Automated Quality Control for Sensor Based Symptom Measurement Performed Outside the Lab
Abstract
1. Introduction
1.1. Related Work
- Pre-specified fixed constraints often appear in the form of: high/low-pass filters; sensor type and sensor location selection procedures; anomaly detection, etc. For example, changes in the orientation of a device introduce unwanted interruptions in sensor data such as those collected from an accelerometer during monitoring (for more information, see Section 2.3.1). This problem is usually approached by applying a high-pass filter to the data with a pre-specified threshold of 0.5 Hz [28]. The threshold value is not learned from the data, but is fixed empirically based on domain knowledge. Although this approach is over-simplifying assumptions about the data, such methods have been shown to be very useful for tackling problems related to hardware and easy to define events.
- In contrast, data-driven quality control techniques look at the structural differences in the data and aim to learn some relation between data that represent the two classes (i.e., good quality vs. bad quality data). For example, hidden Markov models (HMMs) and dynamic Bayesian networks [33,34] have been used in multiple domains to detect anomalous sensor readings and in describing uncertainty associated with sensor readings [33], such as those collected from temperature and conductivity sensors. However, data-driven approaches have not been used before to address more sophisticated experimental setups, which is the case in remote patient monitoring applications.
1.2. Overview
- For simpler quality control problems, we develop a Gaussian mixture model (GMM)-based approach, which attempts to cluster the raw signal into two classes: data adhering or violating the test protocol. Since GMMs ignore the sequential nature of the sensor data, we pass the estimated class indicators through a running median filter to smooth out unrealistic frequent switching between the two classes.
- We also propose a more general solution that involves fitting flexible nonparametric switching autoregressive (AR) models to each of the preprocessed sensor signals. The switching AR model segments the data in an unsupervised manner into random (unknown) numbers of behavioral patterns that are frequently encountered in the data. An additional classifier is then trained to discriminate which of the resulting variable-length segments represent adherence or violation of the test protocol. We demonstrate that a simple multinomial naive Bayes classifier can be trained using a strictly limited amount of