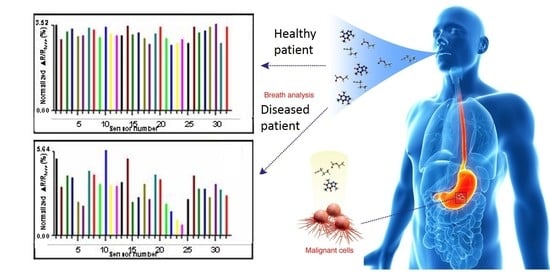
Application of Electronic-Nose Technologies and VOC-Biomarkers for the Noninvasive Early Diagnosis of Gastrointestinal Diseases †
Abstract
1. Introduction
2. Gastrointestinal Disease-Detection Methods
2.1. Biomarker Metabolites
2.2. New Metabolomic Disease-Detection Approaches
2.3. Biomarkers of Microbial GI-Tract Diseases
3. GI-Disease Types and E-Nose Methods for Detection
3.1. Clinical Sample Types for GI E-Nose Analyses
3.2. Importance of QA/QC in E-Nose Disease Detections
3.3. Electronic-Nose Instruments for GI-Disease Detections
3.4. Recent E-Nose GI-Disease Detection Applications
3.5. Combining E-Nose Analyses with Disease Biomarker Data
4. Future E-Nose Developments for GI-Disease Diagnostics
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| ALS | Amyotrophic Lateral Sclerosis |
| ASRDs | Application-Specific Reference Databases |
| BAD | Bile Acid Diarrhea |
| CBPC | Carbon Black Polymer Composite |
| CC | Colon Cancer |
| CD | Celiac Disease |
| CE | Capillary Electrophoresis |
| CGD | Chronic Gastrointestinal Disease |
| CNF | Carbon Nanofiber |
| CP | Conducting Polymer |
| CRC | Colorectal Cancer |
| CRD | Crohn’s Disease |
| EAD | Electronic Aroma Detection |
| EC | Electrochemical |
| ED | Ethyl Dodecanoate (biomarker) |
| E-nose | Electronic-nose |
| EM | Endometriosis |
| FAIMS | Field Asymmetric Ion Mobility Spectroscopy |
| FID | Flame Ionization Detector |
| GC-MS | Gas Chromatography-Mass Spectrometry |
| GC-TOF-MS | Gas Chromatography-Time-Of-Flight-Mass Spectrometry |
| GC/O | GC-Olfactometry |
| GI | Gastrointestinal |
| GIRMs | Gastrointestinal Resident Microbes |
| GNP | Gold Nanoparticle |
| HIA | Hydroxylamine-based Indole Assay |
| HNC | Head and Neck Cancer |
| HS-SPME | Headspace Solid Phase Microextraction |
| ID | Infectious Diarrhea |
| IBD | Inflammatory Bowel Disease |
| IBS | Irritable Bowel Syndrome |
| IMR-MS | Ion Molecule Reaction-Mass Spectrometry |
| IR | Infrared |
| LDA | Linear Discriminant Analysis |
| LD-PCA | Linear Discriminant Principle Component Analyses |
| LOS | Late-Onset Sepsis |
| MIB | 2-methylisoborneol (VOC-metabolite) |
| MOS | Metal Oxide Semiconductors |
| MS | Mass Spectrometry |
| NAFLD | Nonalcoholic Fatty Liver Disease |
| NDIR | Non-Dispersive Infra-red (optical devices) |
| NEC | Necrotizing Enterocolitis |
| NMR | Nuclear Magnetic Resonance |
| NVOMs | Non-Volatile Organic Metabolites |
| PCA | Principal Component Analysis |
| PCR | Polymerase Chain Reaction |
| PID | Photo-Ionization Detector |
| POCT | Point-Of-Care Testing |
| PTR-MS | Proton Transfer Reaction Mass Spectrometry |
| QA/QC | Quality Assurance/Quality Control |
| QCM | Quartz Crystal Microbalance |
| RCC | Renal Cell Carcinoma |
| SAW | Surface Acoustic Wave |
| SESI-MS | Secondary Electrospray Ionization-Mass Spectrometry |
| SIFT-MS | Selected Ion Flow Tube-Mass Spectrometry |
| SPME | Solid Phase Microextraction |
| TB | Tuberculosis |
| TCA | Tricarboxylic Acid (pathway) |
| UC | Ulcerative Colitis |
| UPLC-MS | Ultra-Performance Liquid Chromatography-Mass Spectrometry |
| VE | Viral Enteritis |
| VOCs | Volatile Organic Compounds |
| VOMs | Volatile Organic Metabolites |
References
- Wilson, A.D. Electronic-nose devices—Potential for noninvasive early disease-detection applications. Ann. Clin. Case Rep. 2017, 2, 1401. [Google Scholar]
- Westenbrink, E.; Arasaradnam, R.P.; O’Connell, N.O.; Bailey, C.; Nwokolo, C.; Bardhan, K.D.; Covington, J.A. Development and application of a new electronic nose instrument for the detection of colorectal cancer. Biosens. Bioelectron. 2015, 67, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Biomarker metabolite signatures pave the way for electronic-nose applications in early clinical disease diagnoses. Curr. Metabolom. 2017, 5, 90–101. [Google Scholar] [CrossRef]
- Arasaradnam, R.P.; Covington, J.A.; Harmston, C.; Nwokolo, C.U. Review article: Next generation diagnostic modalities in gastroenterology—Gas phase volatile compound biomarker detection. Aliment. Pharmacol. Ther. 2014, 39, 780–789. [Google Scholar] [CrossRef] [PubMed]
- De Meij, T.G.; Larbi, I.B.; van der Schee, M.P.; Lentferink, Y.E.; Paff, T.; sive Droste, J.S.T.; Mulder, C.J.; van Bodegraven, A.A.; de Boer, N.K. Electronic nose can discriminate colorectal carcinoma and advanced adenomas by fecal volatile biomarker analysis: Proof of principle study. Int. J. Cancer 2014, 134, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- De Groot, E.F.; de Meij, TG.; Berkhout, D.J.; van der Schee, M.P.; de Boer, N.K. Flatography: Detection of gastrointestinal diseases by faecal gas analysis. World J. Gastrointest. Pharmacol. Ther. 2015, 6, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Sagar, N.M.; Cree, I.A.; Covington, J.A.; Arasaradnam, R.P. The interplay of the gut microbiome, bile acids and volatile organic compounds. Gastroent. Res. Pract. 2015, 398585, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Recent progress in the design and clinical development of electronic-nose technologies. Nanobiosens. Dis. Diagn. 2016, 5, 15–27. [Google Scholar] [CrossRef]
