The Role of Proton Transport in Gating Current in a Voltage Gated Ion Channel, as Shown by Quantum Calculations
Abstract
1. Introduction
2. Evidence that Can Be Interpreted without Invoking S4 Motion
3. Temperature Dependence of Gating
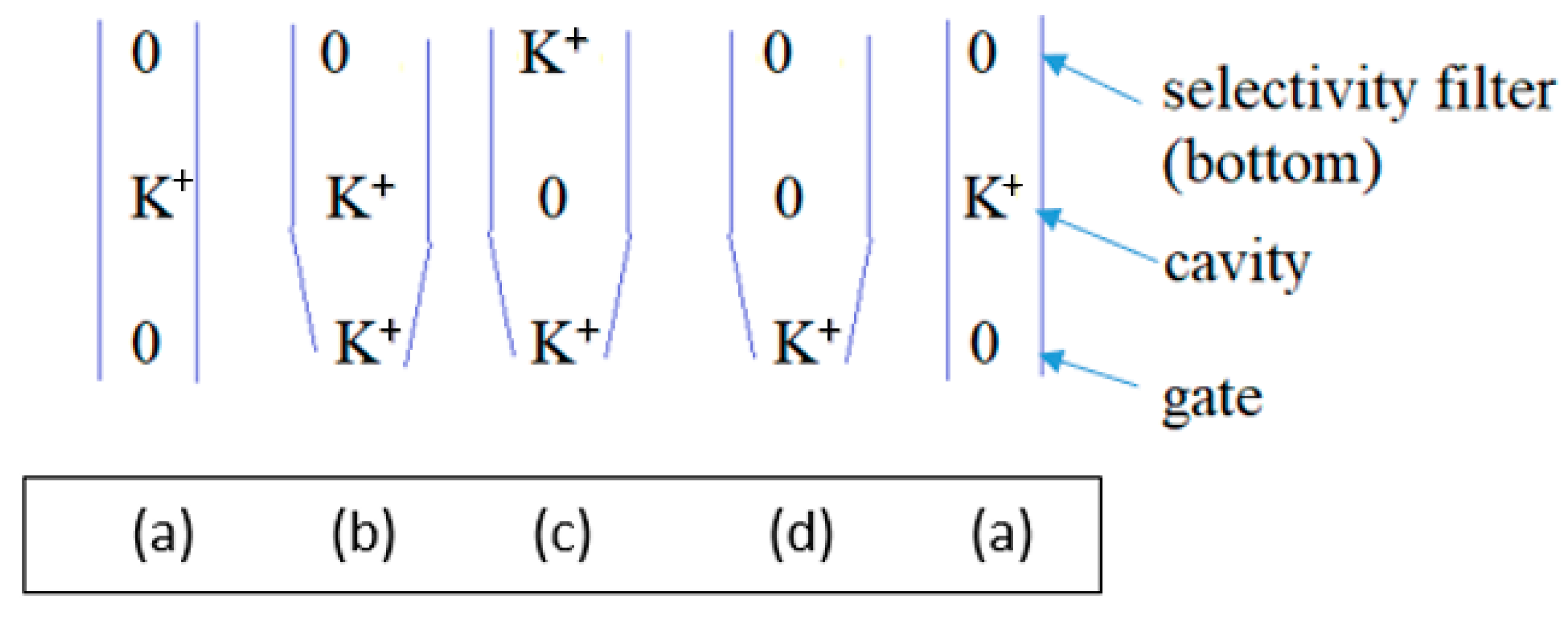
4. K+ Concentration Dependence of Conduction
5. Proton Delocalization and a Transition Threshold
6. Computational Techniques: General Considerations Relating to Choice of Method
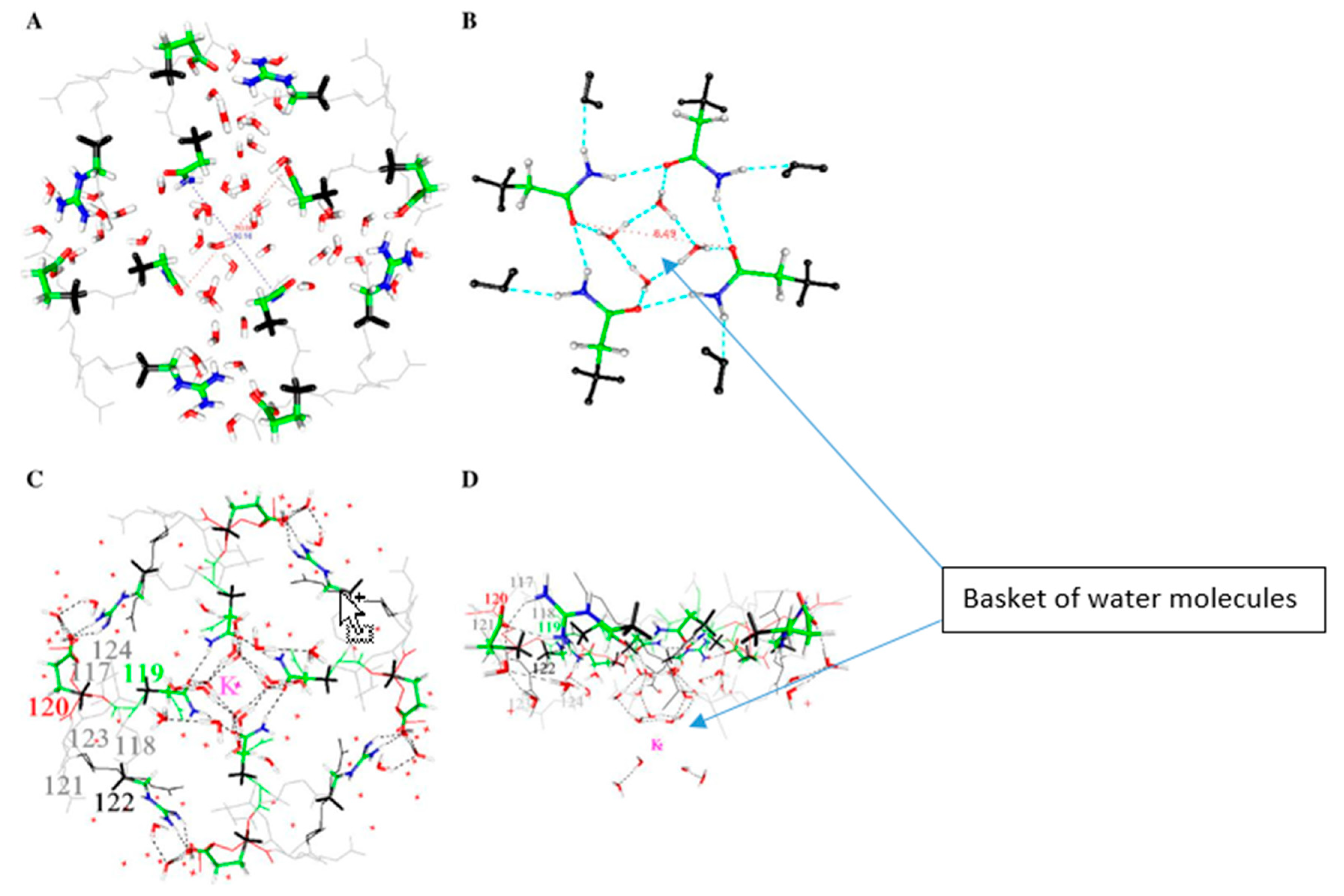
7. Some Interesting Things That Water Does
8. The Absence of a Direct Determination of Temperature Dependence in These Calculations
9. Pressure
10. Results of Recent Quantum Calculations on the VSD
10.1. Choice of System for Quantum Calculations
10.2. Results
11. Gating Model from the Computation
12. The Structure of the Gate
13. Summary on VSD Calculation
14. Specific Computational Methods
15. Summary
- (1)
