Single-Step FRET-Based Detection of Femtomoles DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Sensors
2.3. Bulk FRET Experiments
2.4. Surface-Functionalization of Flow Cell
2.5. pTIRF Sample Preparation, Imaging, and Data Analysis
3. Results and Discussion
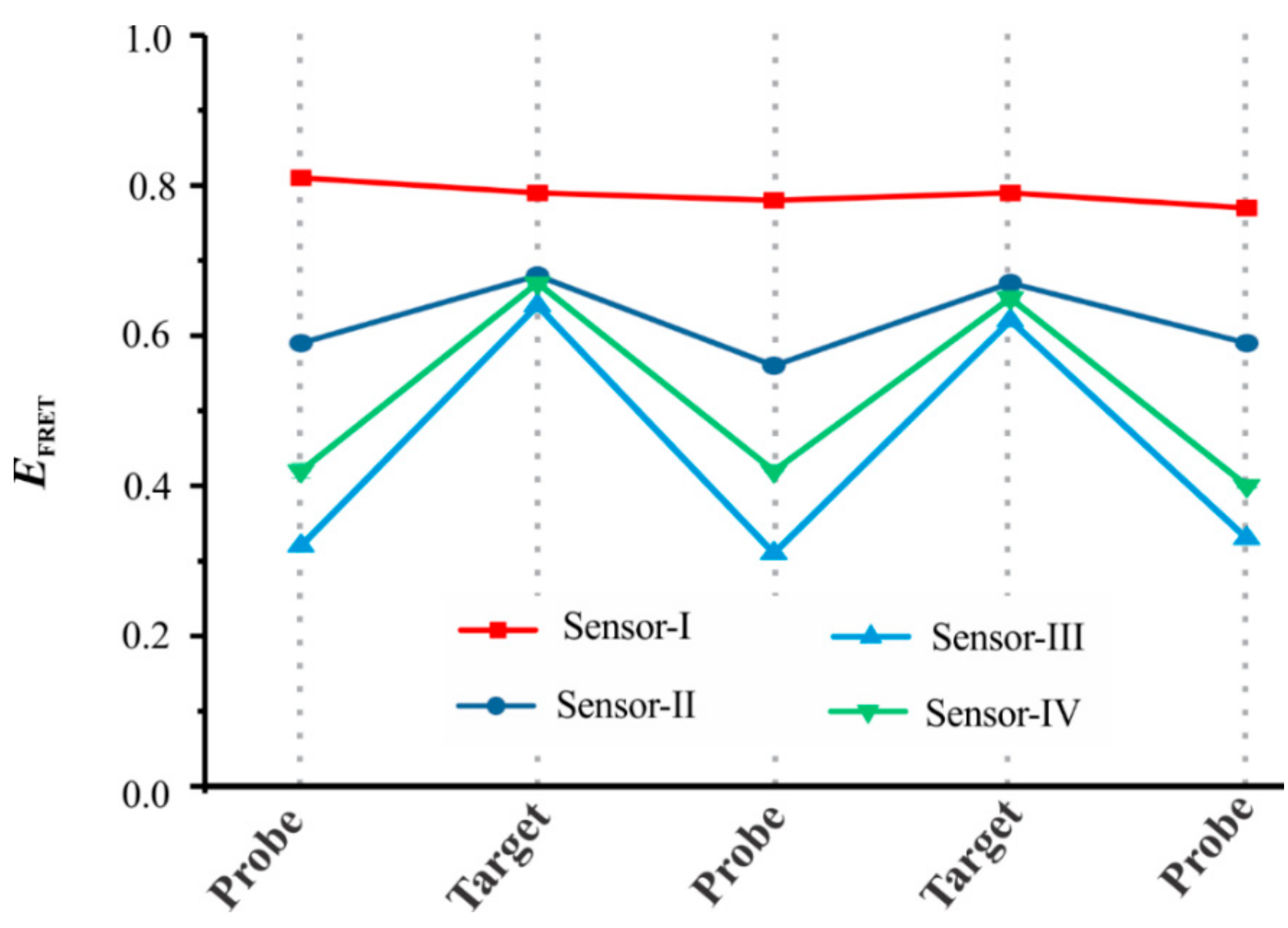
3.1. Sensor Design and Optimization
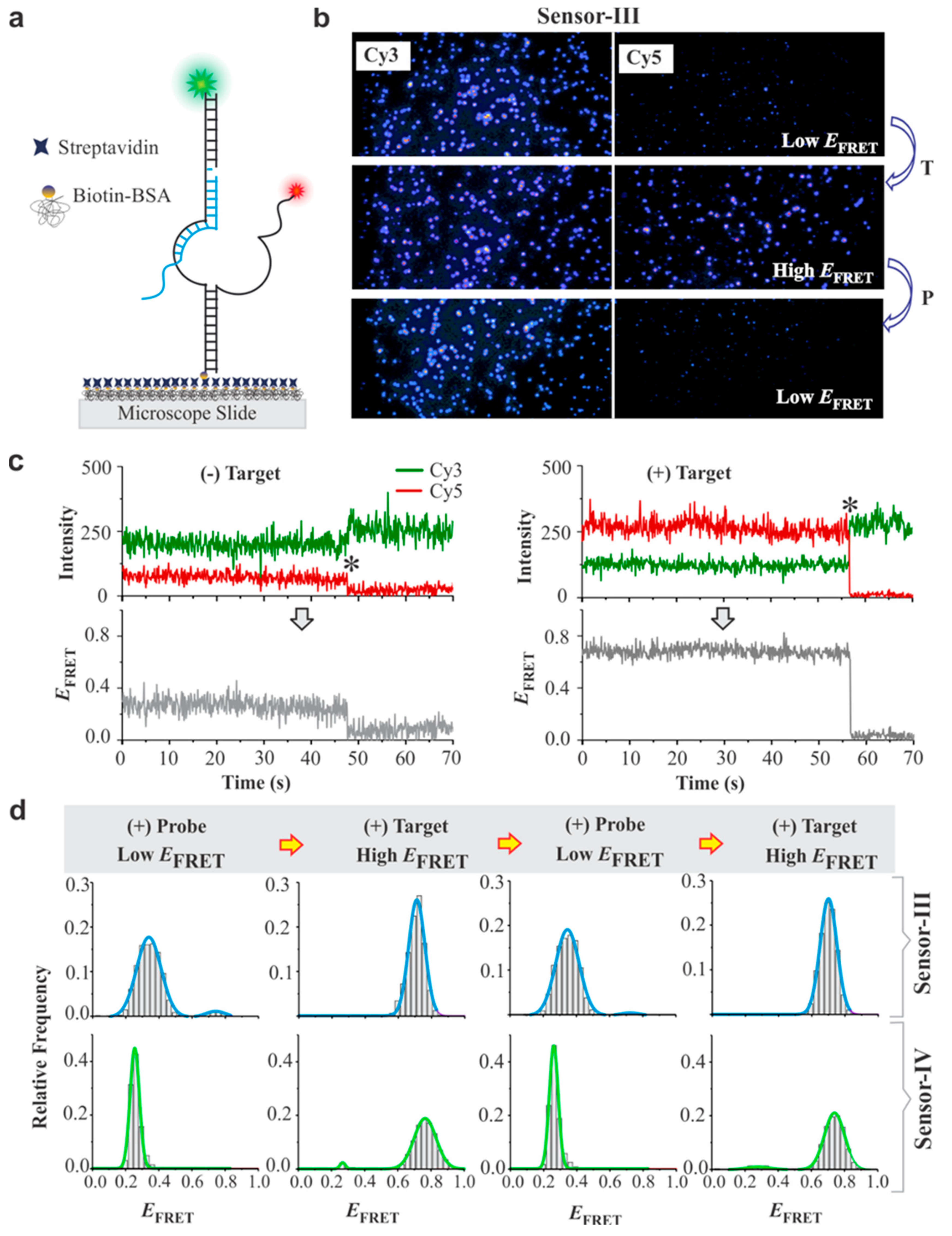
3.2. Single Molecule Analysis of DNA Sensors
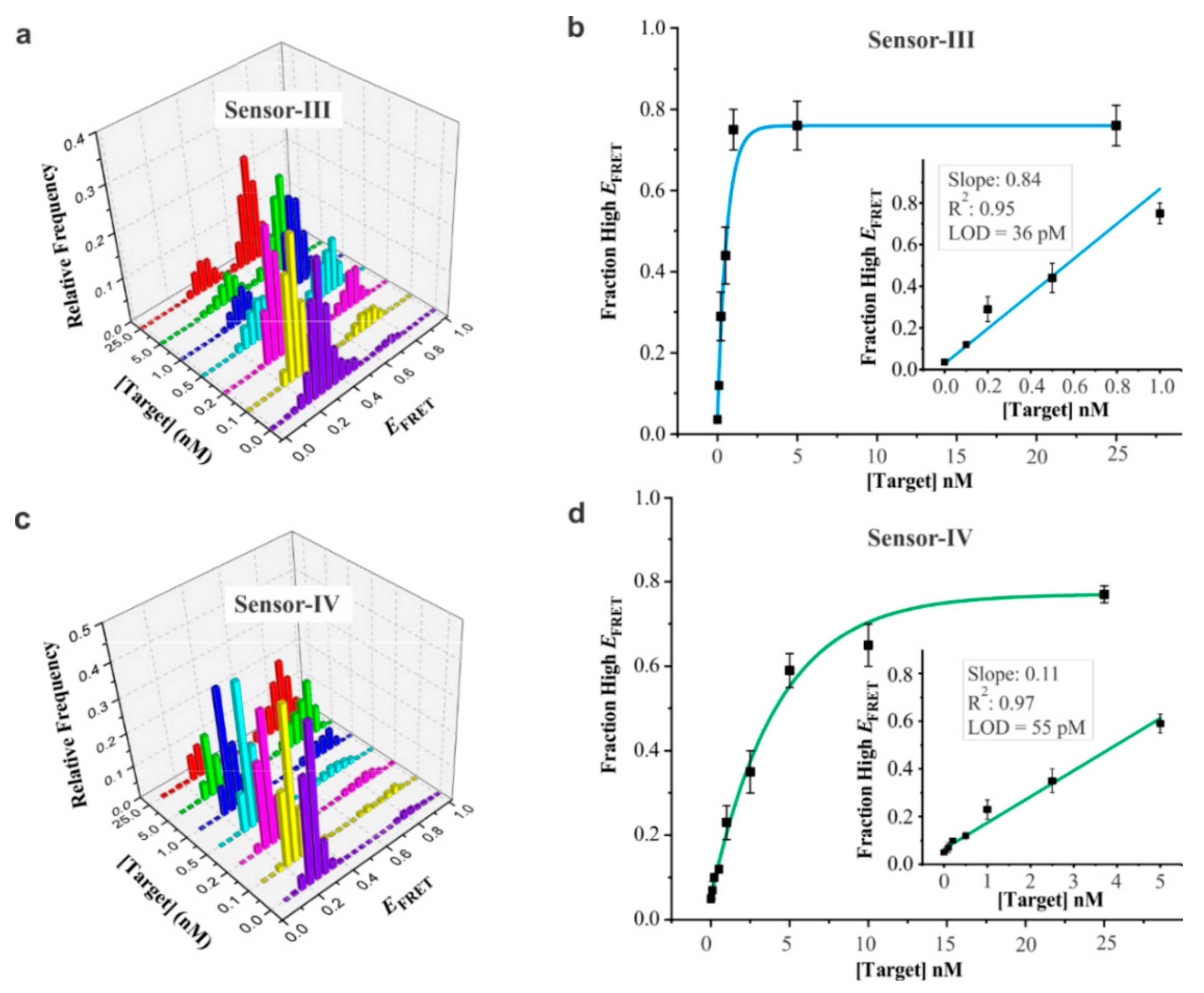
3.3. Analytical Sensitivity of DNA Sensors
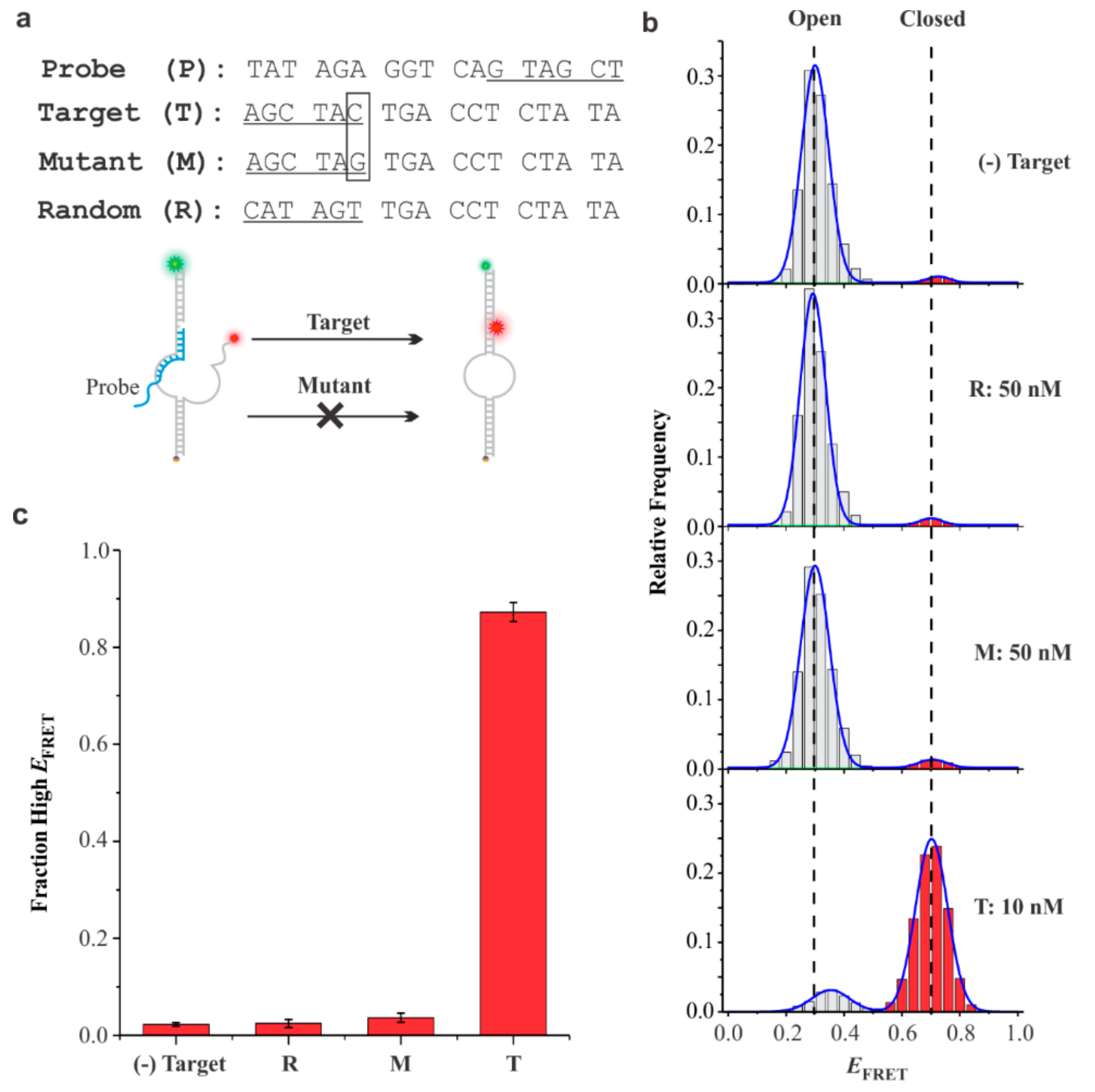
3.4. Specificity of DNA Sensors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu, J.; Cao, Z.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Nimse, S.B.; Sonawane, M.D.; Song, K.-S.; Kim, T. Biomarker Detection Technologies and Future Directions. Analyst 2016, 141, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, N.; Ouldridge, T.E.; Šulc, P.; Schaeffer, J.M.; Yurke, B.; Louis, A.A.; Doye, J.P.K.; Winfree, E. On the Biophysics and Kinetics of Toehold-Mediated DNA Strand Displacement. Nucleic Acids Res. 2013, 41, 10641–10658. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, N.; Parkin, J.; Seelig, G.; Winfree, E.; Soloveichik, D. Enzyme-Free Nucleic Acid Dynamical Systems. Science 2017, 358, eaal2052. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Johnson-Buck, A.; Yang, Y.R.; Shih, W.M.; Yan, H.; Walter, N.G. Exploring the Speed Limit of Toehold Exchange with a Cartwheeling DNA Acrobat. Nat. Nanotechnol. 2018, 13, 723–729. [Google Scholar] [CrossRef]
- Yang, K.; Yang, Y.; Zhang, C. Single-Molecule FRET for Ultrasensitive Detection of Biomolecules. NanoBioImaging 2013, 1, 13–24. [Google Scholar] [CrossRef]