- Santini, G.; Mores, N.; Penas, A.; Capuano, R.; Mondino, C.; Trové, A.; Macagno, F.; Zini, G.; Cattani, P.; Martinelli, E.; et al. Electronic nose and exhaled breath NMR-based metabolomics applications in airways disease. Curr. Top. Med. Chem. 2016, 16, 1610–1630. [Google Scholar] [CrossRef] [PubMed]
- Ruzsanyi, V.; Fischer, L.; Herbig, J.; Ager, C.; Amann, A. Multi-capillary-column proton-transfer-reaction time-of-flight mass spectrometry. J. Chromatogr. A 2013, 1316, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Spaněl, P.; Smith, D. Progress in SIFT-MS: Breath analysis and other applications. Mass Spectrom. Rev. 2011, 30, 236–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sahay, P. Breath analysis using laser spectroscopic techniques: Breath biomarkers, spectral fingerprints, and detection limits. Sensors 2009, 9, 8230–8262. [Google Scholar] [CrossRef] [PubMed]
- Kybert, N.J.; Egan, L.; Waldman, R.Z.; Zeng, X.-N.; Krein, M.; Preti, G.; Stuart, J.A.; Johnson, A.T.C. Analysis of sweat simulant mixtures using multiplexed arrays of DNA-carbon nanotube vapor sensors. J. Forensic Sci. Criminol. 2014, 1, S102. [Google Scholar] [CrossRef]
- Wilson, A.D. Electronic-nose applications in forensic science and for analysis of volatile biomarkers in the human breath. J. Forensic Sci. Criminol. 2014, 1, S103. [Google Scholar] [CrossRef]
- Wilson, A.D.; Baietto, M. Advances in electronic-nose technologies developed for biomedical applications. Sensors 2011, 11, 1105–1176. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Future applications of electronic-nose technologies in healthcare and biomedicine. In Wide Spectra of Quality Control; Akyar, I., Ed.; InTech Publishing: Rijeka, Croatia, 2011; Chapter 15; pp. 267–290. ISBN 978-953-307-683-6. Available online: https://www.intechopen.com/books/wide-spectra-of-quality-control/future-applications-of-electronic-nose-technologies-in-healthcare-and-biomedicine (accessed on 6 August 2018).
- Wilson, A.D. Theoretical and practical considerations for teaching diagnostic electronic-nose technologies to clinical laboratory technicians. Proc. Soc. Behav. Sci. 2012, 31, 262–274. [Google Scholar] [CrossRef]
- Wilson, A.D. Advanced methods for teaching electronic-nose technologies to diagnosticians and clinical laboratory technicians. Proc. Soc. Behav. Sci. 2012, 46, 4544–4554. [Google Scholar] [CrossRef]
- Wilson, A.D.; Lester, D.G.; Oberle, C.S. Development of conductive polymer analysis for the rapid detection and identification of phytopathogenic microbes. Phytopathology 2004, 94, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D.; Baietto, M. Applications and advances in electronic-nose technologies. Sensors 2009, 9, 5099–5148. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Diverse applications of electronic nose technologies in agriculture and forestry. Sensors 2013, 13, 2295–2348. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Review of electronic-nose technologies and algorithms to detect hazardous chemicals in the environment. Procedia Technol. 2012, 1, 453–463. [Google Scholar] [CrossRef]
- Ahmed, I.; Greenwood, R.; de Lacy Costello, B.; Ratcliffe, N.M.; Probert, C.S. An investigation of fecal volatile organic metabolites in irritable bowel syndrome. PLoS ONE 2013, 8, e58204. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.K.; Leggett, C.L.; Wang, K.K. Diagnosing gastrointestinal illnesses using fecal headspace volatile organic compounds. World J. Gastroenterol. 2016, 22, 1639–1649. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Advances in electronic-nose technologies for the detection of volatile biomarker metabolites in the human breath. Metabolites 2015, 5, 140–163. [Google Scholar] [CrossRef] [PubMed]
- De Groot, E.F.J.; de Meij, T.G.J.; van der Schee, M.P.; de Boer, N.K.H. Letter: Volatile metabolomics of exhaled breath or faecal gas? Aliment. Pharmacol. Ther. 2015, 41, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.J. Toward precision medicine and health: Opportunities and challenges in allergic diseases. J. Allergy Clin. Immunol. 2016, 137, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Weston, A.D.; Hood, L. Systems biology, proteomics, and the future of health care: Toward predictive, preventative, and personalized medicine. J. Proteome Res. 2004, 3, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wan, S.; Jiang, Y.; Wang, Y.; Fu, T.; Liu, Q.; Cao, Z.; Qiu, L.; Tan, W. Molecular elucidation of disease biomarkers at the interface of chemistry and biology. J. Am. Chem. Soc. 2017, 139, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Buyse, M.; Sargent, D.J.; Grothey, A.; Matheson, A.; de Gramont, A. Biomarkers and surrogate end points—the challenge of statistical validation. Nat. Rev. Clin. Oncol. 