- There is a standard model in which gating current is provided by the motion perpendicular to the membrane of the S4 TM segment of the VSD, supported principally by:
- (a)
- SCAM studies of this and related channels.
- (b)
- MD simulations.
- (c)
- FRET and LRET studies.
- (2)
- We have offered an alternative interpretation in which the gating current is provided by proton motion. This is supported by:
- (a)
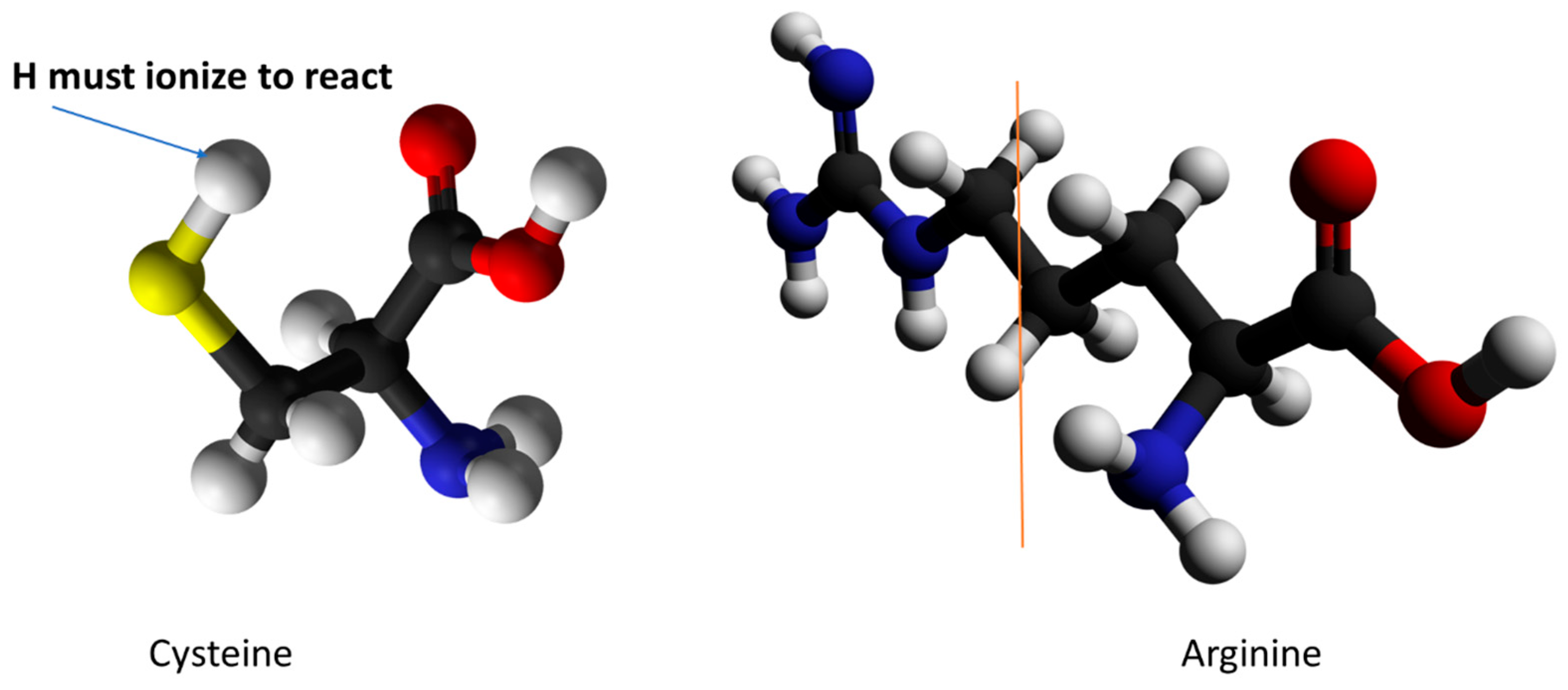
- Quantum calculations showing how a proton can make a local transfer in the Kv1.2 channel VSD; this depends in part on the fact that the side chain of arginine is amphoteric, unlike the side chains of all other amino acids. The amphoteric nature of the arginine side chain is unique among natural amino acids.
- (b)
- Extending these calculations to show that charge transfer is consistent with known gating charge.
- (c)
- Providing a hypothesis that shows much of the remainder of a path that a proton could follow from gate to the extracellular surface of the membrane.