- Ling, Y.; Zhang, X.F.; Chen, X.H.; Liu, L.; Wang, X.H.; Wang, D.S.; Li, N.B.; Luo, H.Q. A Dual-Cycling Biosensor for Target DNA Detection Based on the Toehold-Mediated Strand Displacement Reaction and Exonuclease III Assisted Amplification. New J. Chem. 2018, 42, 4714–4718. [Google Scholar] [CrossRef]
- Deng, B.; Lin, Y.; Wang, C.; Li, F.; Wang, Z.; Zhang, H.; Li, X.-F.; Le, X.C. Aptamer Binding Assays for Proteins: The Thrombin Example—A Review. Anal. Chim. Acta 2014, 837, 1–15. [Google Scholar] [CrossRef]
- Song, Q.; Wang, R.; Sun, F.; Chen, H.; Wang, Z.; Na, N.; Ouyang, J. A Nuclease-Assisted Label-Free Aptasensor for Fluorescence Turn-on Detection of ATP Based on the in Situ Formation of Copper Nanoparticles. Biosens. Bioelectron. 2017, 87, 760–763. [Google Scholar] [CrossRef]
- Labib, M.; Khan, N.; Ghobadloo, S.M.; Cheng, J.; Pezacki, J.P.; Berezovski, M.V. Three-Mode Electrochemical Sensing of Ultralow MicroRNA Levels. J. Am. Chem. Soc. 2013, 135, 3027–3038. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, A.; Zhang, D.; Tam, F.; Halas, N.J. Surface-Enhanced Raman Spectroscopy of DNA. J. Am. Chem. Soc. 2008, 130, 5523–5529. [Google Scholar] [CrossRef] [PubMed]
- Panikkanvalappil, S.R.; Mackey, M.A.; El-Sayed, M.A. Probing the Unique Dehydration-Induced Structural Modifications in Cancer Cell DNA Using Surface Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2013, 135, 4815–4821. [Google Scholar] [CrossRef] [PubMed]
- Law, W.-C.; Yong, K.-T.; Baev, A.; Prasad, P.N. Sensitivity Improved Surface Plasmon Resonance Biosensor for Cancer Biomarker Detection Based on Plasmonic Enhancement. ACS Nano 2011, 5, 4858–4864. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Mani, V.; Wasalathanthri, D.; Kumar, C.V.; Rusling, J.F. Attomolar Detection of a Cancer Biomarker Protein in Serum by Surface Plasmon Resonance Using Superparamagnetic Particle Labels. Angew. Chem. Int. Ed. 2011, 50, 1175–1178. [Google Scholar] [CrossRef] [PubMed]