labeled data annotated by a human expert. Since the instructions in any clinimetric test protocol are limited, whereas the number of potential behavioral violations of the protocol are not, we assume that any previously unseen segments that we detect are a new type of violation of the specified instructions of the protocol.
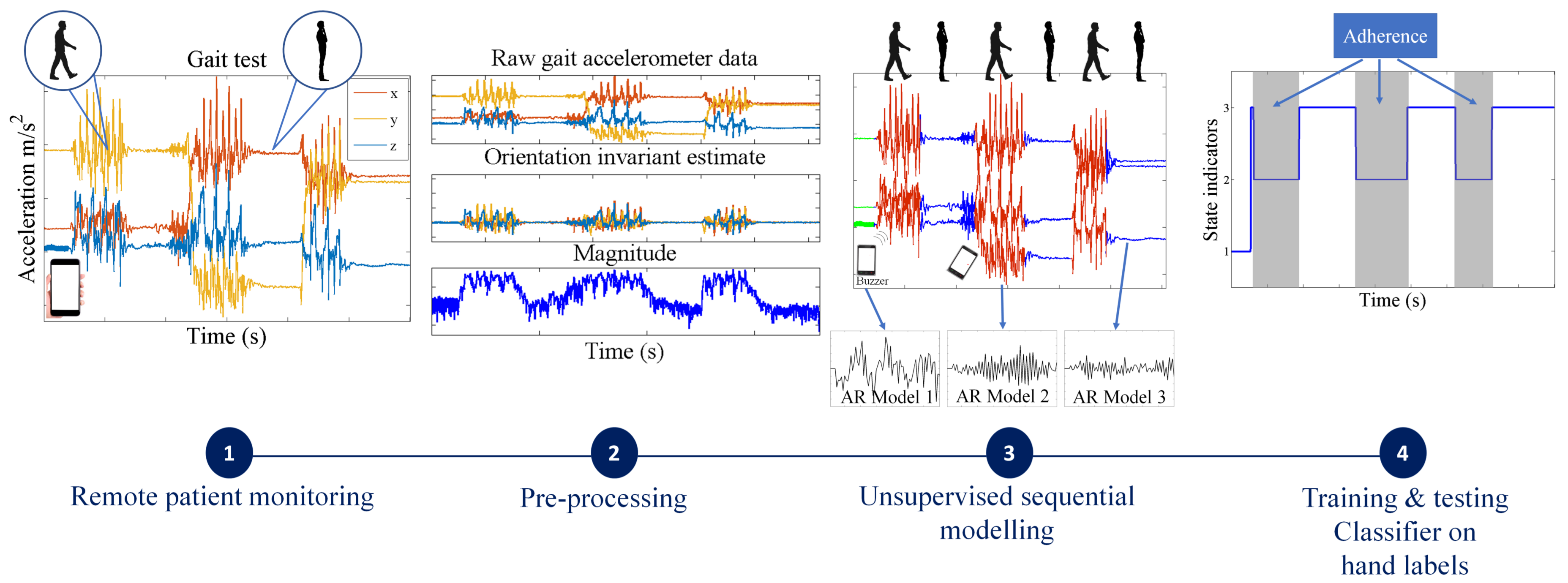
2. Methodology
2.1. Data Collection
2.2. Hand-Labeling for Algorithm Evaluation
2.3. Sensor-Specific Preprocessing
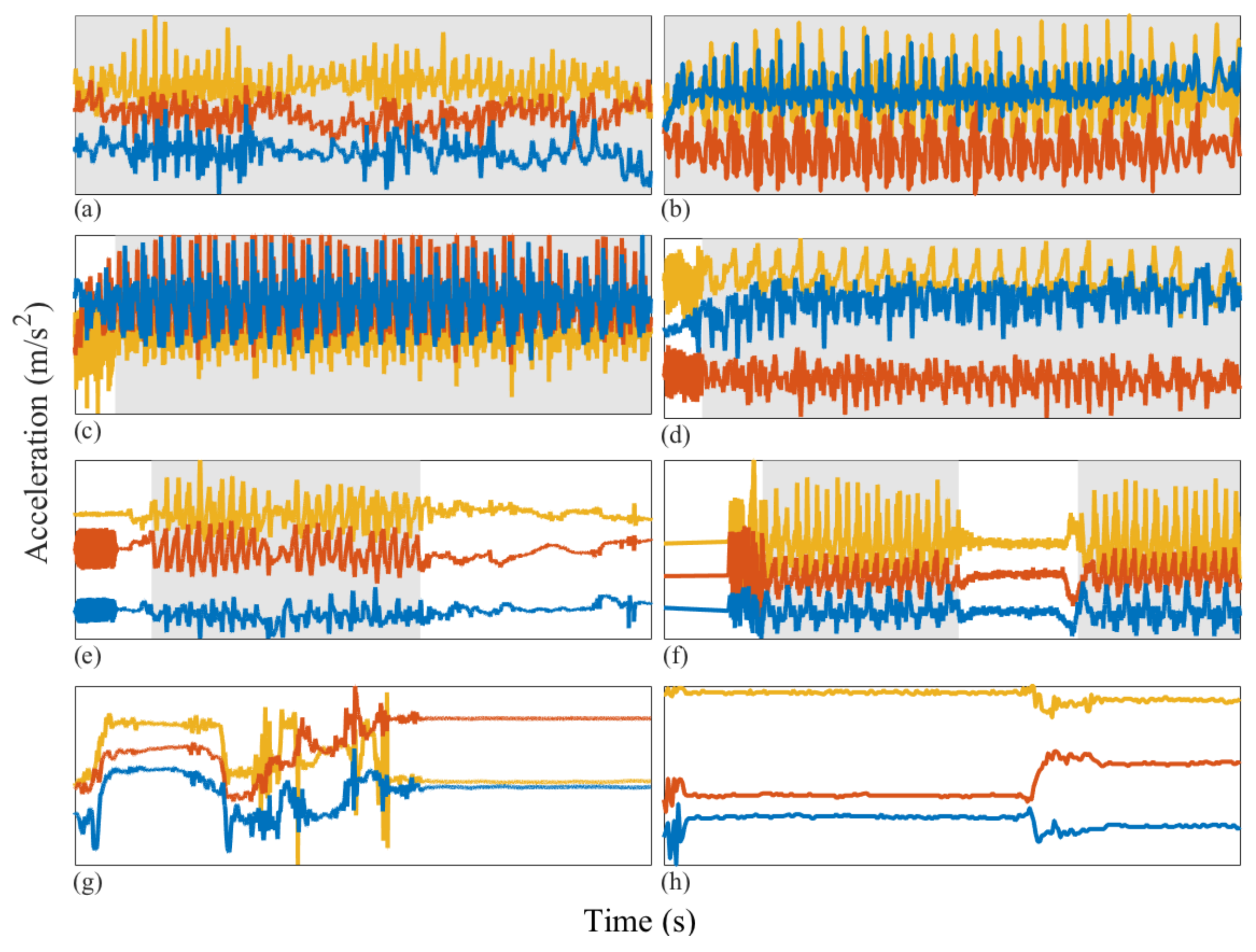
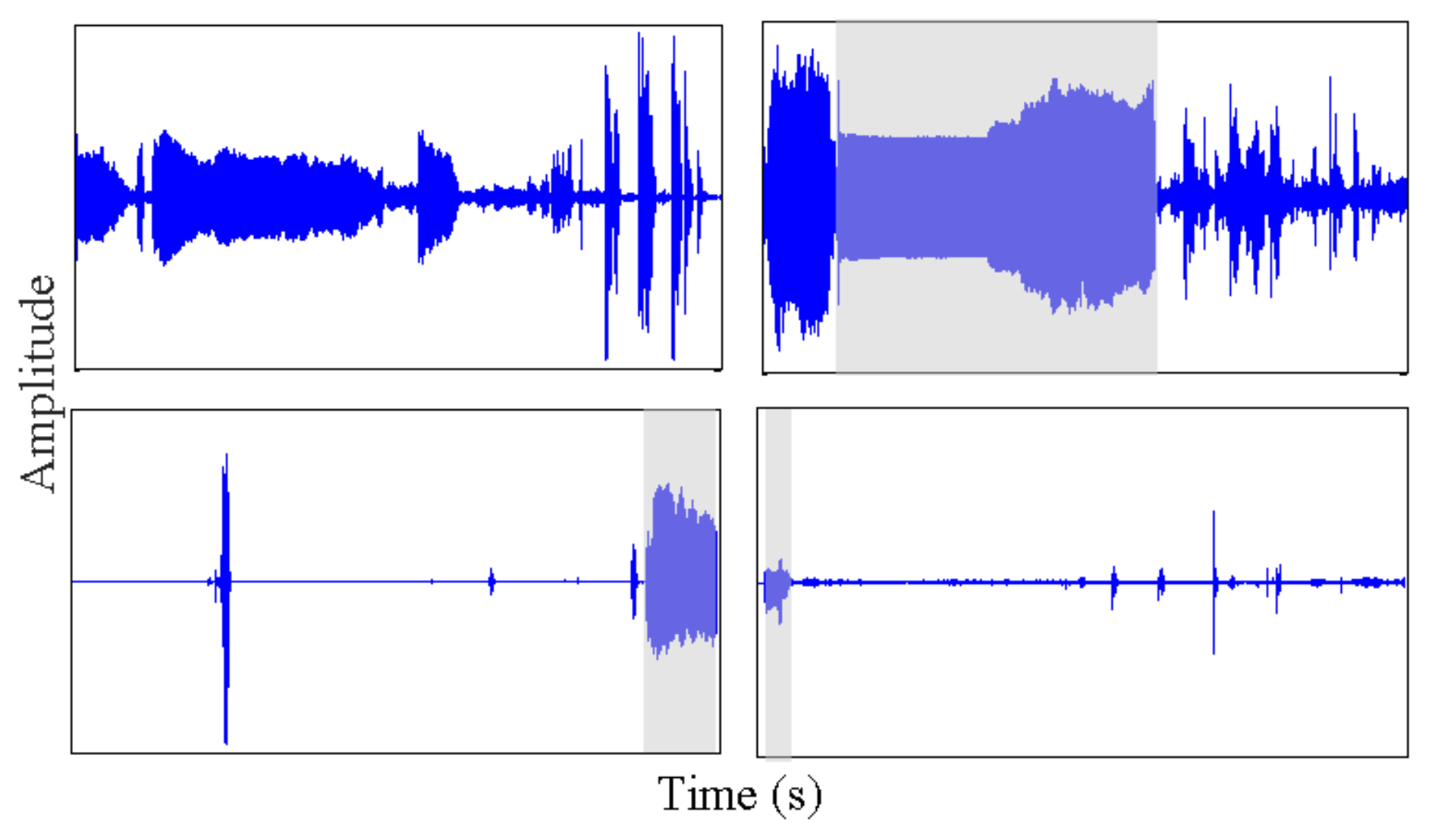
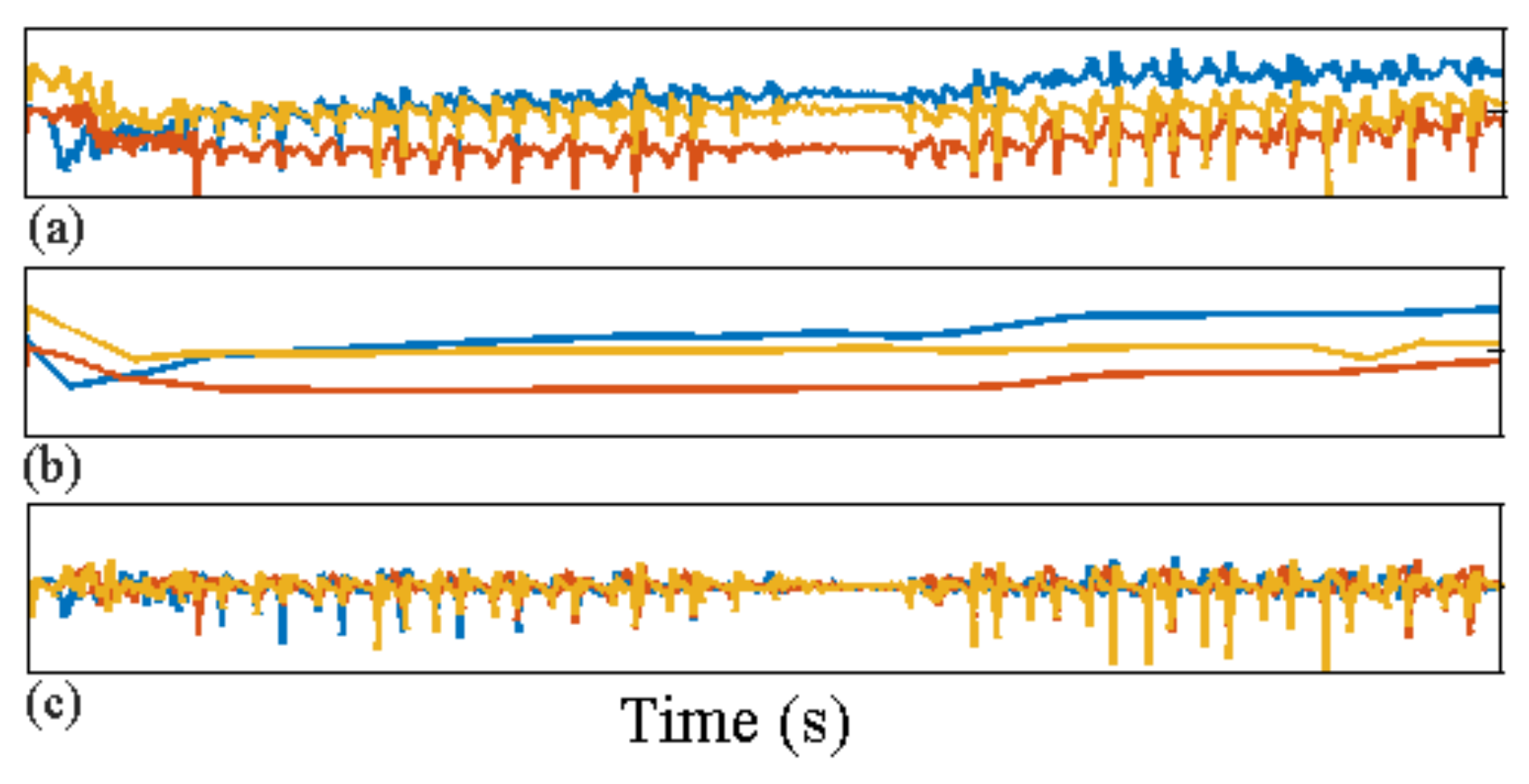
2.3.1. Isolating and Removing Orientation Changes from Accelerometry Data
2.3.2. Feature Extraction
2.3.3. Down-Sampling
2.4. Sequential Behavior Modeling
2.4.1. Unsupervised Behavior Modeling
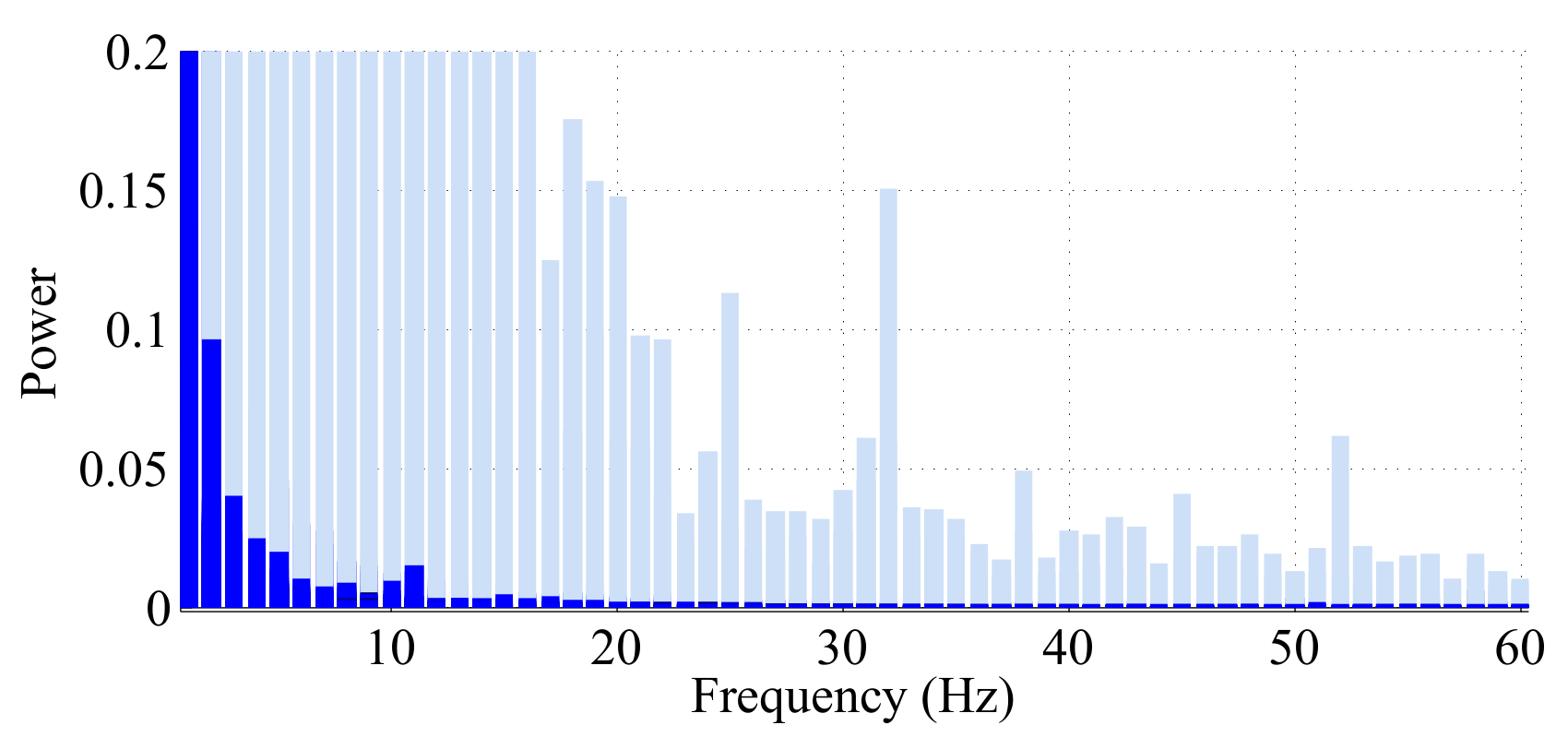
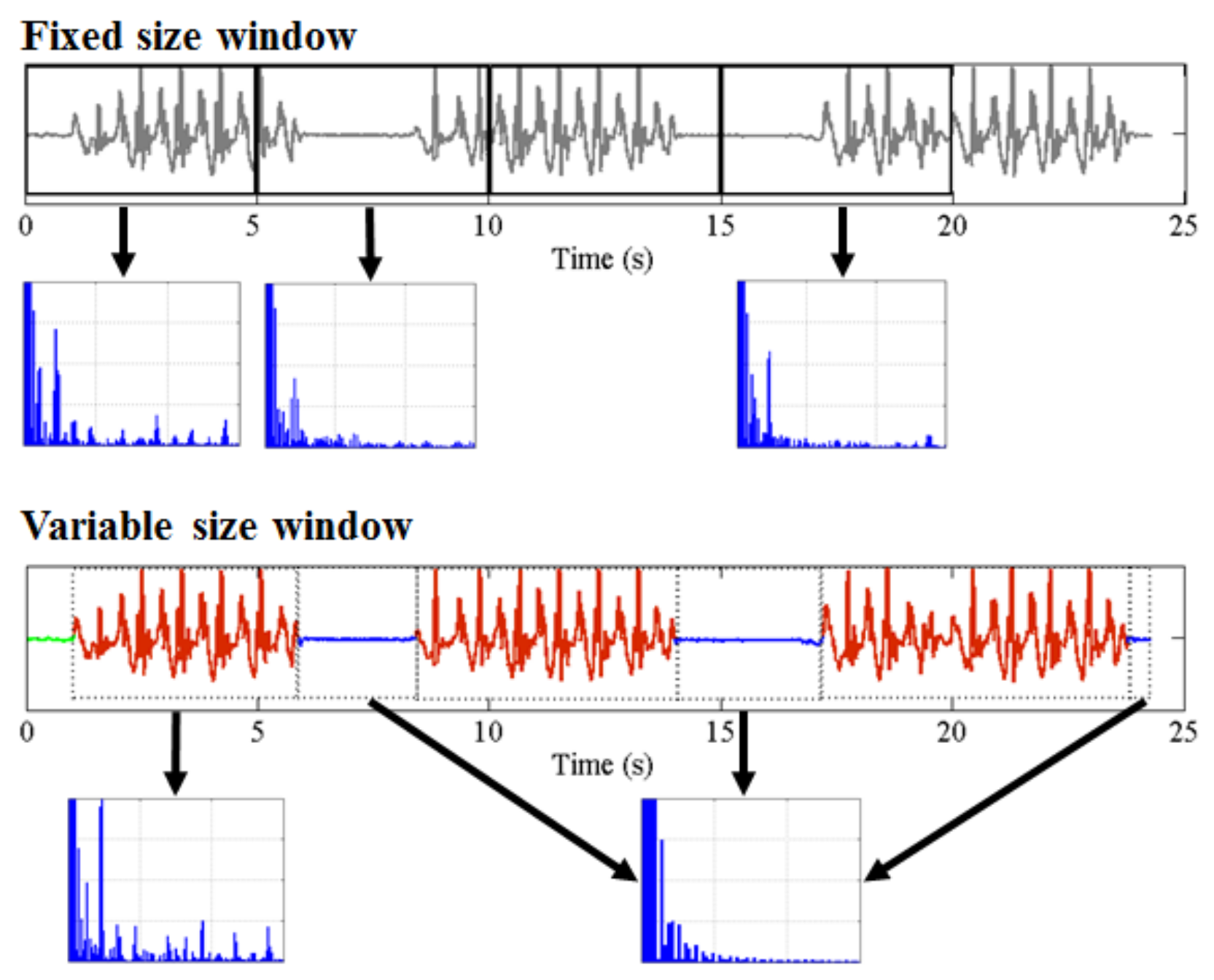
- Any behavior changes that occur in the data within a window cannot be represented (see Figure 6). Due to our inability to model and account for them, they confound the feature values for the window in which they occur. Many of the features used for processing sensor data are some type of frequency domain feature (for example, dominant frequency component; largest magnitude Fourier coefficients; various wavelet