Terahertz Spectroscopic Signatures of Microcystin Aptamer Solution Probed with a Microfluidic Chip
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microfluidic Chip
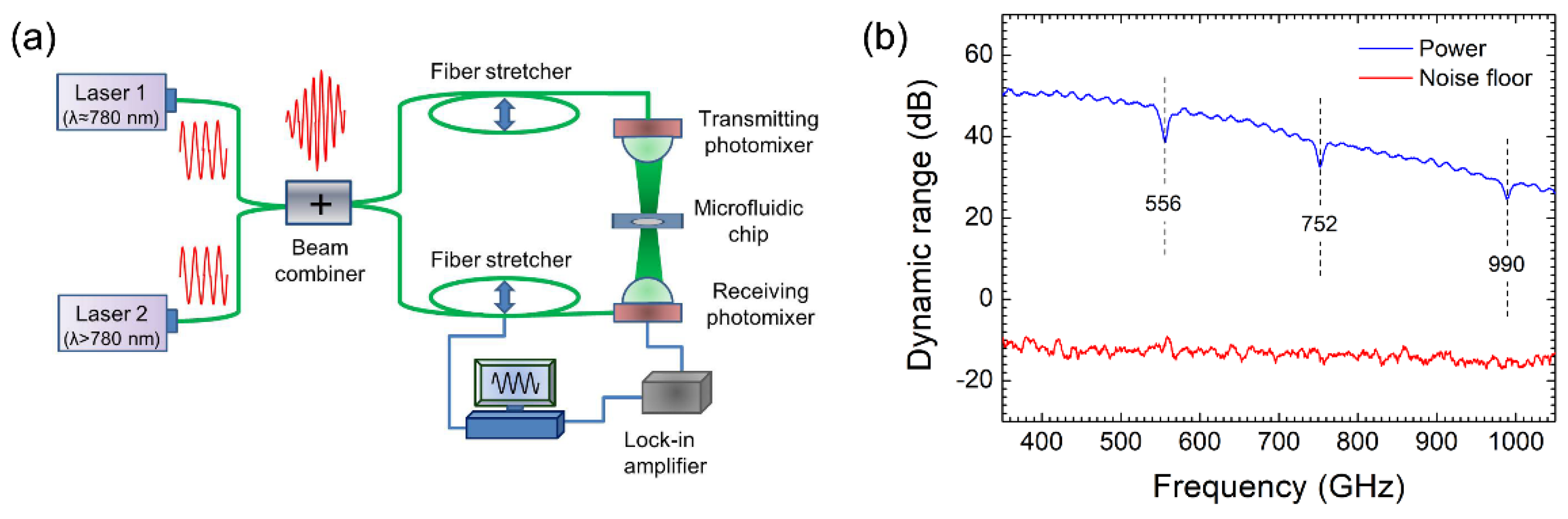
2.2. Coherent Photomixing Spectrometer
2.3. Sample Preparation and Spectrometer Adjustment
2.4. Molecular Dynamics Simulation
3. Results
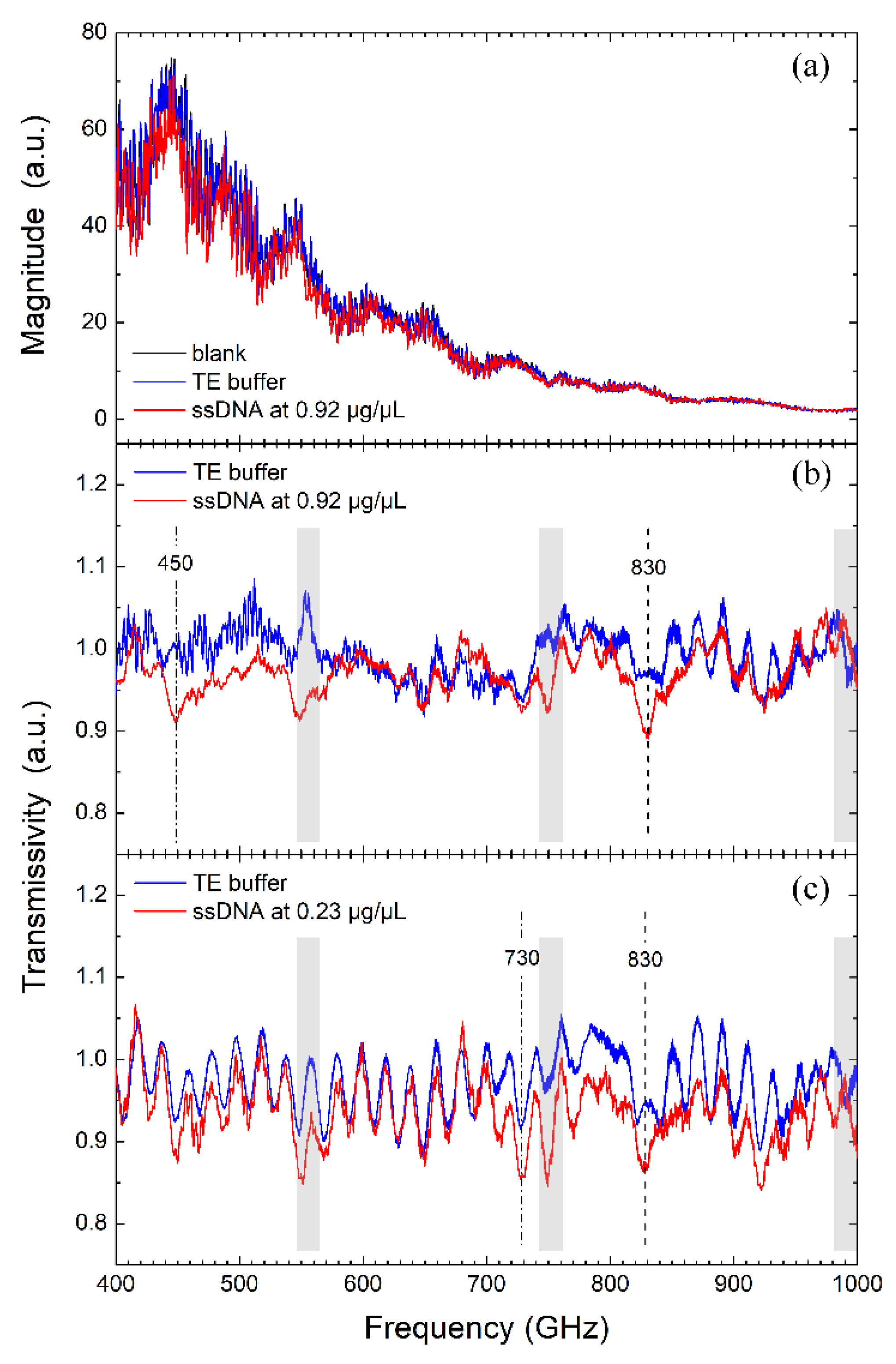
3.1. THz Signature Estimation from Transmission Spectra
3.2. Signature Recognition via Fitting Analysis
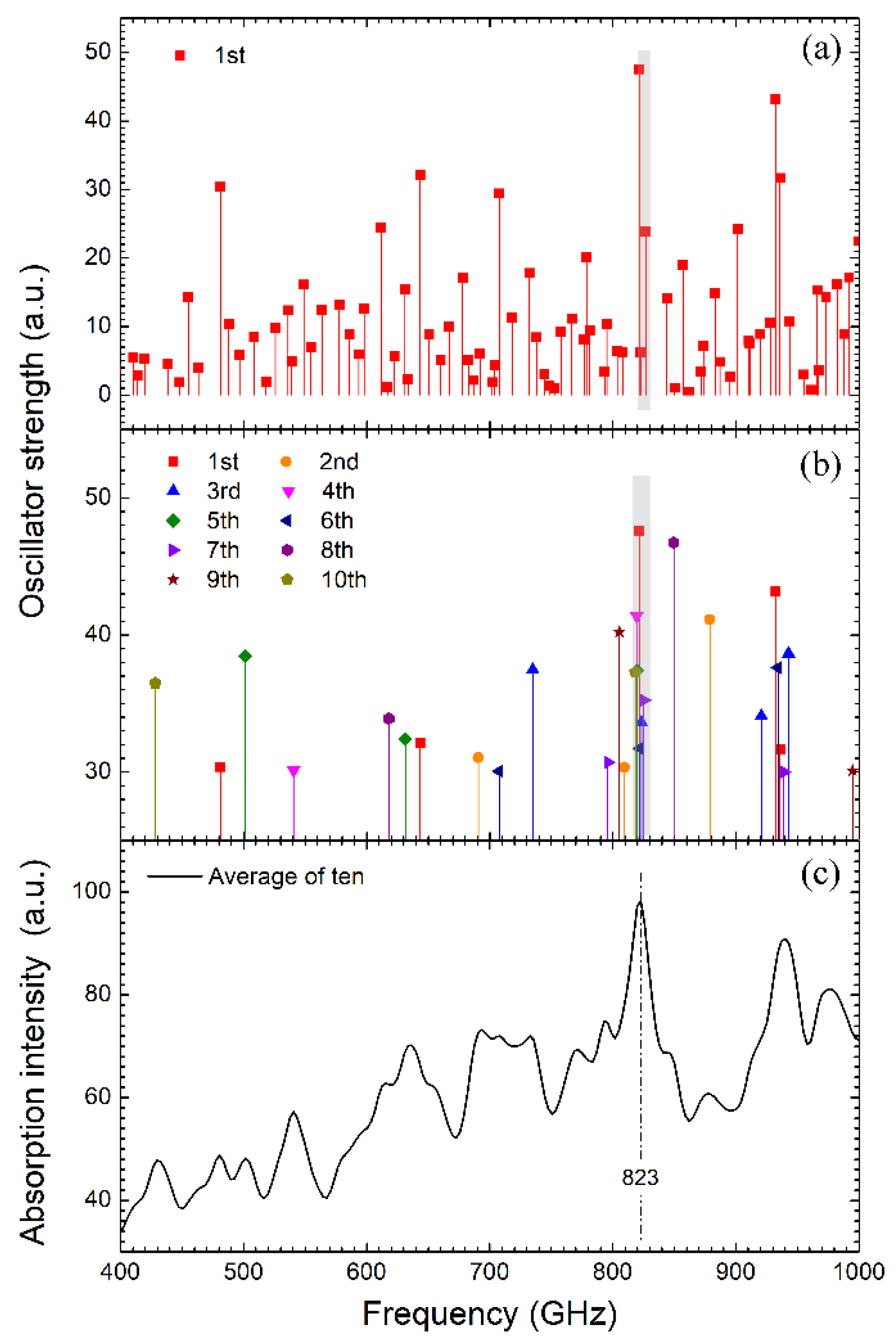
3.3. Resonance Analysis Using Quasi-Harmonic Approximation
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Heyden, M.; Havenith, M. Combining Thz Spectroscopy and Md Simulations to Study Protein-Hydration Coupling. Methods 2010, 52, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Born, B.; Havenith, M.; Gruebele, M. Real-Time Detection of Protein-Water Dynamics Upon Protein Folding by Terahertz Absorption Spectroscopy. Angew. Chem.-Int. Ed. 2008, 47, 6486–6489. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.W.; Zhong, J.L.; Zhang, C.L.; Zuo, J.; Pickwell-MacPherson, E. Label-Free Detection and Characterization of the Binding of Hemagglutinin Protein and Broadly Neutralizing Monoclonal Antibodies Using Terahertz Spectroscopy. J. Biomed. Opt. 2015, 20, 037006. [Google Scholar] [CrossRef] [PubMed]
- Li, M.L.; Chang, T.Y.; Wei, D.S.; Tang, M.J.; Yan, S.H.; Du, C.L.; Cui, H.L. Label-Free Detection of Anti-Estrogen Receptor Alpha and Its Binding with Estrogen Receptor Peptide Alpha by Terahertz Spectroscopy. RSC Adv. 2017, 7, 24338–24344. [Google Scholar] [CrossRef]
- Arora, A.; Luong, T.Q.; Kruger, M.; Kim, Y.J.; Nam, C.H.; Manz, A.; Havenith, M. Terahertz-Time Domain Spectroscopy for the Detection of Pcr Amplified DNA in Aqueous Solution. Analyst 2012, 137, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Stanley, B.; Rahman, A.K. Ultrasensitive Label-Free Detection and Quantitation of DNA Hybridization Via Terahertz Spectrometry. In Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues Viii; Farkas, D.L., Nicolau, D.V., Leif, R.C., Eds.; Spie-Int Soc Optical Engineering: Bellingham, WA, USA, 2010. [Google Scholar]