- Gui, Z.; Wang, Q.B.; Li, J.C.; Zhu, M.C.; Yu, L.L.; Xun, T.; Yan, F.; Ju, H.X. Direct Detection of Circulating Free DNA Extracted from Serum Samples of Breast Cancer Using Locked Nucleic Acid Molecular Beacon. Talanta 2016, 154, 520–525. [Google Scholar] [CrossRef]
- Huang, S.; Qiu, H.; Xiao, Q.; Huang, C.; Su, W.; Hu, B. A Simple QD–FRET Bioprobe for Sensitive and Specific Detection of Hepatitis B Virus DNA. J. Fluoresc. 2013, 23, 1089–1098. [Google Scholar] [CrossRef]
- Bi, S.; Yue, S.; Zhang, S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef]
- Cannon, B.; Campos, A.R.; Lewitz, Z.; Willets, K.A.; Russell, R. Zeptomole Detection of DNA Nanoparticles by Single-Molecule Fluorescence with Magnetic Field-Directed Localization. Anal. Biochem. 2012, 431, 40–47. [Google Scholar] [CrossRef]
- Mayr, R.; Haider, M.; Thünauer, R.; Haselgrübler, T.; Schütz, G.J.; Sonnleitner, A.; Hesse, J. A Microfluidic Platform for Transcription-and Amplification-Free Detection of Zepto-Mole Amounts of Nucleic Acid Molecules. Biosens. Bioelectron. 2016, 78, 1–6. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Li, L.; Wang, J.; Jin, W. Ultra-Sensitive DNA Assay Based on Single-Molecule Detection Coupled with Fluorescent Quantum Dot-Labeling and Its Application to Determination of Messenger RNA. Anal. Chim. Acta 2011, 685, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Hesse, J.; Jacak, J.; Kasper, M.; Regl, G.; Eichberger, T.; Winklmayr, M.; Aberger, F.; Sonnleitner, M.; Schlapak, R.; Howorka, S.; et al. RNA Expression Profiling at the Single Molecule Level. Genome Res. 2006, 16, 1041–1045. [Google Scholar] [CrossRef] [PubMed]