coefficients, etc.). Frequency domain features are only meaningful for signals that have no abrupt changes; Fourier analysis over windows that contain abrupt discontinuities is dominated by unavoidable Gibbs’ phenomena [44]. Unfortunately, behavioral data from clinimetric tests are rife with such discontinuities due to inevitable changes in activities during tests. In the approach we propose, the window sizes and boundaries adapt to the data, since segmentation is learned using a probabilistic model that is specifically designed to capture rapid changes in activity when they occur, but also to model the intricacies of each activity.
- The optimal features to be extracted from each window depend largely on the task/activity occurring in that window. If we are interested in developing a unified framework that works under a realistically wide set of scenarios encountered outside the lab, hand-picking an appropriate set of features for each activity that a clinimetric test might include is not feasible. This issue could be partially overcome if we use “automated” features such as principle component analysis (PCA), but this entails unrealistic assumptions (i.e., linearity). Alternatively, we could use an unsupervised approach for automated feature learning such as layers of restricted Boltzmann machines (RBM) or deep belief networks (DBN). However, these methods require large volumes of data from every behavior (which is unlikely to ever be available from health-impaired users), and sufficient computational power to train, making them unsuitable for deployment in real-time applications on smartphones or other resource-constrained devices. Even so, basic RBMs and DBNs would still need to be trained on features extracted after windowing of the sensor data. Additionally, although the features extracted using deep learning systems have demonstrated highly accurate classification results for many applications, we lack any ability to interpret these models to give a human understanding of what aspects of the data they represent. When dealing with healthcare applications, this lack of interpretability could significantly reduce the explanatory power required to gain confidence in the technique. The system we propose does not rely on extensive feature engineering or inscrutable deep learning algorithms, since we demonstrate sufficiently high performance using a single feature for each of the different sensor types. Of course, the proposed approach can be easily extended to use multiple features per data type, and this could potentially boost performance when appropriate features are chosen.