- (d)
- Considering several experiments that are typically ignored or interpreted in contradictory fashion in order to fit them to the standard model, when more natural interpretations are consistent with proton transport.
- (e)
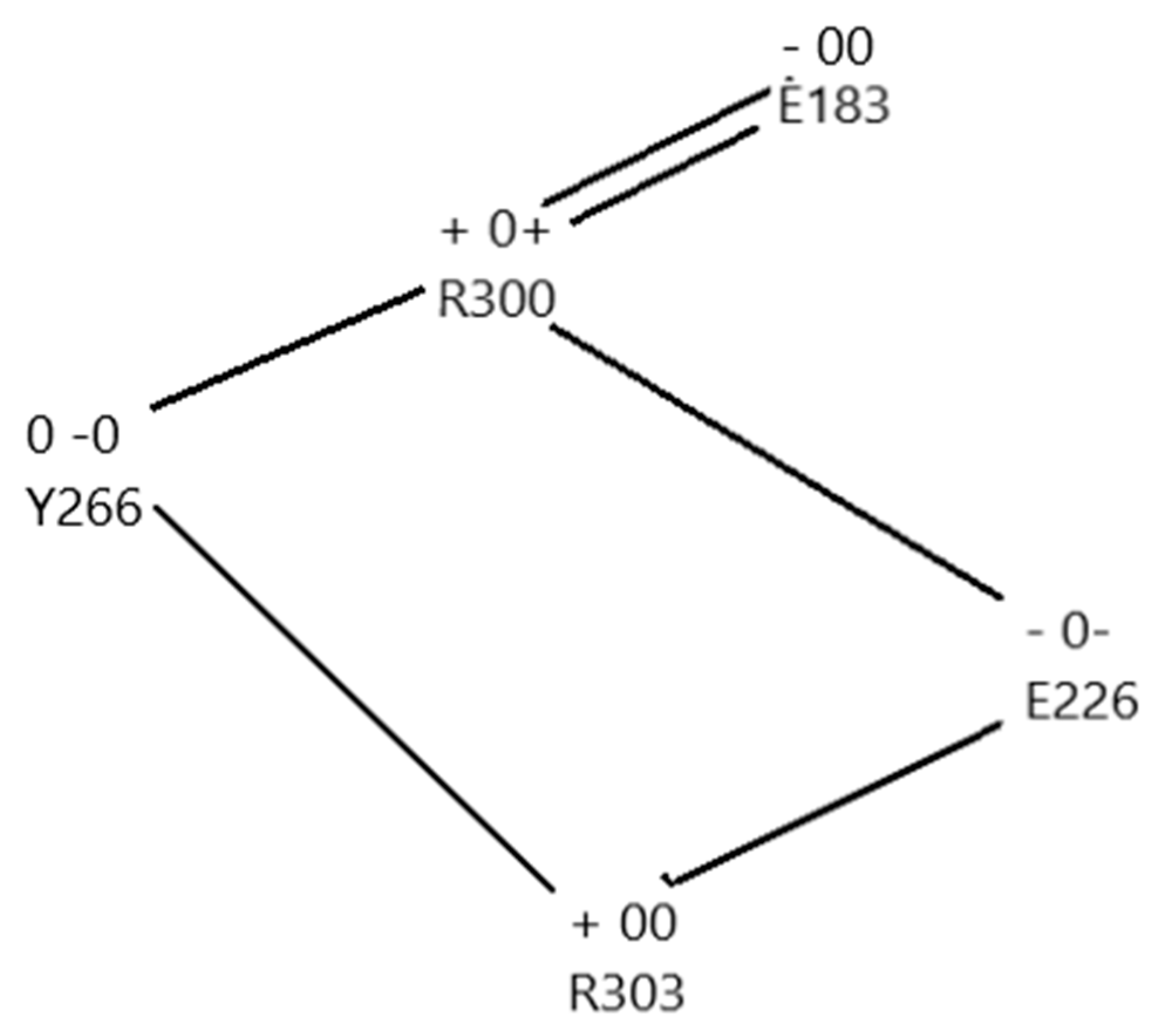
- Consideration of analogous systems, such as Hv1, cytochrome c, bacteriorhodopsin, and the flu M2 channel that are known to transmit protons. It is of particular interest that, except for the M2 channel, all of these, like the VSD of the Kv channel, have the same tyrosine-arginine-glutamate arrangement in the apparent proton path (or equivalent, for example with aspartate in place of glutamate), as well as other similarities (e.g., an RER/RDR triad) that would create a proton path. It would be of interest to investigate other proteins that transmit protons to determine whether this general form of triad is a recurring motif in proton transport in proteins.