2010, 7, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Dalton, W.S.; Friend, S.H. Cancer biomarkers—An invitation to the table. Science 2006, 312, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Gutman, S.; Kessler, L.G. The US Food and Drug Administration perspective on cancer biomarker development. Nat. Rev. Cancer 2006, 6, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Cheng, S.H.; Chui, K.M.; Fok, T.F.; Wong, M.Y.; Wong, W.; Wong, R.P.; Cheung, K.L. Diagnosis of late onset neonatal sepsis with cytokines, adhesion molecule, and C-reactive protein in preterm very low birthweight infants. Arch. Dis. Child Fetal Neonatal Ed. 1997, 77, F221–F227. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Li, K.; Leung, T.F.; Wong, R.P.; Li, G.; Chui, K.M.; Wong, E.; Cheng, F.W.; Fok, T.F. Early prediction of sepsis-induced disseminated intravascular coagulation with interleukin-10, interleukin-6, and RANTES in preterm infants. Clin. Chem. 2006, 52, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Li, K.; Chui, K.M.; Leung, T.F.; Wong, R.P.; Chu, W.C.; Wong, E.; Fok, T.F. IP-10 is an early diagnostic marker for identification of late-onset bacterial infection in preterm infants. Pediatr. Res. 2007, 61, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Turunen, R.; Andersson, S.; Nupponen, I.; Kautiainen, H.; Siitonen, S.; Repo, H. Increased CD11b-density on circulating phagocytes as an early sign of late-onset sepsis in extremely low-birth-weight infants. Pediatr. Res. 2005, 57, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Li, K.; Wong, R.P.; Chui, K.M.; Wong, E.; Fok, T.F. Neutrophil CD64 expression: A sensitive diagnostic marker for late-onset nosocomial infection in very low birthweight infants. Pediatr. Res. 2002, 51, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Mally, P.; Xu, J.; Hendricks-Muñoz, K.D. Biomarkers for neonatal sepsis: Recent developments. Res. Rep. Neonatol. 2014, 4, 157–168. [Google Scholar] [CrossRef]
- Lubes, G.; Goodarzi, M. GC-MS based metabolomics used for the identification of cancer volatile organic compounds as biomarkers. J. Pharmaceut. Biomed. Anal. 2018, 147, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, C.; Ren, M.; Yin, X.; Chi, C.; Guo, L.; Ke, C.; Feng, H.; Li, E. Blood volatile organic compounds as potential biomarkers for amyotrophic lateral sclerosis: An animal study in the SOD1 G93A mouse. J. Mol. Neurosci. 2015, 55, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Peled, N.; Ionescu, R.; Nol, P.; Barash, O.; McCollum, M.; VerCauteren, K.; Koslow, M.; Stahl, R.; Rhyan, J.; Hossam, H. Detection of volatile organic compounds in cattle naturally infected with Mycobacterium bovis. Sens. Actuators B Chem. 2012, 171–172, 588–594. [Google Scholar] [CrossRef]
- Probert, C.S.J.; Jones, P.R.H.; Ratcliffe, N.M. A novel method for rapidly diagnosing the causes of diarrhea. Gut 2004, 53, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Chappell, C.L.; Darkoh, C.; Shimmin, L.; Farhana, N.; Kim, D.K.; Okhuysen, P.C.; Hixson, J. Fecal indole as a biomarker of susceptibility to Cryptosporidium infection. Infect. Immun. 2016, 84, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guan, L.; Zhang, H.; Gao, Y.; Sun, J.; Gong, X.; Li, D.; Chen, P.; Liang, X.; Huang, M.; et al. Endometrium metabolomic profiling reveals potential biomarkers for diagnosis of endometriosis at minimal-mild stages. Reprod. Biol. Endocrinol. 2018, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Taware, R.; Taunk, K.; Pereira, J.A.M.; Dhakne, R.; Kannan, N.; Soneji, D.; Câmara, J.S.; Nagarajaram, H.A.; Rapole, S. Investigation of urinary volatomic alterations in head and neck cancer: A non-invasive approach towards diagnosis and prognosis. Metabolomics 2017, 13, 111. [Google Scholar] [CrossRef]
- Monteiro, M.; Moreira, N.; Pinto, J.; Pires-Luís, A.S.; Henrique, R.; Jerónimo, C.; Bastos, M.L.; Gil, A.M.; Carvalho, M.; Guedes, P.G. GC-MS metabolomics-based approach for the identification of a potential VOC-biomarker panel in the urine of renal cell carcinoma patients. J. Cell. Mol. Med. 2017, 21, 2092–2105. [Google Scholar] [CrossRef] [PubMed]
- Ulanowska, A.; Kowalowski, T.; Hrynkiewicz, K.; Jackowski, M.; Buszewski, B. Determination of volatile organic compounds in human breath for Helicobacter pylori detection by SPME-GC/MS. Biomed. Chromatogr. 2011, 25, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D.; Oberle, C.S.; Oberle, D.F. Detection of off-flavor in catfish using a conducting polymer electronic-nose technology. Sensors 2013, 13, 15968–15984. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D.; Forse, L.B. Discrimination between Pseudogymnoascus destructans, other dermatophytes of cave-dwelling bats, and related innocuous keratinophilic fungi based on electronic-nose/GC signatures of VOC-metabolites produced in culture. In Proceedings of the VIII International Conference on Sensor Device Technologies and Applications, Rome, Italy, 10–14 September 2017; International Academy, Research, and Industry Association (IARIA): Wilmington, DE, USA, 2017; pp. 5–11, ISBN 978-1-61208-581-4. Available online: http://www.thinkmind.org/index.php?view=article&articleid=sensordevices_2017_1_20_20058 (accessed on 6 August 2018).