- Wang, F.; Zhao, D.B.; Dong, H.; Jiang, L.; Liu, Y.F.; Li, S.H. Terahertz Spectra of DNA Nucleobase Crystals: A Joint Experimental and Computational Study. Spectrochim. Acta Part a-Mol. Biomol. Spectrosc. 2017, 179, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.J.; Huang, Q.; Wei, D.S.; Zhao, G.Z.; Chang, T.Y.; Kou, K.; Wang, M.; Du, C.L.; Fu, W.L.; Cui, H.L. Terahertz Spectroscopy of Oligonucleotides in Aqueous Solutions. J. Biomed. Opt. 2015, 20, 095009. [Google Scholar] [CrossRef]
- Baragwanath, A.J.; Swift, G.P.; Dai, D.; Gallant, A.J.; Chamberlain, J.M. Silicon Based Microfluidic Cell for Terahertz Frequencies. J. Appl. Phys. 2010, 108, 013102. [Google Scholar] [CrossRef]
- George, P.A.; Hui, W.; Rana, F.; Hawkins, B.G.; Smith, A.E.; Kirby, B.J. Microfluidic Devices for Terahertz Spectroscopy of Biomolecules. Opt. Express 2008, 16, 1577–1582. [Google Scholar] [CrossRef]
- Liu, L.; Pathak, R.; Cheng, L.J.; Wang, T. Real-Time Frequency-Domain Terahertz Sensing and Imaging of Isopropyl Alcohol-Water Mixtures on a Microfluidic Chip. Sens. Actuators B Chem. 2013, 184, 228–234. [Google Scholar] [CrossRef]
- Saha, S.C.; Grant, J.P.; Ma, Y.; Khalid, A.; Hong, F.; Cumming, D.R.S. Application of Terahertz Spectroscopy to the Characterization of Biological Samples Using Birefringence Silicon Grating. J. Biomed. Opt. 2012, 17, 067006. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.C.; Grant, J.P.; Ma, Y.; Khalid, A.; Hong, F.; Cumming, D.R.S. Terahertz Frequency-Domain Spectroscopy Method for Vector Characterization of Liquid Using an Artificial Dielectric. IEEE Trans. Ter. Sci. Technol. 2012, 2, 113–122. [Google Scholar] [CrossRef]