- (f)
- The center of charge shifts very nearly just as much as one would expect; this result must have a fairly large error bar attached, but it is almost exactly what should be expected.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hodgkin, A.L.; Huxley, A.F. Currents carried by sodium and potassium ions through the giant membrane of Loligo. J. Physiol. 1952, 116, 449–472. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, A.L.; Huxley, A.F. The components of membrane conductance in the giant axon of Loligo. J. Physiol. 1952, 116, 473–496. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, A.L.; Huxley, A.F. The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. J. Physiol. 1952, 116, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, A.L.; Keynes, R.D. The potassium permeability of a giant nerve fibre. J. Physiol. 1955, 128, 61–88. [Google Scholar] [CrossRef] [PubMed]
- Doyle, D.A.; Cabral, J.M.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; MacKinnon, R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Long, S.B.; Campbell, E.B.; MacKinnon, R. Crystal structure of a mammalian voltage-dependent shaker family K+ channel. Science 2005, 309, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Long, S.B.; Campbell, E.B.; MacKinnon, R. Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science 2005, 309, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Ruta, V.; Chen, J.; Lee, A.; MacKinnon, R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature 2003, 423, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.O.; Borhani, D.W.; Lindorff-Larsen, K.; Maragakis, P.; Jogini, V.; Eastwood, M.P.; Dror, R.O.; Shaw, D.E. Principles of conduction and hydrophobic gating in K+ channels. Proc. Natl. Acad. Sci. USA 2010, 107, 5833–5838. [Google Scholar] [CrossRef] [PubMed]
- Ahern, C.A.; Horn, R. Focused electric field across the voltage sensor of potassium channels. Neuron 2005, 48, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Bezanilla, F. Voltage sensor movements. J. Gen. Physiol. 2002, 120, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. Ion channel voltage sensors: Structure, function, and pathophysiology. Neuron 2010, 67, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Delemotte, L.; Tarek, M.; Klein, M.L.; Amaral, C.; Treptow, W. Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2011, 108, 6109–6114. [Google Scholar] [CrossRef] [PubMed]
- Tronin, A.Y.; Nordgren, C.E.; Strzalka, J.W.; Kuzmenko, J.; Worcester, D.L.; Lauter, V.; Freites, J.A.; Tobias, D.J.; Blasie, J.K. Direct Evidence of Conformational Changes Associated with Voltage Gating in a Voltage Sensor Protein by Time-Resolved X-ray/Neutron Interferometry. Langmuir 2014, 30, 4784–4796. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, N.; Goradia, N.; Ohlenschlager, O.; Schonherr, R.; Friedrich, M.; Plass, W.; Kappl, R.; Hoshi, T.; Heinemann, S.H. Heme impairs the ball-and-chain inactivation of potassium channels. Proc. Natl. Acad. Sci. USA 2013, 110, E4036–E4044. [Google Scholar] [CrossRef] [PubMed]
- Hille, B. Ion Channels of Excitable Membranes, 3rd ed.; Sinauer Associates: Sunderland, MA, USA, 2001. [Google Scholar]
- Gazzarrini, S.; Van, E.J.L.; DiFrancesco, D.; Thiel, G.; Moroni, A. Voltage-dependence of virus-encoded miniature K+ channel Kcv. J. Membr. Biol. 2002, 187, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Islas, L.D. Functional diversity of potassium channel voltage-sensing domains. Channels 2016, 10, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Q.; Ni, F.; Ma, J. Structure of the full-length shaker potassium channel Kv1.2 by normal-mode-based x-ray crystallographic refinement. Proc. Natl. Acad. Sci. USA 2010, 107, 11352–11357. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.M.; Bezanilla, F. Charge movement associated with the opening and closing of of the activation gates of the Na channels. J. Gen. Physiol. 1974, 63, 533–552. [Google Scholar] [CrossRef] [PubMed]
- Bezanilla, F.; Villalba-Galea, C.A. The gating charge should not be estimated by fitting a two-state model to a Q-V curve. J. Gen. Physiol. 2013, 142, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Blunck, R. The isolated voltage sensing domain of the shaker potassium channel forms a voltage-gated cation channel. Elife 2016, 5, e18130. [Google Scholar] [CrossRef] [PubMed]
- Minor, D.L., Jr.; Lin, Y.-F.; Mobley, B.C.; Avelar, A.; Jan, Y.N.; Jan, L.Y.; Berger, J.M. The polar t1 interface is linked to conformational changes that open the voltage-gated potassium channel. Cell 2000, 102, 657–670. [Google Scholar] [CrossRef]
- Akabas, M.H.; Stauffer, D.A.; Xu, M.; Karlin, A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science 1992, 258, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Horn, R. Cysteine scanning. In Methods in Enzymology; Conn, P.M., Ed.; Academic Press: New York, NY, USA, 1998; pp. 145–155. [Google Scholar]
- Yang, N.; George, A.L., Jr.; Horn, R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron 1996, 16, 113–122. [Google Scholar] [CrossRef]
- Darby, N.J.; Creighton, T.E. Dissecting the disulfide-coupled folding pathway of bovine pancreatic trypsin inhibitor. Forming the first disulfide bonds in analogs of the reduced protein. J. Mol. Biol. 1993, 232, 873–896. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.P.; Horn, R. Movement and crevices around a sodium channel s3 segment. J. Gen. Physiol. 2002, 120, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, V.; Stack, K.; Boric, K.; Naranjo, D. Reduced voltage sensitivity in a K+-channel voltage sensor by electric field remodeling. Proc. Natl. Acad. Sci. USA 2010, 107, 5178–5183. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Durek, T.; Dang, B.; Finol-Urdaneta, R.K.; Craik, D.J.; Kent, S.B.H.; French, R.J.; Bezanilla, F.; Correa, A.M. Mapping of voltage sensor positions in resting and inactivated mammalian sodium channels by lret. Proc. Natl. Acad. Sci. USA 2017, 114, E1857–E1865. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Lacroix, J.J.; Bezanilla, F.; Correa, A.M. Probing α-310 transitions in a voltage-sensing s4 helix. Biophys. J. 2014, 107, 1117–1128. [Google Scholar] [CrossRef] [PubMed]
