maximum likelihood estimation. Stat. Med. 2019, 38, 4146–4158. [Google Scholar]
- Huang, H.H.; Ma, X.H.; Huang, H.Y.; Zhou, Y.F.; Zhang, W.; Huang, Y.H. A preliminary study on monitoring of dead pine trees caused by pine wilt disease with fixed-wing unmanned aerial vehicle. J. Environ. Entomol. 2018, 40, 306–313. [Google Scholar]
- Li, W.; Shen, S.; He, P.; Hao, D.; Fang, Y.; Tao, L.; Zhang, S. A precisely positioning technique by remote sensing the dead trees in stands with inexpensive small UAV. J. For. Eng. 2014, 28, 102–105. [Google Scholar]
- Tao, H.; Li, C.; Zhou, J.; Huai, H.; Jiang, L.; Li, F. Recognition of red-attack pine trees from UAV imagery based on the HSV threshold method. J. Nanjing For. Univ. Nat. Sci. Ed. 2019, 43, 99–106. [Google Scholar]
- Naik, S.K.; Murthy, C.A. Hue-preserving color image enhancement without gamut problem. IEEE Trans. Image Process. Publ. IEEE Signal Process. Soc. 2003, 12, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, Y.; Katoh, M.; Deng, S.; Cheung, K. Detecting forests damaged by pine wilt disease at the individual tree level using airborne laser data and worldview-2/3 images over two seasons. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, XLII-3/W3, 181–184. [Google Scholar] [CrossRef]
- Du, H.Q.; Ge, H.L.; Fan, W.Y.; Jin, W.; Zhou, Y.F.; Li, J. Study on relationships between total chlorophyll with hyperspectral features for leaves of Pinus masopniana forest. Spectrosc. Spectr. Anal. 2009, 29, 3033–3037. [Google Scholar]
- Zhang, S.; Huang, J.; Qin, L.; Li, H. Ridge regression model for estimating pine wilt disease based on hyperspectral characteristics. Trans. Chin. Soc. Agric. Mach. 2019, 50, 196–202. [Google Scholar]
- Wang, X.T. Research on Dynamic Changes of Pine Wilt Disease Based on Hyperspectral. Master’s Thesis, Nanjing Forestry University, Nanjing, China, 2011. [Google Scholar]
- Ma, Y.; Lv, Q.; Zhao, X.T.; He, B.L.; Liu, H.X.; Zhang, X.Y. Analysis of spectral characteristics of Pinus thunbergii inoculated with pine wood nematode. Shandong Agric. Sci. 2012, 44, 12–16. [Google Scholar]
- Huang, M.X.; Gong, J.H.; Li, S.; Zhang, B.; Hao, Q.T. Study on pine wilt disease hyper-spectral time series and sensitive features. Remote Sens. Technol. Appl. 2012, 27, 954–960. [Google Scholar]
- Torresan, C.; Berton, A.; Carotenuto, F.; Gennaro, S.F.D.; Gioli, B.; Matese, A. Forestry applications of UAVs in Europe: A review. Int. J. Remote Sens. 2017, 38, 2427–2447. [Google Scholar] [CrossRef]
- Tang, L.; Shao, G. Drone remote sensing for forestry research and practices. J. For. Res. 2015, 26, 791–797. [Google Scholar] [CrossRef]
- Torre-Sánchez, J.; Granados-López, F.; Castro, A.I.D. Configuration and specifications of an Unmanned Aerial Vehicle (UAV) for early site specific weed management. PLoS ONE 2013, 8, e58210. [Google Scholar] [CrossRef]
- Lehmann, J.; Nieberding, F.; Prinz, T.; Knoth, C. Analysis of unmanned aerial system-based CIR images in forestry—A new perspective to monitor pest infestation levels. Forests 2015, 6, 594–612. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Gennaro, S.F.D.; Genensio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef]
- Nebiker, S.; Lack, N.; Abächerli, M.; Laderach, S. Light-weight multispectral uav sensors and their capabilities for predicting grain yield and detection plantdiseases. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, XLI-B1, 963–970. [Google Scholar] [CrossRef]
- Severtson, D.; Callow, N.; Flower, K.; Neuhaus, A.; Olejnik, M.; Nansen, C. Unmanned aerial vehicle canopy reflectance data detects potassium deficiency and green peach aphid susceptibility in canola. Precis. Agric. 2016, 17, 659–677. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Sandino, J.; Pegg, G.; Gonzalez, F.; Smith, G. Aerial mapping of forests affected by pathogens using UAVs, hyperspectral sensors, and artificial intelligence. Sensors 2018, 18, 944. [Google Scholar] [CrossRef] [PubMed]
- Behmann, J.; Mahlein, A.K.; Rumpf, T.; Romer, C.; Plumer, L. A review of advanced machine learning methods for the detection of biotic stress in precision crop protection. Precis. Agric. 2015, 16, 239–260. [Google Scholar] [CrossRef]
- Behmann, J.; Acebron, K.; Emin, D.; Bennertz, S.; Matsubara, S.; Thoms, S.; Bohnenkamp, D.; Kuska, M.T.; Jussila, J.; Salo, H.; et al. Specim IQ: Evaluation of a new, miniaturized handheld hyperspectral camera and its application for plant phenotyping and disease detection. Sensors 2018, 18, 441. [Google Scholar] [CrossRef]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Goetz, A.F.H. Three decades of hyperspectral remote sensing of the Earth: A personal view. Remote Sens. Environ. 2009, 113, S5–S16. [Google Scholar] [CrossRef]