- Laurette, S.; Treizebre, A.; Bocquet, B. Co-Integrated Microfluidic and Thz Functions for Biochip Devices. J. Micromech. Microeng. 2011, 21, 065029. [Google Scholar] [CrossRef]
- Kiwa, T.; Oka, S.; Kondo, J.; Kawayama, I.; Yamada, H.; Tonouchi, M.; Tsukada, K. A Terahertz Chemical Microscope to Visualize Chemical Concentrations in Microftuidic Chips. Jpn. J. Appl. Phys. Part 2-Lett. Express Lett. 2007, 46, L1052–L1054. [Google Scholar] [CrossRef]
- Kitagawa, J.; Ohkubo, T.; Onuma, M.; Kadoya, Y. THz Spectroscopic Characterization of Biomolecule/Water Systems by Compact Sensor Chips. Appl. Phys. Lett. 2006, 89, 041114. [Google Scholar] [CrossRef]
- Mendis, R.; Astley, V.; Liu, J.B.; Mittleman, D.M. Terahertz Microfluidic Sensor Based on a Parallel-Plate Waveguide Resonant Cavity. Appl. Phys. Lett. 2009, 95, 171113. [Google Scholar] [CrossRef]
- Duponchel, L.; Laurette, S.; Hatirnaz, B.; Treizebre, A.; Affouard, F.; Bocquet, B. Terahertz Microfluidic Sensor for in Situ Exploration of Hydration Shell of Molecules. Chemom. Intell. Lab. Syst. 2013, 123, 28–35. [Google Scholar] [CrossRef]
- Laurette, S.; Treizebre, A.; Affouard, F.; Bocquet, B. Subterahertz Characterization of Ethanol Hydration Layers by Microfluidic System. Appl. Phys. Lett. 2010, 97, 111904. [Google Scholar] [CrossRef]
- Chang, K.; Pi, Y.; Lu, W.P.; Wang, F.; Pan, F.; Li, F.K.; Jia, S.R.; Shi, J.F.; Deng, S.L.; Chen, M. Label-Free and High-Sensitive Detection of Human Breast Cancer Cells by Aptamer-Based Leaky Surface Acoustic Wave Biosensor Array. Biosens. Bioelectron. 2014, 60, 318–324. [Google Scholar] [CrossRef]
- Zhang, M.K.; Wei, D.S.; Tang, M.J.; Shi, C.C.; Cui, H.L.; Du, C.L. Molecular Dynamics Simulations of Conformation and Chain Length Dependent Terahertz Spectra of Alanine Polypeptides. Mol. Simul. 2016, 42, 398–404. [Google Scholar] [CrossRef]
- Brown, E.R.; Mendoza, E.A.; Kuznetsova, Y.; Neumann, A.; Brueck, S.R.J. Thz Signatures of DNA in Nanochannels under Electrophoretic Control. In Proceedings of the SENSORS, Baltimore, MD, USA, 3–6 November 2013. [Google Scholar]