- Wilson, A.D.; Forse, L.B. Differences in VOC-metabolite profiles of Pseudogymnoascus destructans and related fungi by electronic-nose/GC analyses of headspace volatiles derived from axenic cultures. Sens. Transducers 2018, 220, 9–19. [Google Scholar]
- Fend, R.; Geddes, R.; Lesellier, S.; Vordermeier, H.-M.; Corner, L.A.L.; Gormley, E.; Costello, E.; Hewinson, R.G.; Marlin, D.J.; Woodman, A.C.; et al. Use of an electronic nose to diagnose Mycobacterium bovis infection in badgers and cattle. J. Clin. Microbiol. 2005, 43, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.B.; Wang, S.Z.; Yin, Y.G.; Hoffmann, W.C.; Zheng, X.Z. Using a surface plasmon resonance biosensor for rapid detection of Salmonella typhimurium in chicken carcass. J. Bionic Eng. 2008, 5, 239–246. [Google Scholar] [CrossRef]
- Knobloch, H.; Schroedl, W.; Turner, C.; Chambers, M.; Reinhold, P. Electronic nose responses and acute phase proteins correlate in blood using a bovine model of respiratory infection. Sens. Actuators B Chem. 2010, 144, 81–87. [Google Scholar] [CrossRef]
- Cramp, A.P.; Sohn, J.H.; James, P.J. Detection of cutaneous myiasis in sheep using an ‘electronic nose’. Vet. Parasitol. 2009, 166, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Gerber, N.N.; Lechevalier, H.A. Geosmin, an earthy-smelling compound isolated from actinomycetes. Appl. Microbiol. 1965, 13, 935–938. [Google Scholar] [PubMed]
- Grimm, C.C.; Lloyd, S.W.; Batista, R.; Zimba, P.V. Using microwave distillation-solid-phase-microextraction-gas chromatography-mass spectrometry for analyzing fish tissue. J. Chromatogr. Sci. 2000, 38, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Di Lena, M.; Porcelli, F.; Altomare, D.F. Volatile organic compounds as new biomarkers for colorectal cancer: A review. Colorectal Dis. 2016, 18, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Buijck, M.; Berkhout, D.J.; de Groot, E.F.; Benninga, M.A.; van der Schee, M.P.; Kneepkens, C.M.; de Boer, N.K.; de Meij, T.G. Sniffing out paediatric gastrointestinal diseases: The potential of volatile organic compounds as biomarkers for disease. J. Pediatr. Gastroenterol. Nutr. 2016, 63, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Panebiancoa, C.; Kelmana, E.; Vene, K.; Gioffreda, D.; Tavano, F.; Vilu, R.; Terracciano, F.; Pata, I.; Adamberg, K.; Andriulli, A.; et al. Cancer sniffer dogs: How can we translate this peculiarity in laboratory medicine? Results of a pilot study on gastrointestinal cancers. Clin. Chem. Lab. Med. 2017, 56, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Greenwood, R.; Costello, B.; Ratcliffe, N.; Probert, C.S. Investigation of faecal volatile organic metabolites as novel diagnostic biomarkers in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2016, 43, 596–611. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Alkhouri, N.; Eng, K.; Cikach, F.; Mahajan, L.; Yan, C.; Grove, D.; Rome, E.S.; Lopez, R.; Dweik, R.A. Metabolomic analysis of breath volatile organic compounds reveals unique breathprints in children with inflammatory bowel disease: A pilot study. Aliment. Pharmacol. Ther. 2014, 40, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Arasaradnam, R.P.; McFarlane, M.; Daulton, E.; Skinner, J.; O’Connell, N.; Wurie, S.; Chambers, S.; Nwokolo, C.U.; Bardhan, K.; Savage, R.; et al. Non-invasive exhaled volatile organic biomarker analysis to detect inflammatory bowel disease (IBD). Digest. Liver Dis. 2016, 48, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Arasaradnam, R.P.; McFarlane, M.J.; Ryan-Fisher, C.; Westenbrink, E.; Hodges, P.; Thomas, M.G.; Chambers, S.; O’Connell, N.; Bailey, C.; Harmston, C.; et al. Detection of colorectal cancer (CRC) by urinary volatile organic compound analysis. PLoS ONE 2014, 9, e108750. [Google Scholar] [CrossRef] [PubMed]
- Arasaradnam, R.P.; Westenbrink, E.; McFarlane, M.J.; Harbord, R.; Chambers, S.; O’Connell, N.; Bailey, C.; Nwokolo, C.U.; Bardhan, K.D.; Savage, R.; et al. Differentiating coeliac disease from irritable bowel syndrome by urinary volatile organic compound analysis—A pilot study. PLoS ONE 2014, 9, e107312. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, D.J.C.; Niemarkt, H.J.; Klaas, N.; de Boerd, N.K.H.; Benningaa, M.A.; de Meij, T.G.J. The potential of gut microbiota and fecal volatile organic compounds analysis as early diagnostic biomarker for necrotizing enterocolitis and sepsis in preterm infants. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, S.; Fujimoto, K.; Jang, M.H.; Yang, B.G.; Jung, Y.J.; Nishiyama, M.; Sato, S.; Tsujimura, T.; Yamamoto, M.; Yokota, Y.; et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 2008, 9, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Momose, Y.; Hirayama, K.; Itoh, K. Competition for proline between indigenous Escherichia coli and E. coli O157: H7in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157: H7. Antonie Leeuwenhoek 2008, 94, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Darkoh, C.; Chappell, C.L.; Gonzales, C.; Okhuysen, P. A rapid and specific 507 method for the detection of indole in complex biological samples. Appl. Environ. Microbiol. 2015, 81, 8093–8097. [Google Scholar] [CrossRef] [PubMed]