- Anazawa, T.; Matsunaga, H.; Yeung, E.S. Electrophoretic Quantitation of Nucleic Acids without Amplification by Single-Molecule Imaging. Anal. Chem. 2002, 74, 5033–5038. [Google Scholar] [CrossRef] [PubMed]
- Doré, K.; Dubus, S.; Ho, H.-A.; Lévesque, I.; Brunette, M.; Corbeil, G.; Boissinot, M.; Boivin, G.; Bergeron, M.G.; Boudreau, D.; et al. Fluorescent Polymeric Transducer for the Rapid, Simple, and Specific Detection of Nucleic Acids at the Zeptomole Level. J. Am. Chem. Soc. 2004, 126, 4240–4244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ning, X.; Mao, G.; Ji, X.; He, Z. Fluorescence turn-on detection of target sequence DNA based on silicon nanodot-mediated quenching. Anal. Bioanal. Chem. 2018, 410, 3209–3216. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Nasirian, V.; Mansouri, K.; Barati, A.; Veisi, A.; Kashanian, S. A highly sensitive quantum dots-DNA nanobiosensor based on fluorescence resonance energy transfer for rapid detection of nanomolar amounts of human papillomavirus 18. J. Pharm. Biomed. Anal. 2017, 136, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Joda, H.; Moutsiopoulou, A.; Stone, G.; Daunert, S.; Deo, S. Design of Gaussia luciferase-based bioluminescent stem-loop probe for sensitive detection of HIV-1 nucleic acids. Analyst 2018, 143, 3374–3381. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Hou, C.; Huo, D.; Bao, J.; Fa, H.; Shen, C. An electrochemical DNA biosensor based on nitrogen-doped graphene/Au nanoparticles for human multidrug resistance gene detection. Biosens. Bioelecton. 2016, 85, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Esfandiari, L.; Lorenzini, M.; Kocharyan, G.; Monbouquette, H.G.; Schmidt, J.J. Sequence-Specific DNA Detection at 10 FM by Electromechanical Signal Transduction. Anal. Chem. 2014, 86, 9638–9643. [Google Scholar] [CrossRef]