- Nanazashvili, M.; Sanchez-Rodriguez, J.E.; Fosque, B.; Bezanilla, F.; Sackin, H. Lret determination of molecular distances during ph gating of the mammalian inward rectifier kir1.1b. Biophys. J. 2018, 114, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Olejnik, J.; Brzezinski, B.; Zundel, G. A proton pathway with large proton polarizability and the proton pumping mechanism in bacteriorhodopsin-fourier transform difference spectra of photoproducts of bacteriorhodopsin and of its pentademethyl analog. J. Mol. Struct. 1992, 271, 157–173. [Google Scholar] [CrossRef]
- Blunck, R.; Starace, D.M.; Correa, A.M.; Bezanilla, F. Detecting rearrangements of shaker and nachbac in real-time with fluorescence spectroscopy in patch-clamped mammalian cells. Biophys. J. 2004, 86, 3966–3980. [Google Scholar] [CrossRef] [PubMed]
- Chanda, B.; Asamoah, O.K.; Blunck, R.; Roux, B.; Bezanilla, F. Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature 2005, 436, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Sandtner, W.; Bezanilla, F.; Correa, A.M. In Vivo measurement of intramolecular distances using genetically encoded reporters. Biophys. J. 2007, 93, L45–L47. [Google Scholar] [CrossRef] [PubMed]
- Villalba-Galea, C.A.; Sandtner, W.; Starace, D.M.; Bezanilla, F. S4-based voltage sensors have three major conformation. Proc. Natl. Acad. Sci. USA 2008, 105, 17600–17607. [Google Scholar] [CrossRef] [PubMed]
- Posson, D.J.; Ge, P.; Miller, C.; Bezanilla, F.; Selvin, P.R. Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature 2005, 436, 848–851. [Google Scholar] [CrossRef] [PubMed]
- Asamoah, O.K.; Wuskell, J.P.; Loew, L.M.; Bezanilla, F. A fluorometric approach to local electric field measurements in a voltage-gated ion channel. Neuron 2003, 37, 85–97. [Google Scholar] [CrossRef]
- Cha, A.; Snyder, G.; Selvin, P.R.; Bezanilla, F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 1999, 402, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Ishida, I.G.; Rangel-Yescas, G.E.; Carrasco-Zanini, J.; Islas, L.D. Voltage-dependent gating and gating charge measurements in the Kv1.2 potassium channel. J. Gen. Physiol. 2015, 145, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.-N.; Gong, H. Simulating the activation of voltage sensing domain for a voltage gated sodium channel using polarizable force field. J. Phys. Chem. Lett. 2017, 8, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Kariev, A.M.; Green, M.E. Voltage gated ion channel function: Gating, conduction, and the role of water and protons. Int. J. Mol. Sci. 2012, 13, 1680–1709. [Google Scholar] [CrossRef] [PubMed]
- Kariev, A.M.; Green, M.E. Caution is required in interpretation of mutations in the voltage sensing domain of voltage gated channels as evidence for gating mechanisms. Int. J. Mol. Sci. 2015, 16, 1627–1643. [Google Scholar] [CrossRef] [PubMed]
- Kariev, A.M.; Green, M.E. Quantum calculations of a large section of the voltage sensing domain of the Kv1.2 channel show that proton transfer, not S4 motion, provides the gating current. bioRxiv 2017. [Google Scholar] [CrossRef]
- Lu, J.; Yin, J.; Green, M.E. A model for ion channel voltage gating with static S4 segments. Ferroelectrics 1999, 220, 249–271. [Google Scholar] [CrossRef]
- Sapronova, A.; Bystrov, V.S.; Green, M.E. Water, proton transfer, and hydrogen bonding in ion channel gating. Front. Biosci. 2003, 8, s1356–s1370. [Google Scholar] [PubMed]
- Sapronova, A.V.; Bystrov, V.S.; Green, M.E. Ion channel gating and proton transport. J. Mol. Struct. 2003, 630, 297–307. [Google Scholar] [CrossRef]
- Decoursey, T.E. Voltage-gated proton channels. Compr. Physiol. 2012, 2, 1355–1385. [Google Scholar] [PubMed]
- DeCoursey, T.E. Voltage-gated proton channels: Molecular biology, physiology, and pathophysiology of the hv family. Physiol. Rev. 2013, 93, 599–652. [Google Scholar] [CrossRef] [PubMed]
- DeCoursey, T.E. The voltage-gated proton channel: A riddle, wrapped in a mystery, inside an enigma. Biochemistry 2015, 54, 3250–3268. [Google Scholar] [CrossRef] [PubMed]
- Okamura, Y. Biodiversity of voltage sensor domain proteins. Pflug. Arch.-Eur. J. Physiol. 2007, 454, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Takagi, M.; Okamura, Y. A voltage-sensor-domain protein is a voltage-gated proton channel. Science 2006, 312, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, I.S.; Moran, M.M.; Chong, J.A.; Clapham, D.E. A voltage-gated proton-selective channel lacking the pore domain. Nature 2006, 440, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Mony, L.; Berger, T.K.; Isacoff, E.Y. A specialized molecular motion opens the hv1 voltage-gated proton channel. Nat. Struct. Mol. Biol. 2015, 22, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Goldschen-Ohm, M.P.; Chanda, B. How to open a proton pore-more than S4? Nat. Struct. Mol. Biol. 2015, 22, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Starace, D.; Stefani, E.; Bezanilla, F. Histidine scanning mutagenesis indicates full translocation of two charges of the shaker k channel voltage sensor. Biophys. J. 1998, 74, A215. [Google Scholar]
- Starace, D.M.; Bezanilla, F. Histidine scanning mutagenesis of uncharged residues of the shaker K+ channel S4 segment. Biophys. J. 2001, 80, 217A. [Google Scholar]
- Starace, D.M.; Bezanilla, F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature 2004, 427, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Starace, D.M.; Bezanilla, F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J. Gen. Physiol. 2001, 117, 469–490. [Google Scholar] [CrossRef] [PubMed]
- Villalba-Galea, C.A.; Frezza, L.; Sandtner, W.; Bezanilla, F. Sensing charges of the ciona intestinalis voltage-sensing phosphatase. J. Gen. Physiol. 2013, 142, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Sakata, S.; Jinno, Y.; Kawanabe, A.; Okamura, Y. Voltage-dependent motion of the catalytic region of voltage-sensing phosphatase monitored by a fluorescent amino acid. Proc. Natl. Acad. Sci. USA 2016, 113, 7521–7526. [Google Scholar] [CrossRef] [PubMed]