- Bond, A.; Vernon, A.; Reade, S.; Mayor, A.; Minetti, C.; Wastling, J.; Lamden, K.; Probert, C. Investigation of volatile organic compounds emitted from faeces for the diagnosis of Giardiasis. J. Gastrointestin. Liver Dis. 2015, 24, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Garner, C.E.; Smith, S.; Elviss, N.C.; Humphrey, T.J.; White, P.; Ratcliffe, N.M.; Probert, C.S. Identification of Campylobacter infection in chickens from volatile faecal emissions. Biomarkers 2008, 13, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Bean, H.D.; Kuo, Y.M.; Hill, J.E. Fast detection of volatile organic compounds from bacterial cultures by secondary electrospray ionization-mass spectrometry. J. Clin. Microbiol. 2010, 48, 4426–4431. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, B.L.D.; Li, J.; Sanford, J.A.; Kim, Y.-M.; Kronewitter, S.R.; Jones, M.B.; Peterson, C.T.; Peterson, S.N.; Frank, B.C.; Purvine, S.O.; et al. A multi-omic view of host-pathogen-commensal interplay in Salmonella-mediated intestinal infection. PLoS ONE 2013, 8, e67155. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Shi, H.; Tang, C.; Zhang, R. Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry. J. Basic Microbiol. 2017, 57, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Covington, J.A.; Westenbrink, E.W.; Ouaret, N.; Harbord, R.; Bailey, C.; O’Connell, N.; Cullis, J.; Williams, N.; Nwokolo, C.U.; Bardhan, K.D.; et al. Application of a novel tool for diagnosing bile acid diarrhoea. Sensors 2013, 13, 11899–11912. [Google Scholar] [CrossRef] [PubMed]
- Monasta, L.; Pierobon, C.; Princivalle, A.; Martelossi, S.; Marcuzzi, A.; Pasini, F.; Perbellini, L. Inflammatory bowel disease and patterns of volatile organic compounds in the exhaled breath of children: A case-control study using ion molecule reaction-mass spectrometry. PLoS ONE 2017, 12, e0184118. [Google Scholar] [CrossRef] [PubMed]
- Baranska, A.; Mujagic, Z.; Smolinska, A.; Dallinga, J.W.; Jonkers, D.M.A.E.; Tigchelaar, E.F.; Dekens, J.; Zhernakova, A.; Ludwig, T.; Masclee, A.A.M.; et al. Volatile organic compounds in breath as markers for irritable bowel syndrome: A metabolomic approach. Aliment Pharmacol. Ther. 2016, 44, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Embleton, N.D.; Marrs, E.C.L.; Smith, D.P.; Fofanova, T.; Nelson, A.; Skeath, T.; Perry, J.D.; Petrosino, J.F.; Berrington, J.E.; et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 2017, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Garner, C.E.; Ewer, A.K.; Elasouad, A.K.; Power, F.; Greenwood, R.; Ratcliffe, N.M.; Costello Bde, L.; Probert, C.S. Analysis of faecal volatile organic compounds in preterm infants who develop necrotising enterocolitis: A pilot study. J. Pediatr. Gastroenterol. Nutr. 2009, 49, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Arasaradnam, R.P.; Covington, J.; Nwokolo, C.U. Editorial: Metabolomics analysis of breath volatile organic compounds—A new scent for inflammatory bowel disease. Aliment. Pharmacol. Ther. 2014, 40, 732–733. [Google Scholar] [CrossRef] [PubMed]
- Buszewski, B.; Ulanowska, A.; Ligor, T.; Jackowski, M.; Klodzinska, E.; Szeliga, J. Identification of volatile organic compounds secreted from cancer tissues and bacterial cultures. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008, 868, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Couch, R.D.; Navarro, K.; Sikaroodi, M.; Gillevet, P.; Forsyth, C.B.; Mutlu, E.; Engen, P.A.; Keshavarzian, A. The approach to sample acquisition and its impact on the derived human fecal microbiome and VOC metabolome. PLoS ONE 2013, 8, e81163. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Young, C.; Neu, J. Molecular modulation of intestinal epithelial barrier: Contribution of microbiota. J. Biomed. Biotechnol. 2010, 2010, 305879. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; De Angelis, M.; De Pasquale, I.; Ndagijimana, M.; Vernocchi, P.; Ricciuti, P.; Gagliardi, F.; Laghi, L.; Crecchio, C.; Guerzoni, M.E.; et al. Duodenal and faecal microbiota of celiac children: Molecular, phenotype and metabolome characterization. BMC Microbiol. 2011, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P.; et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef] [PubMed]
- Walton, C.; Fowler, D.P.; Turner, C.; Jia, W.; Whitehead, R.N.; Griffiths, L.; Dawson, C.; Waring, R.H.; Ramsden, D.B.; Cole, J.A.; et al. Analysis of volatile organic compounds of bacterial origin in chronic gastrointestinal diseases. Inflamm. Bowel Dis. 2013, 19, 2069–2078. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, M.; Zhang, L.; Zhang, K.; Ahmetagic, A.; Wei, M.Q. Volatile organic compounds as novel markers for the detection of bacterial infections. Clin. Microbiol. 2014, 3, 151. [Google Scholar] [CrossRef]
- Probert, C.S.J.; Ahmed, I.; Khalid, T.; Johnson, E.; Smith, S.; Ratcliffe, N. Volatile organic compounds as diagnostic biomarkers in gastrointestinal and liver diseases. Gastrointestin. Liver Dis. 2009, 18, 337–343. [Google Scholar]
- Bomers, M.K.; Menke, F.P.; Savage, R.S.; Vandenbroucke-Grauls, C.M.J.E.; van Agtmael, M.A.; Covington, J.A.; Smulders, Y.M. Rapid, accurate, and on-site detection of C. difficile in stool samples. Am. J. Gastroenterol. 2015, 110, 588–594. [Google Scholar] [CrossRef] [PubMed]