- Bo, W.; Chen, C.C.; Kechadi, T.M.; Sun, L.Y. A comparative evaluation of filter-based feature selection methods for hyper-spectral band selection. Int. J. Remote Sens. 2013, 34, 7974–7990. [Google Scholar]
- Hagen, N.A.; Kester, R.T.; Gao, L.S.; Tkaczyk, T.S. Snapshot advantage: A review of the light collection improvement for parallel high-dimensional measurement systems. Opt. Eng. 2012, 51, 1371–1379. [Google Scholar] [CrossRef]
- Aasen, H.; Burkart, A.; Bolten, A.; Baerth, G. Generating 3D hyperspectral information with lightweight UAV snapshot cameras for vegetation monitoring: From camera calibration to quality assurance. ISPRS J. Photogramm. Remote Sens. 2015, 108, 245–259. [Google Scholar] [CrossRef]
- Mahlein, A. Plant disease detection by imaging sensors-parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 2016, 2, 241–251. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Detection of nutrition deficiencies in plants using proximal images and machine learning: A review. Comput. Electron. Agric. 2019, 162, 482–492. [Google Scholar] [CrossRef]
- Damm, A.; Guanter, L.; Verhoef, W.; Schlapfer, D. Impact of varying irradiance on vegetation indices and chlorophyll fluorescence derived from spectroscopy data. Remote Sens. Environ. 2015, 156, 202–215. [Google Scholar] [CrossRef]
- Pinto, F.; Damm, A.; Schickling, A.; Panigada, C.; Cogliati, S.; Muller-Linow, M.; Balvora, A.; Rascher, U. Sun-induced chlorophyll fluorescence from high-resolution imaging spectroscopy data to quantify spatio-temporal patterns of photosynthetic function in crop canopies. Plant Cell Environ. 2016, 39, 1500–1512. [Google Scholar] [CrossRef] [PubMed]
- Milton, E.J.; Schaepman, M.E.; Anderson, K.; Kneubuhler, M.; Fox, N. Progress in field spectroscopy. Remote Sens. Environ. 2009, 113, S92–S109. [Google Scholar] [CrossRef]
- Vigneau, N.; Ecarnot, M.; Rabatel, G.; Roumet, P. Potential of field hyperspectral imaging as a non destructive method to assess leaf nitrogen content in Wheat. Field Crops Res. 2011, 122, 25–31. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Paulus, S.; Kuhlmann, H.; Oerke, E.C.; Plumer, L. Calibration of hyperspectral close-range pushbroom cameras for plant phenotyping. ISPRS J. Photogramm. Remote Sens. 2015, 106, 172–182. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Sarkar, S.; Singh, A.K.; Singh, A.; Ganapathysubrmanian, B. Hyperspectral band selection using genetic algorithm and support vector machines for early identification of charcoal rot disease in soybean stems. Plant Methods 2018, 14, 86. [Google Scholar] [CrossRef]
- Thomas, S.; Wahabzada, M.; Kuska, M.T.; Rascher, U.; Mahlein, A.K. Observation of plant pathogen interaction by simultaneous hyperspectral imaging reflection and transmission measurements. Funct. Plant Biol. 2017, 44, 23. [Google Scholar] [CrossRef]
- Elvidge, C.D.; Keith, D.M.; Tuttle, B.T.; Baugh, K. Spectral Identification of Lighting Type and Character. Sensors 2010, 10, 3961–3988. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods 2019, 15, 98. [Google Scholar] [CrossRef]
- Yao, Z.; Lei, Y.; He, D. Early visual detection of wheat stripe rust using visible/Near-Infrared hyperspectral imaging. Sensors 2019, 19, 952. [Google Scholar] [CrossRef] [PubMed]
- Jarolmasjed, S.; Khot, L.; Sankaran, S. Hyperspectral imaging and spectrometry-derived spectral features for bitter pit detection in storage apples. Sensors 2018, 18, 1561. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wei, K.; Liu, Q.; Pan, L.; Tu, K. Classification and discrimination of different fungal diseases of three infection levels on peaches using hyperspectral reflectance imaging analysis. Sensors 2018, 18, 1295. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.D.; de Lourdes Bueno Trindade Galo, M.; Vieira, B.S. Detecting and mapping root-knot nematode infection in coffee crop using remote sensing measurements. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 5395–5403. [Google Scholar] [CrossRef]
- Liu, Y.L.; Qiang, L.; He, S.L.; Yi, S.L.; Liu, X.F.; Zheng, Y.Q.; Deng, L. Prediction of nitrogen and phosphorus contents in citrus leaves based on hyperspectral imaging. Int. J. Agric. Biol. Eng. 2015, 8, 80–88. [Google Scholar]