- Perozo, E.; Cuello, L.G.; Cortes, D.M.; Liu, Y.S.; Sompornpisut, P. Epr approaches to ion channel structure and function. Novartis Found. Symp. 2002, 245, 146–164. [Google Scholar] [PubMed]
- Perozo, E. New structural perspectives on K+ channel gating. Structure 2002, 10, 1027–1029. [Google Scholar] [CrossRef]
- Basak, S.; Chatterjee, S.; Chakrapani, S. Site directed spin labeling and epr spectroscopic studies of pentameric ligand-gated ion channels. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed]
- Chakrapani, S. Epr studies of gating mechanisms in ion channels. Methods Enzymol. 2015, 557, 279–306. [Google Scholar] [PubMed]
- Li, Q.; Wanderling, S.; Paduch, M.; Medovoy, D.; Singharoy, A.; McGreevy, R.; Villalba-Galea, C.A.; Hulse, R.E.; Roux, B.; Schulten, K.; et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 2014, 21, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Wikstrom, M.; Hummer, G. Kinetic gating of the proton pump in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2009, 106, 13707–13712. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Voth, G.A. Expanding the view of proton pumping in cytochrome c oxidase through computer simulation. Biochim. Biophys. Acta Bioenergy 2012, 1817, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Woelke, A.L.; Wagner, A.; Galstyan, G.; Meyer, T.; Knapp, E.-W. Proton transfer in the k-channel analog of b-type cytochrome c oxidase from thermus thermophilus. Biophys. J. 2014, 107, 2177–2184. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Gunner, M.R. Unraveling the mechanism of proton translocation in the extracellular half-channel of bacteriorhodopsin. Proteins Struct. Funct. Bioinf. 2016, 84, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Koyama, K. Proton transfer mechanism of retinal proteins studied by a photoelectrochemical method. Seibutsu Butsuri 2000, 40, 385–390. [Google Scholar] [CrossRef]
- Schobert, B.; Brown, L.S.; Lanyi, J.K. Crystallographic structures of the M and N intermediates of bacteriorhodopsin: Assembly of a hydrogen-bonded chain of water molecules between Asp-96 and the retinal schiff base. J. Mol. Biol. 2003, 330, 553–570. [Google Scholar] [CrossRef]
- Gianti, E.; Carnevale, V.; DeGrado, W.F.; Klein, M.L.; Fiorin, G. Hydrogen-bonded water molecules in the M2 channel of the influenza a virus guide the binding preferences of ammonium-based inhibitors. J. Phys. Chem. B 2015, 119, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fu, R.; Nishimura, K.; Zhang, L.; Zhou, H.-X.; Busath, D.D.; Vijayvergiya, V.; Cross, T.A. Histidines, heart of the hydrogen ion channel from influenza a virus: Toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. USA 2006, 103, 6865–6870. [Google Scholar] [CrossRef] [PubMed]
- Ruta, V.; Chen, J.; MacKinnon, R. Calibrated measurement of gating-charge arginine displacement in the kvap voltage-dependent K+ channel. Cell 2005, 123, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Ruta, V.; Jiang, Y.; Lee, A.; Chen, J.; MacKinnon, R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature 2003, 422, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Stefani, E.; Bezanilla, F. Voltage dependence of the early events in voltage gating. Biophys. J. 1997, 72, A131. [Google Scholar]
- Stefani, E.; Sigg, D.; Bezanilla, F. Correlation between the early component of gating current and total gating current in shaker k channels. Biophys. J. 2000, 78, 7A. [Google Scholar]
- Sigg, D.; Bezanilla, F.; Stefani, E. Fast gating in the shaker K+ channel and the energy landscape of activation. Proc. Natl. Acad. Sci. USA 2003, 100, 7611–7615. [Google Scholar] [CrossRef] [PubMed]
- Fatade, A.; Snowhite, J.; Green, M.E. A resonance model gives the response to membrane potential for an ion channel: II. Simplification of the calculation, and prediction of stochastic resonance. J. Theor. Biol. 2000, 206, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Green, M.E. A resonance model gives the response to membrane potential for an ion channel. J. Theor. Biol. 1998, 193, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Fohlmeister, J.F.; Adelman, J.W.J. Gating current harmonics II: Model simulations of axonal gating currents. Biophys. J. 1985, 48, 391–400. [Google Scholar] [CrossRef]
- Fohlmeister, J.F.; Adelman, W.J., Jr. Gating current harmonics. I. Sodium channel activation gating in dynamic steady states. Biophys. J. 1985, 48, 375–390. [Google Scholar] [CrossRef]
- Schmidt, D.; Jiang, Q.-X.; MacKinnon, R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature 2006, 444, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; MacKinnon, R. Voltage-dependent K+ channel gating and voltage sensor toxin sensitivity depend on the mechanical state of the lipid membrane. Proc. Natl. Acad. Sci. USA 2008, 105, 19276–19281. [Google Scholar] [CrossRef] [PubMed]
- Suh, B.-C.; Hille, B. PIP2 is a necessary cofactor for ion channel function: How and why? Annu. Rev. Biophys. 2008, 37, 175–195. [Google Scholar] [CrossRef] [PubMed]
- Falkenburger, B.H.; Jensen, J.B.; Dickson, E.J.; Suh, B.-C.; Hille, B. Phosphoinositides: Lipid regulators of membrane proteins. J. Physiol. 2010, 588, 3179–3185. [Google Scholar] [CrossRef] [PubMed]