- De Lacy Costello, B.; Ewer, A.K.; Garner, C.E.; Probert, C.S.J.; Ratcliffe, N.M.; Smith, S. An analysis of volatiles in the headspace of the faeces of neonates. J. Breath Res. 2008, 2, 037023. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, D.J.C.; Niemarkt, H.J.; Buijck, M.; van Weissenbruch, M.M.; Brinkman, P.; Benninga, M.A.; van Kaam, A.H.; Kramer, B.W.; Andriessen, P.; de Boer, N.K.H.; et al. Detection of sepsis in preterm infants by fecal volatile organic compounds analysis: A proof of principle study. J. Pediatr. Gastroenterol. Nutr. 2017, 65, e47–e52. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Recent applications of electronic-nose technologies for the noninvasive early diagnosis of gastrointestinal diseases. Proceedings 2018, 2, 147. [Google Scholar] [CrossRef]
- Wojnowski, W.; Partyka, A.; Dymerski, T.; Gebicki, J.; Namiesnik, J. Electronic noses in medical diagnostics. Curr. Med. Chem. 2018, 25, 1–9. [Google Scholar] [CrossRef]
- Peng, G.; Hakim, M.; Broza, Y.Y.; Billan, S.; Abdah-Bortnyak, R.; Kuten, A.; Tisch, U.; Haick, H. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br. J. Cancer 2010, 103, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Arasaradnam, R.P.; Ouaret, N.; Thomas, M.G.; Quraishi, N.; Heatherington, E.; Nwokolo, C.U.; Bardhan, K.D.; Covington, J.A. A novel tool for noninvasive diagnosis and tracking of patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2013, 19, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Covington, J.A.; Harbord, R.; Westenbrink, E.W.; Bailey, C.; Nicola O’connell, N.; Dhaliwal, A.; Nwokolo, C.; Foley, A.; Marya, N.B.; Baptista, V.; et al. Detection of urinary volatile organic compounds in patients with inflammatory bowel disease and controls by an electronic nose—A Transatlantic Study. Gastroenterology 2014, 146, S795–S796. [Google Scholar] [CrossRef]
- De Meij, T.G.J.; de Boer, N.K.H.; Benninga, M.A.; Lentferink, Y.E.; de Groot, E.F.J.; van de Velde, M.E.; van Bodegraven, A.A.; van der Scheea, M.P. Faecal gas analysis by electronic nose as a novel, non-invasive method for assessment of active and quiescent paediatric inflammatory bowel disease: Proof of principle study. J. Crohns Colitis 2014, 8, 91–106. [Google Scholar] [CrossRef]
- Shepherd, S.F.; McGuire, N.D.; de Lacy Costello, B.P.; Ewen, R.J.; Jayasena, D.H.; Vaughan, K.; Ahmed, I.; Probert, C.S.; Ratcliffe, N.M. The use of a gas chromatograph coupled to a metal oxide sensor for rapid assessment of stool samples from irritable bowel syndrome and inflammatory bowel disease patients. J. Breath Res. 2014, 8, 026001. [Google Scholar] [CrossRef] [PubMed]
- McGuire, N.D.; Ewen, R.J.; Costello, C.D.; Garner, C.E.; Probert, C.S.J.; Vaughan, K.; Ratcliffe, N.M. Towards point of care testing for C. difficile infection by volatile profiling, using the combination of a short multi-capillary gas chromatography column with metal oxide detection. Meas. Sci. Technol. 2014, 25, 065108. [Google Scholar] [CrossRef] [PubMed]
- De Meij, T.G.; van der Schee, M.P.; Berkhout, D.J.; van de Velde, M.E.; Jansen, A.E.; Kramer, B.W.; van Weissenbruch, M.M.; van Kaam, A.H.; Andriessen, P.; van Goudoever, J.B.; et al. Early detection of necrotizing enterocolitis by fecal volatile organic compounds analysis. J. Pediatr. 2015, 167, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Esfahani, S.; Covington, J.A. Low cost optical electronic nose for biomedical applications. Proceedings 2017, 1, 589. [Google Scholar] [CrossRef]
- Bunge, M.; Araghipour, N.; Mikoviny, T.; Dunkl, J.; Schnitzhofer, R.; Hansel, A.; Schinner, F.; Wisthaler, A.; Margesin, R.; Märk, T.D. On-line monitoring of microbial volatile metabolites by proton transfer reaction-mass spectrometry. Appl. Environ. Microbiol. 2008, 74, 2179–2186. [Google Scholar] [CrossRef] [PubMed]
- Carroll, W.; Lenney, W.; Wang, T.; Spanel, P.; Alcock, A.; Smith, D. Detection of volatile compounds emitted by Pseudomonas aeruginosa using selected ion flow tube mass spectrometry. Pediatr. Pulmonol. 2005, 39, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Smith, D.; Spanel, P. Selected ion flow tube, SIFT, studies of the reactions of H3O+, NO+ and O2+ with compounds released by Pseudomonas and related bacteria. Int. J. Mass Spectrom. 2004, 233, 245–251. [Google Scholar] [CrossRef]
- Chang, D.-Y.; Lee, C.-C.; Shiea, J. Detecting large biomolecules from high-salt solutions by fused-droplet electrospray ionization mass spectrometry. Anal. Chem. 2002, 74, 2465–2469. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wortmann, A.; Zenobi, R. Neutral desorption sampling coupled to extractive electrospray ionization mass spectrometry for rapid differentiation of biosamples by metabolomic fingerprinting. J. Mass Spectrom. 2007, 42, 1123–1135. [Google Scholar] [CrossRef] [PubMed]
- Cooks, R.G.; Ouyang, Z.; Takats, Z.; Wiseman, J.M. Ambient mass spectrometry. Science 2006, 311, 1566–1570. [Google Scholar] [CrossRef] [PubMed]
- Shieh, I.-F.; Lee, C.-Y.; Shiea, J. Eliminating the interferences from TRIS buffer and SDS in protein analysis by fused-droplet electrospray ionization mass spectrometry. J. Proteome Res. 2005, 4, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Takáts, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Electrosonic spray ionization. A gentle technique for generating folded proteins and protein complexes in the gas phase and for studying ion-molecule reactions at atmospheric pressure. Anal. Chem. 2004, 76, 4050–4058. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Hill, H.H., Jr.; Wittmer, D.P. Analytical merit of electrospray ion mobility spectrometry as a chromatographic detector. J. Microcolumn Sep. 1994, 6, 515–524. [Google Scholar] [CrossRef]