- Gooding, J.J.; Gaus, K. Single-Molecule Sensors: Challenges and Opportunities for Quantitative Analysis. Angew. Chem. Int. Ed. 2016, 55, 11354–11366. [Google Scholar] [CrossRef]
- Kaur, A.; Sapkota, K.; Dhakal, S. Multiplexed Nucleic Acid Sensing with Single-Molecule FRET. ACS Sens. 2019, 4, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Wesner, D.; Schönherr, H.; Nöll, G. Molecular Beacon Modified Sensor Chips for Oligonucleotide Detection with Optical Readout. Langmuir 2014, 30, 14360–14367. [Google Scholar] [CrossRef] [PubMed]
- Mahani, M.; Mousapour, Z.; Divsar, F.; Nomani, A.; Ju, H. A Carbon Dot and Molecular Beacon Based Fluorometric Sensor for the Cancer Marker MicroRNA-21. Microchim. Acta 2019, 186, 132. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kannegulla, A.; Wu, B.; Cheng, L.-J. Quantum Dot Fullerene-Based Molecular Beacon Nanosensors for Rapid, Highly Sensitive Nucleic Acid Detection. ACS Appl. Mater. Interfaces 2018, 10, 18524–18531. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.-L.; Chan, H.-M.; Ha, A.W.-Y.; Wong, R.N.-S.; Li, H.-W. Direct Quantification of Circulating MiRNAs in Different Stages of Nasopharyngeal Cancerous Serum Samples in Single Molecule Level with Total Internal Reflection Fluorescence Microscopy. Anal. Chem. 2014, 86, 9880–9886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Y.; Chao, S.-Y.; Wang, T.-H. Comparative Quantification of Nucleic Acids Using Single-Molecule Detection and Molecular Beacons. Analyst 2005, 130, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Mo, M.-H.; Chen, L.; Fu, Y.; Wang, W.; Fu, S.W. Cell-Free Circulating MiRNA Biomarkers in Cancer. J. Cancer 2012, 3, 432–448. [Google Scholar] [CrossRef]