- Ahmadi, P.; Muharam, F.M.; Ahmad, K.; Mansor, S.; Seman, L.A. Early Detection of Ganoderma Basal Stem Rot of Oil Palms Using Artificial Neural Network Spectral Analysis. Plant Dis. 2017, 101, 1009–1016. [Google Scholar] [CrossRef]
- Arellano, P.; Tansey, K.; Balzter, H.; Boyd, D.S. Field spectroscopy and radiative transfer modelling to assess impacts of petroleum pollution on biophysical and biochemical parameters of the Amazon rainforest. Environ. Earth Sci. 2017, 76, 217. [Google Scholar] [CrossRef]
- Ghamisi, P.; Yokoya, N.; Li, J.; Liao, W.Z.; Liu, S.; Plaza, J.; Rasti, B.; Plaza, A. Advances in hyperspectral image and signal processing: A comprehensive overview of the state of the art. IEEE Geosci. Remote Sens. Mag. 2018, 5, 37–78. [Google Scholar] [CrossRef]
- Zhang, Z.G. The Early Identification of Remote Sensing about Bursaphelenchus xylophilus Based on Process Model. Master’s Thesis, Beijing Forestry University, Beijing, China, 2011. [Google Scholar]
- Dennison, P.E.; Halligan, K.Q.; Roberts, D.A. A comparison of error metrics and constraints for multiple endmember spectral mixture analysis and spectral angle mapper. Remote Sens. Environ. 2004, 93, 359–367. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Lu, K.K. Prediction of the pine wood nematode based on artificial neural network and hyperspectral data. Master’s Thesis, Zhejiang A&F University, Hangzhou, China, 2016. [Google Scholar]
- He, Q. Neural Network and Its Application in IR; Graduate School of Library and Information Science, University of Illinois at Urbana-Champaign Spring: Champaign, IL, USA, 1999; pp. 1–31. [Google Scholar]
- Kimes, D.S.; Nelson, R.F.; Manry, M.T.; Fung, A.K. Review article: Attributes of neural networks for extracting continuous vegetation variables from optical and radar measurements. Int. J. Remote Sens. 1998, 19, 2639–2663. [Google Scholar] [CrossRef]
- Narendra, K.S.; Parthasarathy, K. Identification and control of dynamical systems using neural networks. IEEE Trans. Neural Netw. 1990, 1, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yu, L.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Xu, H.C.; Luo, Y.Q.; Zhang, Y.Y.; Shi, Y.J. Changes of Reflectance Spectra of Pine Needles in Different Stage after Being Infected by Pine Wood Nematode. Spectrosc. Spectr. Anal. 2011, 31, 1352–1356. [Google Scholar]
- Basheer, I.A.; Hajmeer, M. Artificial neural networks: Fundamentals, computing, design, and application. J. Microbiol. Methods 2000, 43, 3–31. [Google Scholar] [CrossRef]
- Huang, G.B.; Lei, C. Convex incremental extreme learning machine. Neurocomputing. 2007, 70, 3056–3062. [Google Scholar] [CrossRef]
- Kumar, S.P.; Sriraam, N.; Benakop, P.G.; Jinaga, B.C. Entropies based detection of epileptic seizures with artificial neural network classifiers. Expert Syst. Appl. 2010, 37, 3284–3291. [Google Scholar] [CrossRef]
- Liu, W.Y. Hyperspectral estimation model for physiological parameters of pine tree under Bursaphelenchus xylophilus stress. Master’s Thesis, Nanjing Forestry University, Nanjing, China, 2017. [Google Scholar]
- Pal, M.; Foody, G.M. Feature Selection for Classification of Hyperspectral Data by SVM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2297–2307. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Liu, F.; Nie, P.C.; He, Y. Rice seed cultivar identification using near-infrared hyperspectral imaging and multivariate data analysis. Sensors 2013, 13, 8916–8927. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Zhang, Z.; Zheng, L.; Han, C.; Wang, X.; Xu, J.; Wang, X. Research Progress on the Early Monitoring of Pine Wilt Disease Using Hyperspectral Techniques. Sensors 2020, 20, 3729. https://doi.org/10.3390/s20133729
Wu W, Zhang Z, Zheng L, Han C, Wang X, Xu J, Wang X. Research Progress on the Early Monitoring of Pine Wilt Disease Using Hyperspectral Techniques. Sensors. 2020; 20(13):3729. https://doi.org/10.3390/s20133729
Chicago/Turabian StyleWu, Weibin, Zhenbang Zhang, Lijun Zheng, Chongyang Han, Xiaoming Wang, Jian Xu, and Xinrong Wang. 2020. "Research Progress on the Early Monitoring of Pine Wilt Disease Using Hyperspectral Techniques" Sensors 20, no. 13: 3729. https://doi.org/10.3390/s20133729
APA StyleWu, W., Zhang, Z., Zheng, L., Han, C., Wang, X., Xu, J., & Wang, X. (2020). Research Progress on the Early Monitoring of Pine Wilt Disease Using Hyperspectral Techniques. Sensors, 20(13), 3729. https://doi.org/10.3390/s20133729