- Lee, C.Y.; Shiea, J. Gas chromatography connected to multiple channel electrospray ionization mass spectrometry for the detection of volatile organic compounds. Anal. Chem. 1998, 70, 2757–2761. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Lozano, P.; Rus, J.; de la Mora, G.F.; Hernández, M. Secondary electrospray ionization (SESI) of ambient vapors for explosive detection at concentrations below parts per trillion. J. Am. Soc. Mass Spectrom. 2009, 20, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Siems, W.F.; Hill, H.H. Secondary electrospray ionization ion mobility spectrometry/mass spectrometry of illicit drugs. Anal. Chem. 2000, 72, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Lozano, P.; Fernández de la Mora, J. Direct analysis of fatty acid vapors in breath by electrospray ionization and atmospheric pressure ionization-mass spectrometry. Anal. Chem. 2008, 80, 8210–8215. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Lozano, P.; Fernández de la Mora, J. Electrospray ionization of volatiles in breath. Int. J. Mass Spectrom. 2007, 265, 68–72. [Google Scholar] [CrossRef]
- Martínez-Lozano, P.; Fernández de la Mora, J. On-line detection of human skin vapors. J. Am. Soc. Mass Spectrom. 2009, 20, 1060–1063. [Google Scholar] [CrossRef] [PubMed]
- Baietto, M.; Wilson, A.D.; Bassi, D.; Ferrini, F. Evaluation of three electronic noses for detecting incipient wood decay. Sensors 2010, 10, 1062–1092. [Google Scholar] [CrossRef] [PubMed]
- Baietto, M.; Wilson, A.D. Electronic-nose applications for fruit identification, ripeness and quality grading. Sensors 2015, 15, 899–931. [Google Scholar] [CrossRef] [PubMed]
- El Manouni El Hassani, S.; Berkhout, D.J.C.; Bosch, S.; Benninga, M.A.; de Boer, N.K.H.; de Meij, T.G.J. Application of fecal volatile organic compound analysis in clinical practice: Current state and future perspectives. Chemosensors 2018, 6, 29. [Google Scholar] [CrossRef]
- Bosch, S.; El Manouni El Hassani, S.; Covington, J.A.; Wicaksono, A.N.; Bomers, M.K.; Benninga, M.A.; Mulder, C.J.J.; de Boer, N.K.H.; de Meij, T.G.J. Optimized sampling conditions for fecal volatile organic compound analysis by means of field asymmetric ion mobility spectrometry. Anal. Chem. 2018, 90, 7972–7981. [Google Scholar] [CrossRef] [PubMed]
| Technology Types 1 | Evaluation Criteria | Advantages 2 | Disadvantages |
|---|---|---|---|
| Metabolomic | Analysis costs | Yields more chemistry information and identity of volatile organic compounds (VOCs) in sample | Expensive operating and maintenance costs |
| Clinical and field application | Most useful for confirmation of diagnoses by more rapid disease-detection methods; not portable (immobile) | Not suitable due to untimely results and low-sample throughput | |
| Data analysis | Potentially provides indications of disease mechanisms, host pathways affected, and identity of specific chemical disease biomarkers (more detailed chemical information) | Highly complex, time-consuming, requires sophisticated software and/or models; complex interpretations | |
| Difficulty level | Provides more details of pathophysiology and metabolic conditions of patient; inter-device data comparisons possible | Labor intensive, requires highly trained operating personnel | |
| Time requirements to diagnoses | More detailed chemistry information may yield clues for more accurate diagnoses | Slow diagnostic results, not real-time | |
| Reproducibility | High for clinical samples when prepared with standardized methods & patient histories; low sensor drift over time | Numerous factors may affect volatile organic metabolites (VOMs) and biomarkers identified | |
| Electronic-nose | Analysis costs | Relatively inexpensive (low costs); yields simpler collective signature (profile) of all VOC-metabolites present in sample | Individual VOCs not identified (except with combination-technology e-nose instruments) |
| Clinical and field application | Highly applicable for clinical use, high sample throughput possible, portable for clinical, patient room or field use | Mobility may be limited by power, weight or space requirements | |
| Data analysis | Simpler, more straight-forward analyses with easier interpretations of results | Level of sample discrimination is critical | |
| Difficulty level | Relatively easy to operate and obtain results based on VOC profiles (compared to libraries of reference database) | Inter-device data comparisons (of results) not usually possible | |
| Time requirements to diagnoses | Relatively rapid preliminary diagnoses; reliability greatly improved with ASRDs, real-time results | Confirmations with other diagnostic data may be required | |
| Reproducibility | Precision in sensor outputs generally is an asset of e-nose technologies, but results may vary without adequate QA/QC | Sensor drift over time affects reproducibility; sensor poisoning possible |
| Disease 1 | Pathogen/Cause | Clinical Sample | Biomarker | Chemical Class 2 | Molecular Structure | Reference |
|---|---|---|---|---|---|---|