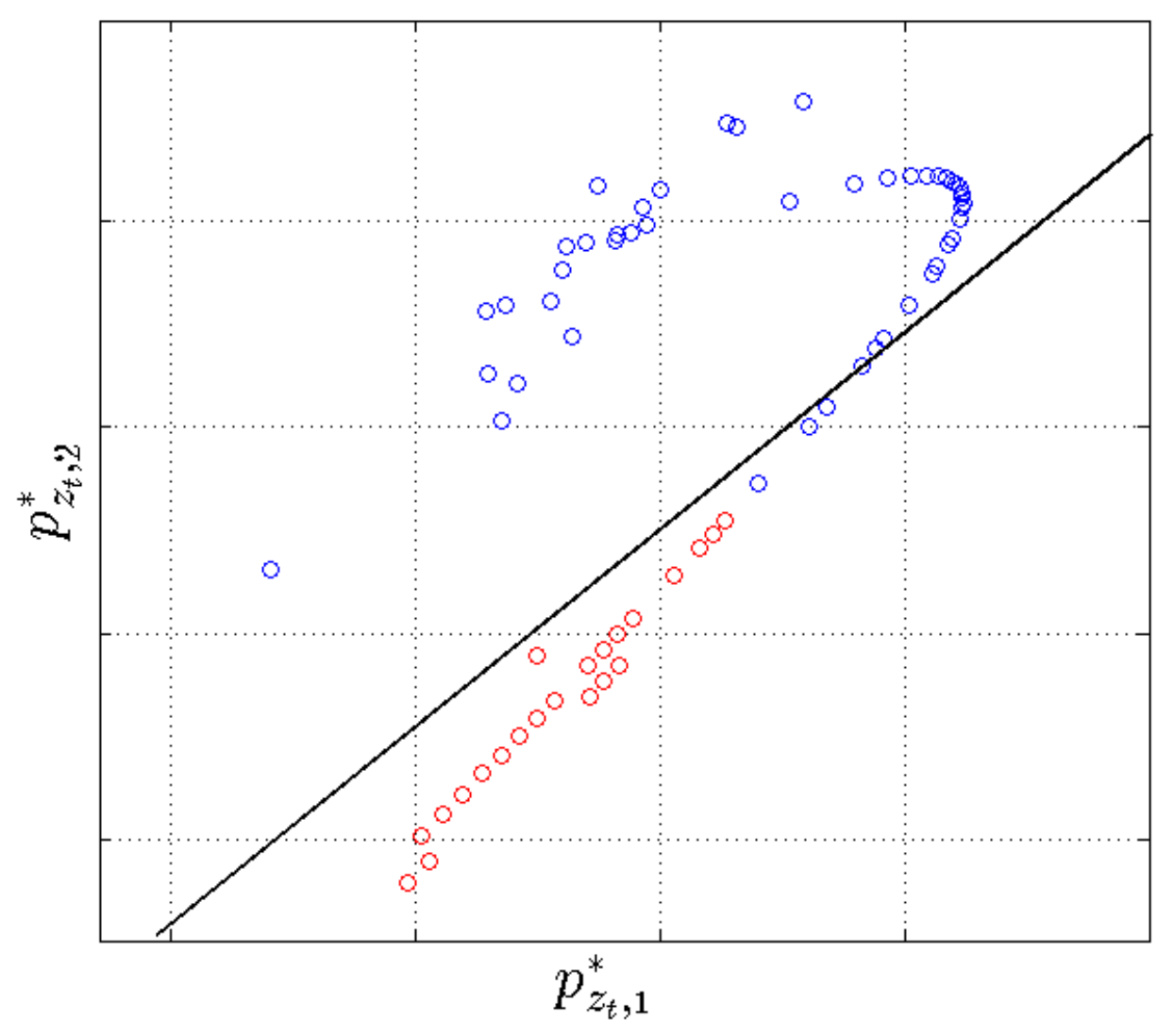
2.4.2. Segmentation with GMMs
2.4.3. Segmentation with the Switching AR Model
2.4.4. Segmentation Context Mapping
3. Results and Discussion
3.1. Future Work
3.1.1. Simultaneous Multimodal Sensing
3.1.2. Real-Time Deployment
3.1.3. Contextual Learning
3.2. Limitations
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sha, K.; Zhan, G.; Shi, W.; Lumley, M.; Wiholm, C.; Arnetz, B. SPA: A smart phone assisted chronic illness self-management system with participatory sensing. In Proceedings of the 2nd International Workshop on Systems and Networking Support for Health Care and Assisted Living Environments, Breckenridge, CO, USA, 17 June 2008; pp. 5:1–5:3. [Google Scholar]
- Oliver, N.; Flores-Mangas, F. HealthGear: Automatic sleep apnea detection and monitoring with a mobile phone. J. Commun. 2007, 2, 1–9. [Google Scholar] [CrossRef]
- Maisonneuve, N.; Stevens, M.; Niessen, M.E.; Steels, L. NoiseTube: Measuring and mapping noise pollution with mobile phones. In Information Technologies in Environmental Engineering, Proceedings of the 4th International ICSC Symposium, Thessaloniki, Greece, 28–29 May 2009; Springer: Berlin, Germany, 2009; pp. 215–228. [Google Scholar]
- Mun, M.; Reddy, S.; Shilton, K.; Yau, N.; Burke, J.; Estrin, D.; Hansen, M.; Howard, E.; West, R.; Boda, P. PEIR, the personal environmental impact report, as a platform for participatory sensing systems research. In Proceedings of the 7th International Conference on Mobile Systems, Applications, and Services, Kraków, Poland, 22–25 June 2009; pp. 55–68. [Google Scholar]
- Thiagarajan, A.; Ravindranath, L.; LaCurts, K.; Madden, S.; Balakrishnan, H.; Toledo, S.; Eriksson, J. VTrack: Accurate, energy-aware road traffic delay estimation using mobile phones. In Proceedings of the 7th ACM Conference on Embedded Networked Sensor Systems, Berkeley, CA, USA, 4–6 November 2009; pp. 85–98. [Google Scholar]
- Zhan, A.; Little, M.A.; Harris, D.A.; Abiola, S.O.; Dorsey, E.R.; Saria, S.; Terzis, A. High Frequency Remote Monitoring of Parkinson’s Disease via Smartphone: Platform Overview and Medication Response Detection. arXiv, 2016; arXiv:abs/1601.00960. [Google Scholar]
- Arora, S.; Venkataraman, V.; Donohue, S.; Biglan, K.M.; Dorsey, E.R.; Little, M.A. High accuracy discrimination of Parkinson’s disease participants from healthy controls using smartphones. In Proceedings of the 2014 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Florence, Italy, 4–9 May 2014; pp. 3641–3644. [Google Scholar]
- Joundi, R.A.; Brittain, J.S.; Jenkinson, N.; Green, A.L.; Aziz, T. Rapid tremor frequency assessment with the iPhone accelerometer. Parkinsonism Relat. Disord. 2011, 17, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Kostikis, N.; Hristu-Varsakelis, D.; Arnaoutoglou, M.; Kotsavasiloglou, C. Smartphone-based evaluation of parkinsonian hand tremor: Quantitative measurements vs clinical assessment scores. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 906–909. [Google Scholar]
- Hosseini, A.; Buonocore, C.M.; Hashemzadeh, S.; Hojaiji, H.; Kalantarian, H.; Sideris, C.; Bui, A.A.; King, C.E.; Sarrafzadeh, M. Feasibility of a Secure Wireless Sensing Smartwatch Application for the Self-Management of Pediatric Asthma. Sensors 2017, 17, 1780. [Google Scholar] [CrossRef] [PubMed]
- Andrzejewski, K.L.; Dowling, A.V.; Stamler, D.; Felong, T.J.; Harris, D.A.; Wong, C.; Cai, H.; Reilmann, R.; Little, M.A.; Gwin, J.; et al. Wearable sensors in Huntington disease: A pilot study. J. Huntingt. Dis. 2016, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Lorincz, K.; Hughes, R.; Huggins, N.; Growdon, J.; Standaert, D.; Akay, M.; Dy, J.; Welsh, M.; Bonato, P. Monitoring motor fluctuations in patients with Parkinson’s disease using wearable sensors. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Maetzler, W.; Domingos, J.; Srulijes, K.; Ferreira, J.J.; Bloem, B.R. Quantitative wearable sensors for objective assessment of Parkinson’s disease. Mov. Disord. 2013, 28, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Han, F.; Liu, H. Challenges of big data analysis. Natl. Sci. Rev. 2014, 1, 293–314. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Smith, D.V.; Timms, G. A novel machine learning approach toward quality assessment of sensor data. IEEE Sens. J. 2014, 14, 1035–1047. [Google Scholar] [CrossRef]
- Bulling, A.; Blanke, U.; Schiele, B. A Tutorial on Human Activity Recognition Using Body-worn Inertial Sensors. ACM Comput. Surv. 2014, 46, 33:1–33:33. [Google Scholar] [CrossRef]