- Falkenburger, B.H.; Jensen, J.B.; Hille, B. Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 2010, 135, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Green, M.E. A possible role for phosphate in complexing the arginines of S4 in voltage gated channels. J. Theor. Biol. 2005, 233, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Green, M.E. Consequences of phosphate-arginine complexes in voltage gated ion channels. Channels 2008, 2, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, P.; Ghose, R.; Green, M.E. Voltage gating and anions, especially phosphate: A model system. Biochem. Biophys. Acta 2005, 1717, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Freites, J.A.; Tobias, D.J.; von Heijne, G.; White, S.H. Interface connections of a transmembrane voltage sensor. Proc. Natl. Acad. Sci. USA 2005, 102, 15059–15064. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, J.K.; Sikdar, S.K. Temperature-dependent conformational changes in a voltage-gated potassium channel. Eur. Biophys. J. 1999, 28, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Kuno, M.; Ando, H.; Morihata, H.; Sakai, H.; Mori, H.; Sawada, M.; Oiki, S. Temperature dependence of proton permeation through a voltage-gated proton channel. J. Gen. Physiol. 2009, 134, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.M.; Sigg, D.; Bezanilla, F. Voltage gating of shaker K+ channels. The effect of temperature on ionic and gating currents. J. Gen. Physiol. 1998, 112, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Chanda, B. Free-energy relationships in ion channels activated by voltage and ligand. J. Gen. Physiol. 2013, 141, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Raddatz, N.; Castillo, J.P.; Gonzalez, C.; Alvarez, O.; Latorre, R. Temperature and voltage coupling to channel opening in transient receptor potential melastatin 8 (trpm8). J. Biol. Chem. 2014, 289, 35438–35454. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.P.; Zaccai, N.R.; Fleming, P.J.; Gessmann, D.; Fleming, K.G. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc. Natl. Acad. Sci. USA 2013, 110, 4285–4290. [Google Scholar] [CrossRef] [PubMed]
- LeMasurier, M.; Heginbotham, L.; Miller, C. Kcsa: It’s a potassium channel. J. Gen. Physiol. 2001, 118, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Kariev, A.M.; Green, M.E. Quantum effects in a simple ring with hydrogen bonds. J. Phys. Chem. B 2015, 119, 5962–5969. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Fiorin, G.; DeGrado, W.F.; Klein, M.L. Proton release from the histidine-tetrad in the M2 channel of the influenza a virus. J. Phys. Chem. B 2014, 118, 12644–12651. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Green, M.E. Intermolecular proton transfer between two methylamine molecules with an external electric field in the gas phase. J. Phys. Chem. A 1998, 102, 7181–7190. [Google Scholar] [CrossRef]
- Eisenberg, B. Ionic channels in biological membranes: Electrostatic analysis of a natural nanotube. arXiv, 2016; arXiv:1610.04123. [Google Scholar] [CrossRef]
- Kaufman, I.K.; McClintock, P.V.E.; Eisenberg, R.S. Coulomb blockade model of permeation and selectivity in biological ion channels. New J. Phys. 2015, 17, 083021. [Google Scholar] [CrossRef]
- Liu, J.-L.; Eisenberg, B. Numerical methods for a poisson-nernst-planck-fermi model. arXiv, 2015; arXiv:1506.05953. [Google Scholar]
- Luchinsky, D.G.; Gibby, W.A.T.; Kaufman, I.; Timucin, D.A.; Eisenberg, R.S.; McClintock, P.V.E. Statistical theory of selectivity and conductivity in biological channels. arXiv, 2016; arXiv:1604.05758. [Google Scholar]
- Ahmadi, S.; Barrios Herrera, L.; Chehelamirani, M.; Hostas, J.; Jalife, S.; Salahub, D.R. Multiscale modeling of enzymes: QM-cluster, QM/MM, and QM/MM/MD: A tutorial review. Int. J. Quantum Chem. 2018, 118, e25558. [Google Scholar] [CrossRef]
- Delemotte, L.; Klein, M.L.; Tarek, M. Molecular dynamics simulations of voltage-gated cation channels: Insights on voltage-sensor domain function and modulation. Front. Pharmacol. Ion Channels Channelopathies 2012, 3, 97. [Google Scholar] [CrossRef] [PubMed]
- Deyawe, A.; Kasimova, M.A.; Delemotte, L.; Tarek, M.; Loussouarn, G.; Tarek, M. Studying Kv channels function using computational methods. Methods Mol. Biol. 2018, 1684, 321–341. [Google Scholar] [PubMed]
- Jensen, M.O.; Jogini, V.; Borhani, D.W.; Leffler, A.E.; Dror, R.O.; Shaw, D.E. Mechanism of voltage gating in potassium channels. Science 2012, 336, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Fowler, P.W.; Sansom, M.S.P. The pore of voltage-gated potassium ion channels is strained when closed. Nat. Commun. 2013, 4, 1872. [Google Scholar] [CrossRef] [PubMed]
- Labro, A.J.; Raes, A.L.; Grottesi, A.; Van Hoorick, D.; Sansom, M.S.P.; Snyders, D.J. Kv channel gating requires a compatible S4–S5 linker and bottom part of S6, constrained by non-interacting residues. J. Gen. Physiol. 2008, 132, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Mokrab, Y.; Sansom, M.S.P. Interaction of diverse voltage sensor homologs with lipid bilayers revealed by self-assembly simulations. Biophys. J. 2011, 100, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ngo, V.; Da Silva, M.C.; Salahub, D.R.; Callahan, K.; Roux, B.; Noskov, S.Y. Representation of ion-protein interactions using the drude polarizable force-field. J. Phys. Chem. B 2015, 119, 9401–9415. [Google Scholar] [CrossRef] [PubMed]
- Kariev, A.M.; Green, M.E. Quantum calculations on the Kv1.2 channel voltage sensing domain show H+ transfer provides the gating current. arXiv, 2017; arXiv:1712.02866. [Google Scholar]
- Svensson, M.; Humbel, S.; Froese, R.D.J.; Matsubara, T.; Sieber, S.; Morokuma, K. Oniom: A multilayered integrated MO + MM method for geometry optimizations and single point energy predictions. A test for diels-alder reactions and Pt(P(t-Bu)3)2 + H2 oxidative addition. J. Phys. Chem. 1996, 100, 19357–19363. [Google Scholar] [CrossRef]
- Dudev, T.; Grauffel, C.; Lim, C. Influence of the selectivity filter properties on proton selectivity in the influenza a M2 channel. J. Am. Chem. Soc. 2016, 138, 13038–13047. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Mazmanian, K.; Lim, C. Factors controlling the selectivity for Na+ over Mg2+ in sodium transporters and enzymes. Phys. Chem. Chem. Phys. 2016, 18, 16986–16997. [Google Scholar] [CrossRef] [PubMed]
- Varma, S.; Rempe, S.B. Quantum-Chemical Investigations of Ion Selectivity Mechanisms by K-Channels and Valinomycin; American Chemical Society: Washington, DC, USA, 2007; p. 102. [Google Scholar]
- Varma, S.; Sabo, D.; Rempe, S.B. K+/Na+ selectivity in k channels and valinomycin: Over-coordination versus cavity-size constraints. J. Mol. Biol. 2008, 376, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Green, M.E. Quantum calculations on salt bridges with water: Potentials, structure, and properties. Comput. Theor. Chem. 2011, 963, 207–214. [Google Scholar] [CrossRef]