- Brown, E.R.; Mendoza, E.A.; Xia, D.Y.; Brueck, S.R.J. Narrow Thz Spectral Signatures through an Rna Solution in Nanofluidic Channels. IEEE Sens. J. 2010, 10, 755–759. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, Z.; Tang, M.; Yan, S.; Wei, D.; Cui, H.; Du, C. The Properties, Preparation Approaches and Uses of Microfluidic Channels for Terahertz Absorption Signatures Detection in Aqueous. Paper Presented at the 6th IEEE International Conference on Manipulation, Manufacturing and Measurement on the Nanoscale, IEEE 3M-NANO 2016, Chongqing, China, 18–22 July 2016. [Google Scholar]
- Brown, E.R.; Zhang, W.; Viveros, L.K.; Mendoza, E.A.; Kuznetsova, Y.; Brueck, S.R.J.; Burris, K.P.; Millwood, R.J.; Stewart, C.N. High-Resolution Thz Spectroscopy of Biomolecules and Bioparticles: Concentration Methods. In Terahertz and Mid Infrared Radiation: Detection of Explosives and Cbrn (Using Terahertz); Springer: Berlin/Heidelberg, Germany, 2014; pp. 7–18. [Google Scholar]
- Brown, E.R.; Mendoza, E.A.; Kuznetsova, Y.; Neumann, A.; Brueck, S.R.J. High-Resolution Thz Spectroscopy to Measure Strong Thz Absorption Signatures of Si-Rna in Solution. In Terahertz and Mid Infrared Radiation: Generation, Detection and Applications; Perieira, M.F., Shulika, O., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 15–22. [Google Scholar]
- Ng, A.; Chinnappan, R.; Eissa, S.; Liu, H.C.; Tlili, C.; Zourob, M. Selection, Characterization, and Biosensing Application of High Affinity Congener-Specific Microcystin-Targeting Aptamers. Environ. Sci. Technol. 2012, 46, 10697–10703. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Ng, A.; Siaj, M.; Zourob, M. Label-Free Voltammetric Aptasensor for the Sensitive Detection of Microcystin-Lr Using Graphene-Modified Electrodes. Anal. Chem. 2014, 86, 7551–7557. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Cheng, R.J.; Shi, H.J.; Tang, B.; Xiao, H.S.; Zhao, G.H. A Simple Highly Sensitive and Selective Aptamer-Based Colorimetric Sensor for Environmental Toxins Microcystin-Lr in Water Samples. J. Hazard. Mater. 2016, 304, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.; Kowal, J. Selection of Glass, Anodic Bonding Conditions and Material Compatibility for Silicon-Glass Capacitive Sensors. Sens. Actuators a-Phys. 1995, 46, 113–120. [Google Scholar] [CrossRef]
- Roggenbuck, A.; Schmitz, H.; Deninger, A.; Mayorga, I.C.; Hemberger, J.; Gusten, R.; Gruninger, M. Coherent Broadband Continuous-Wave Terahertz Spectroscopy on Solid-State Samples. New J. Phys. 2010, 12, 043017. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Zgarbová, M.; Otyepka, M.; Sponer, J.; Mládek, A.; Banáš, P.; Cheatham III, T.E.; Jurečka, P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar]
- Li, T.P.; Hassanali, A.A.P.; Kao, Y.T.; Zhong, D.P.; Singer, S.J. Hydration Dynamics and Time Scales of Coupled Water-Protein Fluctuations. J. Am. Chem. Soc. 2007, 129, 3376–3382. [Google Scholar] [CrossRef]
- Zhang, W.; Brown, E.R.; Rahman, M.; Norton, M.L. Observation of Terahertz Absorption Signatures in Microliter DNA Solutions. Appl. Phys. Lett. 2013, 102, 023701. [Google Scholar] [CrossRef]
- Heugen, U.; Schwaab, G.; Brundermann, E.; Heyden, M.; Yu, X.; Leitner, D.M.; Havenith, M. Solute-Induced Retardation of Water Dynamics Probed Directly by Terahertz Spectroscopy. Proc. Natl. Acad. Sci. USA 2006, 103, 12301–12306. [Google Scholar] [CrossRef] [PubMed]
- Leitner, D.M.; Gruebele, M.; Havenith, M. Solvation Dynamics of Biomolecules: Modeling and Terahertz Experiments. Hfsp J. 2008, 2, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Murthy, U.M.S.; Vij, J.K. Submillimeter Wave Spectroscopy of 4-Normal-Alkyl-4′-Cyano Biphenyl Liquid-Crystals. Liq. Cryst. 1989, 4, 529–542. [Google Scholar] [CrossRef]
- Pereverzev, A.; Sewell, T.D. Terahertz Normal Mode Relaxation in Pentaerythritol Tetranitrate. J. Chem. Phys. 2011, 134, 014513. [Google Scholar] [CrossRef]