- Bartels, C.L.; Tsongalis, G.J. MicroRNAs: Novel Biomarkers for Human Cancer. Clin. Chem. 2009, 55, 623–631. [Google Scholar] [CrossRef]
- Shin, V.Y.; Siu, J.M.; Cheuk, I.; Ng, E.K.O.; Kwong, A. Circulating Cell-Free MiRNAs as Biomarker for Triple-Negative Breast Cancer. Br. J. Cancer 2015, 112, 1751–1759. [Google Scholar] [CrossRef]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum MicroRNAs Are Promising Novel Biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Bond, D.T.; McKee, M.; Skog, J.; Păunescu, T.G.; Da Silva, N.; Brown, D.; Russo, L.M. Nucleic Acids within Urinary Exosomes/Microvesicles Are Potential Biomarkers for Renal Disease. Kidney Int. 2010, 78, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Kolpashchikov, D.M. Split DNA Enzyme for Visual Single Nucleotide Polymorphism Typing. J. Am. Chem. Soc. 2008, 130, 2934–2935. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Platella, C.; Riccardi, C.; Moccia, F.; Montesarchio, D. Fluorescence Sensing Using DNA Aptamers in Cancer Research and Clinical Diagnostics. Cancers 2017, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yao, D.; He, M.; Xiao, S.; Liang, H. Optimizing the Toehold Strategy of On-Chip Nucleic Acid Hybridization Probe for the Discrimination of Single Nucleotide Polymorphism. Langmuir 2018, 34, 14811–14816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Chen, S.X.; Yin, P. Optimizing the Specificity of Nucleic Acid Hybridization. Nat. Chem. 2012, 4, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Sato, Y.; Akaike, T.; Maruyama, A. Cationic Comb-Type Copolymers for DNA Analysis. Nat. Mater. 2003, 2, 815–820. [Google Scholar] [CrossRef]
- Huh, Y.S.; Lowe, A.J.; Strickland, A.D.; Batt, C.A.; Erickson, D. Surface-Enhanced Raman Scattering Based Ligase Detection Reaction. J. Am. Chem. Soc. 2009, 131, 2208–2213. [Google Scholar] [CrossRef]