| ALS | Neurodegenerative | Blood | butylated hydroxytoluene | Phenol deriv. |  | [40] |
| Bovine TB | Mycobacterium bovis | Breath | 2,2-dimethyl undecane | Methylated alkane |  | [41] |
| Cholera | Vibrio cholerae | Feces | p-menth-1-en-8-ol | Monoterpene alcohol |  | [42] |
| Cryptosporidiosis | Cryptosporidium parvum | Feces | indole | Benzopyrrole |  | [43] |
| Endometriosis | Unknown | Endometrial tissue | hypoxanthine | Oxypurine |  | [44] |
| HNC | Cancer | Urine | 2,6-dimethyl-7-octen-2-ol | Terpenoid |  | [45] |
| RCC | Cancer | Urine | 2,5,8-trimethyl-1,2,3,4- tetrahydronaphthalene-1-ol | Benzenoid PAH |  | [46] |
| Stomach ulcer | Helicobacter pylori | Breath | 2-butanone | Aliphatic ketone |  | [47] |
| GI-Disease 1 | Pathogen/Cause | N = | Method 2 | Sample | VOCs | Biomarker VOC-Metabolites (Chemical Class Abbrev.) 3 | Ref. |
|---|---|---|---|---|---|---|---|
| Amoebic dysentery | Cryptosporidium parvum | 50 | HIA | Feces | 1 | Indole (low levels, bpy) | [43,70] |
| Giardia duodenalis | 33 | GC-MS | Feces | 9 | 2,2,4,4-tetramethyloctane (ma) 1-propanol (alc) Acetic acid (ca) 2,2,4,6,6-pentamethylheptane (ma) Acetone (ket) | [71] | |
| Bacterial infections (intestinal) | Campylobacter jejuni | 5 | GC-MS | Feces | 3+ | phenols (bd) indoles (bpy) organic acids (ca) | [42] |
| 71 | GC-MS | Feces | 6 | Hexanal (ald) (E)-2-octenal (ald) Pyrrole (az) Ethyl ethanoate (es) Methanol (alc) 2-heptanone (ket) | [72] | ||
| Clostridium difficile | 44 | GC-MS | Feces | 8 | Acetic acid (ca) Butanoic acid (ca) 2-furancarboxaldehyde (ald) 5-methyl-2-furan-carboxaldehyde (ald) Methyl furancarboxylate (fad) 2-hydoxy benzaldehyde (ald) 4-methyl phenol (phed) 2-methoxy phenol (phed) | [42] | |
| Escherichia coli | 45 | SESI-MS | Culture | 3 | Acetonitrile (nit) Ethanol (alc) Indole (bpy) | [73] | |
| Salmonella typhimurium | 20 | GC-MS | Gut | 5 | Lactose (ds) Melibiose (ds) Raffinose (ts) Fucose (ms) Galactinol (sa) | [74] | |
| Shigella flexneri | 80 | GC-MS | Culture | 2 | 1-decanol (alc) 1-octanol (alc) | [75] | |
| BAD | Digestive dysfunction | 110 | FAIMS, GC-MS | Urine | 2 | 2-propanol (alc) Acetamide (amd) | [76] |
| Cholera | Vibrio cholerae | 41 | GC-MS | Feces | 1 | 2-(4-methyl-3-cyclohexen-1-yl)-2-propanol (mta) | [42] |
| Coeliac | Gluten sensitive enteropathy | 47 | FAIMS, GC-MS | Urine | 1 | 1, 3, 5, 7 cyclooctatetraene (cod) | [64] |
| CRC | Cancer | 133 | FAIMS | Urine | 26 | Complex mixture | [63] |
| CRD | Unknown cause of bowel inflammation | 201 | IMR-MS | Breath | 21 | NOx compounds (no) Methane (alk) Ammonia (am) Acetaldehyde (ald) | [77] |
| IBD | Immune-induced inflammation | 117 | SIFT-MS | Breath | 6 | 1-octene (ao) 1-decene (alke) (E)-2-nonene (alke) | [61] |
| IBS | Unknown cause of bowel disorder | 323 | GC-TOF-MS | Breath | 4 | 1,4-cyclohexadiene (trp) Unidentified VOC Aziridine (azi) n-heptane (alk) n-hexane (alk) | [78] |
| LOS | Neonatal bacterial infections | 35 | UPLC-MS | Feces | 10,11-dihydro-12R-hydroxy-leukotriene E4 (leu) * Phylloquinone (nqd) * Ascorbic acid (kaad) * | [79] | |
| NEC | Injury-induced intestinal necrosis | 65 | GC-MS | Feces | 4 | Absent (present in controls) 2-ethylhexyl acetoate (es) Ethyl decanoate (es) Ethyl dodecanoate (es) Ethyl hexadecanoate (es) | [80] |
| UC | Abnormal immune response | 200 | IMR-MS | Breath | 21 | NO (no) Methane (alk) Ammonia (am) Acetaldehyde (ald) | [77] |
| VE | Astrovirus, Adenovirus, Norwalk virus | 1, 5, 9 | GC-MS | Feces | 2 | Ammonia (am) Ethyl dodecanoate (absent, es) | [42] |
| Rotavirus | 5 | GC-MS | Feces | 3 | Ethyl dodecanoate (es) Propyl dodecanoate (es) Dodecanoic acid (ca) | [42] |
| Disease 1 | Location | Sample | N = | E-Nose Model | Sensor Type/No. 2 | References |
|---|---|---|---|---|---|---|
| BAD | BAD | Urine | 110 | Fox 4000 | MOS 18 | [76] |
| Cancer | Colon | Breath | 26 | Experimental | GNP 14 | [98] |
| Colon | Fecal | 157 | Cyranose 320 | CBPC 32 | [5] | |
| CRC/IBD | Colon | Urine | 92 | WOLF | EC 8, NDIR 2, PID 1 | [2] |
| IBD | Intestine | Urine | 62 | Owlstone | FAIMS | [99,100] |
| Colon | Fecal | 83 | Cyranose 320 | CBPC 32 | [101] | |
| IBS | Colon | Fecal | 182 | Experimental | GC-MOS 1 | [102] |
| Colon | Breath | 234 | V&F Airsense | IMR-MS | [77] | |
| ID | Colon | Fecal | 100 | Experimental | GC-MOS 1 | [103] |
| LOS | Systemic | Fecal | 76 | Cyranose 320 | CBPC 32 | [95] |
| NEC | Colon | Fecal | 27 | Cyranose 320 | CBPC 32 | [104] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, A.D. Application of Electronic-Nose Technologies and VOC-Biomarkers for the Noninvasive Early Diagnosis of Gastrointestinal Diseases. Sensors 2018, 18, 2613. https://doi.org/10.3390/s18082613
Wilson AD. Application of Electronic-Nose Technologies and VOC-Biomarkers for the Noninvasive Early Diagnosis of Gastrointestinal Diseases. Sensors. 2018; 18(8):2613. https://doi.org/10.3390/s18082613
Chicago/Turabian StyleWilson, Alphus Dan. 2018. "Application of Electronic-Nose Technologies and VOC-Biomarkers for the Noninvasive Early Diagnosis of Gastrointestinal Diseases" Sensors 18, no. 8: 2613. https://doi.org/10.3390/s18082613
APA StyleWilson, A. D. (2018). Application of Electronic-Nose Technologies and VOC-Biomarkers for the Noninvasive Early Diagnosis of Gastrointestinal Diseases. Sensors, 18(8), 2613. https://doi.org/10.3390/s18082613