- Levy, R.M.; Karplus, M.; Kushick, J.; Perahia, D. Evaluation of the Configurational Entropy for Proteins—Application to Molecular-Dynamics Simulations of an Alpha-Helix. Macromolecules 1984, 17, 1370–1374. [Google Scholar] [CrossRef]
- Wittlin, A.; Genzel, L.; Kremer, F.; Haseler, S.; Poglitsch, A.; Rupprecht, A. Far-Infrared Spectroscopy on Oriented Films of Dry and Hydrated DNA. Phys. Rev. A 1986, 34, 493–500. [Google Scholar] [CrossRef]


| Resonant Center (GHz) | Oscillator Strength (cm−2) | Background Attenuation (cm−1) | FWHM (GHz) | |
|---|---|---|---|---|
| Figure 4a | 449.1 ± 0.1 | 890.1 ± 109.0 | −345.9 ± 64.7 | 27.6 ± 1.7 |
| Figure 4b | 829.3 ± 0.1 | 377.3 ± 46.1 | 94.3 ± 38.7 | 16.7 ± 1.3 |
| Figure 4c | 728.5 ± 0.1 | 890.4 ± 93.7 | −346.2 ± 80.8 | 18.4 ± 1.0 |
| Figure 4d | 828.3 ± 0.2 | 600.1 ± 144.6 | 111.0 ± 106.1 | 20.5 ± 2.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Yang, Z.; Tang, M.; Wang, D.; Wang, H.; Yan, S.; Wei, D.; Cui, H.-L. Terahertz Spectroscopic Signatures of Microcystin Aptamer Solution Probed with a Microfluidic Chip. Sensors 2019, 19, 534. https://doi.org/10.3390/s19030534
Zhang M, Yang Z, Tang M, Wang D, Wang H, Yan S, Wei D, Cui H-L. Terahertz Spectroscopic Signatures of Microcystin Aptamer Solution Probed with a Microfluidic Chip. Sensors. 2019; 19(3):534. https://doi.org/10.3390/s19030534
Chicago/Turabian StyleZhang, Mingkun, Zhongbo Yang, Mingjie Tang, Deqiang Wang, Huabin Wang, Shihan Yan, Dongshan Wei, and Hong-Liang Cui. 2019. "Terahertz Spectroscopic Signatures of Microcystin Aptamer Solution Probed with a Microfluidic Chip" Sensors 19, no. 3: 534. https://doi.org/10.3390/s19030534
APA StyleZhang, M., Yang, Z., Tang, M., Wang, D., Wang, H., Yan, S., Wei, D., & Cui, H.-L. (2019). Terahertz Spectroscopic Signatures of Microcystin Aptamer Solution Probed with a Microfluidic Chip. Sensors, 19(3), 534. https://doi.org/10.3390/s19030534