- Wang, X.; Lou, X.; Wang, Y.; Guo, Q.; Fang, Z.; Zhong, X.; Mao, H.; Jin, Q.; Wu, L.; Zhao, H.; et al. QDs-DNA Nanosensor for the Detection of Hepatitis B Virus DNA and the Single-Base Mutants. Biosens. Bioelectron. 2010, 25, 1934–1940. [Google Scholar] [CrossRef]
- Howorka, S.; Cheley, S.; Bayley, H. Sequence-Specific Detection of Individual DNA Strands Using Engineered Nanopores. Nat. Biotechnol. 2001, 19, 636–639. [Google Scholar] [CrossRef]
- Kohli, P.; Harrell, C.C.; Cao, Z.; Gasparac, R.; Tan, W.; Martin, C.R. DNA-Functionalized Nanotube Membranes with Single-Base Mismatch Selectivity. Science 2004, 305, 984–986. [Google Scholar] [CrossRef] [PubMed]
- Sobrino, B.; Brión, M.; Carracedo, A. SNPs in Forensic Genetics: A Review on SNP Typing Methodologies. Forensic Sci. Int. 2005, 154, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.R.; Dhakal, S. Single-Molecule Imaging Reveals Conformational Manipulation of Holliday Junction DNA by the Junction Processing Protein RuvA. Biochemistry 2018, 57, 3616–3624. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Hohng, S.; Ha, T. A Practical Guide to Single-Molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Ha, T. Single-Molecule Fluorescence Resonance Energy Transfer. Methods 2001, 25, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.R.; Kaur, A.; Megalathan, A.; Sapkota, K.; Dhakal, S. Build Your Own Microscope: Step-By-Step Guide for Building a Prism-Based TIRF Microscope. Methods Protoc. 2018, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- Megalathan, A.; Cox, B.D.; Wilkerson, P.D.; Kaur, A.; Sapkota, K.; Reiner, J.E.; Dhakal, S. Single-Molecule Analysis of i-Motif within Self-Assembled DNA Duplexes and Nanocircles. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Aitken, C.E.; Marshall, R.A.; Puglisi, J.D. An Oxygen Scavenging System for Improvement of Dye Stability in Single-Molecule Fluorescence Experiments. Biophys. J. 2008, 94, 1826–1835. [Google Scholar] [CrossRef]