- Sauguet, L.; Poitevin, F.; Murail, S.; Van, R.C.; Moraga-Cid, G.; Malherbe, L.; Thompson, A.W.; Koehl, P.; Corringer, P.-J.; Baaden, M.; et al. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 2013, 32, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Kariev, A.M.; Znamenskiy, V.S.; Green, M.E. Quantum mechanical calculations of charge effects on gating the KcsA channel. Biochem. Biophys. Acta 2007, 1768, 1218–1229. [Google Scholar] [CrossRef] [PubMed]
- Schauf, C.L.; Bullock, J.O. Modifications of sodium channel gating in myxicola giant axons by deuterium oxide, temperature, and internal cations. Biophys. J. 1979, 27, 193–208. [Google Scholar] [CrossRef]
- Schauf, C.L.; Bullock, J.O. Solvent substitution as a probe of channel gating in myxicola: Differential effects of D2O on some components of membrane conductance. Biophys. J. 1980, 30, 295–306. [Google Scholar] [CrossRef]
- Schauf, C.L.; Bullock, J.O. Solvent substitution as a probe of channel gating in myxicola. Biophys. J. 1982, 37, 441–452. [Google Scholar] [CrossRef]
- DeCoursey, T.E.; Cherny, V.V. Deuterium isotope effects on permeation and gating of proton channels in rat alveolar epithelium. J. Gen. Physiol. 1997, 109, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Alicata, D.A.; Rayner, M.A.; Starkus, J.A. Sodium channel activation mechanisms: Insights from deuterium oxide substitution. Biophys. J. 1990, 57, 745–758. [Google Scholar] [CrossRef]
- Green, M.E. Electrorheological effects and gating of membrane channels. J.Theor. Biol. 1989, 138, 413–428. [Google Scholar] [CrossRef]
- Heinemann, S.H.; Conti, F.; Stuehmer, W.; Neher, E. Effects of hydrostatic pressure on membrane processes. Sodium channels, calcium channels, and exocytosis. J. Gen. Physiol. 1987, 90, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Zimmerberg, J.; Bezanilla, F.; Parsegian, V.A. Solute inaccessible aqueous volume changes during opening of the potassium channel of the squid giant axon. Biophys. J. 1990, 57, 1049–1064. [Google Scholar] [CrossRef]
- Rayner, M.D.; Starkus, J.G.; Ruben, P.C.; Alicata, D.A. Voltage-sensitive and solvent-sensitive processes in ion channel gating. Kinetic effects of hyperosmolar media on activation and deactivation of sodium channels. Biophys. J. 1992, 61, 96–108. [Google Scholar] [CrossRef]
- Bartok, A.; Feher, K.; Bodor, A.; Rakosi, K.; Toth, G.K.; Kover, K.E.; Panyi, G.; Varga, Z. An engineered scorpion toxin analogue with improved Kv1.3 selectivity displays reduced conformational flexibility. Sci. Rep. 2015, 5, 18397. [Google Scholar] [CrossRef] [PubMed]
- Li-Smerin, Y.; Swartz, K.J. Gating modifier toxins reveal a conserved structural motif in voltage-gated Ca2+ and K+ channels. Proc. Natl. Acad. Sci. USA 1998, 95, 8585–8589. [Google Scholar] [CrossRef] [PubMed]
- Olamendi-Portugal, T.; Csoti, A.; Jimenez-Vargas, J.M.; Gomez-Lagunas, F.; Panyi, G.; Possani, L.D. Pi5 and Pi6, two undescribed peptides from the venom of the scorpion pandinus imperator and their effects on K+-channels. Toxicon 2017, 133, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Shem-Ad, T.; Yifrach, O. Using hierarchical thermodynamic linkage analysis to study ion channel gating. J. Gen. Physiol. 2013, 141, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Wyman, J.J. Linked functions and reciprocal effects in hemoglobin: A second look. Adv. Protein Chem. 1964, 19, 223–286. [Google Scholar] [PubMed]
- Sadovsky, E.; Yifrach, O. Principles underlying energetic coupling along an allosteric communiction trajectory of a voltage-activeated K+ channel. Proc. Natl. Acad. Sci. USA 2007, 104, 19813–19818. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Chanda, B. Thermodynamics of electromechanical coupling in voltage-gated ion channels. J. Gen. Physiol. 2012, 140, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Chanda, B. Estimating the voltage-dependent free energy change of ion channels using the median voltage for activation. J. Gen. Physiol. 2012, 139, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.-L.; Qi, Z.; Zhang, X.-E.; Bi, L.-J.; Jin, G. Regulatory role of the extreme C-terminal end of the S6 inner helix in C-terminal-truncated Kv1.2 channel activation. Cell Biol. Int. 2010, 34, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Cushman, S.J.; Nanao, M.H.; Jahng, A.W.; DeRubeis, D.; Choe, S.; Pfaffinger, P.J. Voltage dependent activation of potassium channels is coupled to T1 domain structure. Nat. Struct. Biol. 2000, 7, 403–407. [Google Scholar] [PubMed]
- Oliver, A.E.; Deamer, D.W. Α-helical hydrophobic polypeptides form proton-selective channels in lipid bilayers. Biophys. J. 1994, 66, 1364–1379. [Google Scholar] [CrossRef]
- Wang, G.; Covarrubias, M. Voltage-dependent gating rearrangements in the intracellular T1-T1 interface of a K+ channel. J. Gen. Physiol. 2006, 127, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Faure, E.; Starek, G.; McGuire, H.; Berneche, S.; Blunck, R. A limited 4 radial displacement of the S4–S5 linker is sufficient for internal gate closing in Kv channels. J. Biol. Chem. 2012, 287, 40091–40098. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kariev, A.M.; Green, M.E. The Role of Proton Transport in Gating Current in a Voltage Gated Ion Channel, as Shown by Quantum Calculations. Sensors 2018, 18, 3143. https://doi.org/10.3390/s18093143
Kariev AM, Green ME. The Role of Proton Transport in Gating Current in a Voltage Gated Ion Channel, as Shown by Quantum Calculations. Sensors. 2018; 18(9):3143. https://doi.org/10.3390/s18093143
Chicago/Turabian StyleKariev, Alisher M., and Michael E. Green. 2018. "The Role of Proton Transport in Gating Current in a Voltage Gated Ion Channel, as Shown by Quantum Calculations" Sensors 18, no. 9: 3143. https://doi.org/10.3390/s18093143
APA StyleKariev, A. M., & Green, M. E. (2018). The Role of Proton Transport in Gating Current in a Voltage Gated Ion Channel, as Shown by Quantum Calculations. Sensors, 18(9), 3143. https://doi.org/10.3390/s18093143