- Zhang, R.; Peng, Z.; Wu, L.; Yao, B.; Guan, Y. Fault Diagnosis from Raw Sensor Data Using Deep Neural Networks Considering Temporal Coherence. Sensors 2017, 17, 549. [Google Scholar] [CrossRef] [PubMed]
- Hand, D.J. Classifier technology and the illusion of progress. Stat. Sci. 2006, 21, 1–14. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. In The Handbook of Brain Theory and Neural Networks; MIT Press: Cambridge, MA, USA, 1995; Volume 3361. [Google Scholar]
- Ho, T.K. Random decision forests. In Proceedings of the Third International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 1, pp. 278–282. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Kubota, K.J.; Chen, J.A.; Little, M.A. Machine learning for large-scale wearable sensor data in Parkinson’s disease: Concepts, promises, pitfalls, and futures. Mov. Disord. 2016, 31, 1314–1326. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Ozdalga, E.; Ozdalga, A.; Ahuja, N. The Smartphone in Medicine: A Review of Current and Potential Use Among Physicians and Students. J. Med. Internet Res. 2012, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Mosa, A.S.M.; Yoo, I.; Sheets, L. A systematic review of healthcare applications for smartphones. BMC Med. Inform. Decis. Mak. 2012, 12, 67. [Google Scholar] [CrossRef] [PubMed]
- Newell, S.A.; Girgis, A.; Sanson-Fisher, R.W.; Savolainen, N.J. The accuracy of self-reported health behaviors and risk factors relating to cancer and cardiovascular disease in the general population 1. Am. J. Prev. Med. 1999, 17, 211–229. [Google Scholar] [CrossRef]
- García-Magariño, I.; Medrano, C.; Plaza, I.; Oliván, B. A smartphone-based system for detecting hand tremors in unconstrained environments. Pers. Ubiquitous Comput. 2016, 20, 959–971. [Google Scholar] [CrossRef]
- Hammerla, N.Y.; Fisher, J.; Andras, P.; Rochester, L.; Walker, R.; Plötz, T. PD Disease State Assessment in Naturalistic Environments Using Deep Learning. In Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; pp. 1742–1748. [Google Scholar]
- Reimer, J.; Grabowski, M.; Lindvall, O.; Hagell, P. Use and interpretation of on/off diaries in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2004, 75, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Hoff, J.; van den Plas, A.; Wagemans, E.; van Hilten, J. Accelerometric assessment of levodopa-induced dyskinesias in Parkinson’s disease. Mov. Disord. 2001, 16, 58–61. [Google Scholar] [CrossRef]
- Cole, B.T.; Roy, S.H.; Luca, C.J.D.; Nawab, S.H. Dynamic neural network detection of tremor and dyskinesia from wearable sensor data. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 6062–6065. [Google Scholar]
- Giuffrida, J.P.; Riley, D.E.; Maddux, B.N.; Heldman, D.A. Clinically deployable Kinesia technology for automated tremor assessment. Mov. Disord. 2009, 24, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Timms, G.; De Souza, P.; DEste, C. A Bayesian framework for the automated online assessment of sensor data quality. Sensors 2012, 12, 9476–9501. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.J.; Minsker, B.S.; Amir, E. Real-time Bayesian anomaly detection in streaming environmental data. Water Resour. Res. 2009, 45. [Google Scholar] [CrossRef]
- Zwartjes, D.G.; Heida, T.; Van Vugt, J.P.; Geelen, J.A.; Veltink, P.H. Ambulatory monitoring of activities and motor symptoms in Parkinson’s disease. IEEE Trans. Biomed. Eng. 2010, 57, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Salarian, A.; Russmann, H.; Vingerhoets, F.J.; Burkhard, P.R.; Aminian, K. Ambulatory monitoring of physical activities in patients with Parkinson’s disease. IEEE Trans. Biomed. Eng. 2007, 54, 2296–2299. [Google Scholar] [CrossRef] [PubMed]
- Tzallas, A.T.; Tsipouras, M.G.; Rigas, G.; Tsalikakis, D.G.; Karvounis, E.C.; Chondrogiorgi, M.; Psomadellis, F.; Cancela, J.; Pastorino, M.; Waldmeyer, M.T.A.; et al. PERFORM: A system for monitoring, assessment and management of patients with Parkinson’s disease. Sensors 2014, 14, 21329–21357. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, E.H.; De La Torre, F.; Hebert, M. Temporal segmentation and activity classification from first-person sensing. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops (CVPR Workshops 2009), Miami, FL, USA, 20–25 June 2009; pp. 17–24. [Google Scholar]