- Fu, J.; Yang, Y.R.; Dhakal, S.; Zhao, Z.; Liu, M.; Zhang, T.; Walter, N.G.; Yan, H. Assembly of Multienzyme Complexes on DNA Nanostructures. Nat. Protoc. 2016, 11, 2243–2273. [Google Scholar] [CrossRef]
- Wang, X.; Zou, M.; Huang, H.; Ren, Y.; Li, L.; Yang, X.; Li, N. Gold Nanoparticle Enhanced Fluorescence Anisotropy for the Assay of Single Nucleotide Polymorphisms (SNPs) Based on Toehold-Mediated Strand-Displacement Reaction. Biosens. Bioelectron. 2013, 41, 569–575. [Google Scholar] [CrossRef]
- Long, J.-B.; Liu, Y.-X.; Cao, Q.-F.; Guo, Q.-P.; Yan, S.-Y.; Meng, X.-X. Sensitive and Enzyme-Free Detection for Single Nucleotide Polymorphism Using Microbead-Assisted Toehold-Mediated Strand Displacement Reaction. Chin. Chem. Lett. 2015, 26, 1031–1035. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapkota, K.; Kaur, A.; Megalathan, A.; Donkoh-Moore, C.; Dhakal, S. Single-Step FRET-Based Detection of Femtomoles DNA. Sensors 2019, 19, 3495. https://doi.org/10.3390/s19163495
Sapkota K, Kaur A, Megalathan A, Donkoh-Moore C, Dhakal S. Single-Step FRET-Based Detection of Femtomoles DNA. Sensors. 2019; 19(16):3495. https://doi.org/10.3390/s19163495
Chicago/Turabian StyleSapkota, Kumar, Anisa Kaur, Anoja Megalathan, Caleb Donkoh-Moore, and Soma Dhakal. 2019. "Single-Step FRET-Based Detection of Femtomoles DNA" Sensors 19, no. 16: 3495. https://doi.org/10.3390/s19163495
APA StyleSapkota, K., Kaur, A., Megalathan, A., Donkoh-Moore, C., & Dhakal, S. (2019). Single-Step FRET-Based Detection of Femtomoles DNA. Sensors, 19(16), 3495. https://doi.org/10.3390/s19163495