- Gupta, P.; Dallas, T. Feature selection and activity recognition system using a single triaxial accelerometer. IEEE Trans. Biomed. Eng. 2014, 61, 1780–1786. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Nurmi, P.; Hammerla, N.; Plötz, T. Using unlabeled data in a sparse-coding framework for human activity recognition. Pervasive Mob. Comput. 2014, 15, 242–262. [Google Scholar] [CrossRef]
- Guo, T.; Yan, Z.; Aberer, K. An Adaptive Approach for Online Segmentation of Multi-dimensional Mobile Data. In Proceedings of the Eleventh ACM International Workshop on Data Engineering for Wireless and Mobile Access, Scottsdale, AZ, USA, 20 May 2012; pp. 7–14. [Google Scholar]
- Hodrick, R.J.; Prescott, E.C. Postwar U.S. Business Cycles: An Empirical Investigation. J. Money Credit Bank. 1997, 29, 1–16. [Google Scholar] [CrossRef]
- Antonsson, E.K.; Mann, R.W. The frequency content of gait. J. Biomech. 1985, 18, 39–47. [Google Scholar] [CrossRef]
- Little, M.A.; Jones, N.S. Generalized methods and solvers for noise removal from piecewise constant signals. I. Background theory. Proc. R. Soc. Lond. A Math. Phys. Eng. Sci. 2011, 467, 3088–3114. [Google Scholar] [CrossRef] [PubMed]
- Arce, G.R. Nonlinear Signal Processing: A Statistical Approach; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005. [Google Scholar]
- Rabiner, L.R. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 1989, 77, 257–286. [Google Scholar] [CrossRef]
- Gao, D.; Reiter, M.K.; Song, D. Behavioral distance measurement using hidden Markov models. In Recent Advances in Intrusion Detection, Proceedings of the International Workshop on Recent Advances in Intrusion Detection, RAID 2006, Hamburg, Germany, 20–22 September 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 19–40. [Google Scholar]
- Raykov, Y.P.; Ozer, E.; Dasika, G.; Boukouvalas, A.; Little, M.A. Predicting room occupancy with a single passive infrared (PIR) sensor through behavior extraction. In Proceedings of the 2016 ACM International Joint Conference on Pervasive and Ubiquitous Computing, Heidelberg, Germany, 12–16 September 2016; pp. 1016–1027. [Google Scholar]
- Chung, P.C.; Liu, C.D. A daily behavior enabled hidden Markov model for human behavior understanding. Pattern Recognit. 2008, 41, 1572–1580. [Google Scholar] [CrossRef]
- Juang, H.; Rabiner, L.R. Hidden Markov models for speech recognition. Technometrics 1991, 33, 251–272. [Google Scholar] [CrossRef]
- Gales, M.; Young, S. The application of hidden Markov models in speech recognition. Found. Trends Signal Process. 2007, 1, 195–304. [Google Scholar] [CrossRef]
- Oh, S.M.; Rehg, J.M.; Balch, T.; Dellaert, F. Learning and inferring motion patterns using parametric segmental switching linear dynamic systems. Int. J. Comput. Vis. 2008, 77, 103–124. [Google Scholar] [CrossRef]
- Jilkov, V.P.; Rong, X.L. Online Bayesian estimation of transition probabilities for Markovian jump systems. IEEE Trans. Signal Process. 2004, 52, 1620–1630. [Google Scholar] [CrossRef]
- Li, C.; Andersen, S.V. Efficient blind system identification of non-Gaussian autoregressive models with HMM modeling of the excitation. IEEE Trans. Signal Process. 2007, 55, 2432–2445. [Google Scholar] [CrossRef]
- Chiang, J.; Wang, Z.J.; McKeown, M.J. A hidden Markov, multivariate autoregressive (HMM-mAR) network framework for analysis of surface EMG (sEMG) data. IEEE Trans. Signal Process. 2008, 56, 4069–4081. [Google Scholar] [CrossRef]
- Goldwater, S.; Griffiths, T. A fully Bayesian approach to unsupervised part-of-speech tagging. In Proceedings of the 45th Annual Meeting of the Association of Computational Linguistics, Prague, Czech Republic, 25–27 June 2007; pp. 744–751. [Google Scholar]
- Johnson, M.; Duvenaud, D.K.; Wiltschko, A.; Adams, R.P.; Datta, S.R. Composing graphical models with neural networks for structured representations and fast inference. In Advances in Neural Information Processing Systems 29; Neural Information Processing Systems: Montréal, QC, Canada, 2016; pp. 2946–2954. [Google Scholar]
- Fox, E.; Sudderth, E.B.; Jordan, M.I.; Willsky, A.S. Nonparametric Bayesian learning of switching linear dynamical systems. In Advances in Neural Information Processing Systems 21; Neural Information Processing Systems: Montréal, QC, Canada, 2009; pp. 457–464. [Google Scholar]
- Teh, Y.W.; Jordan, M.I.; Beal, M.J.; Blei, D.M. Sharing clusters among related groups: Hierarchical Dirichlet processes. In Advances in Neural Information Processing Systems 17; Neural Information Processing Systems: Montréal, QC, Canada, 2005; pp. 1385–1392. [Google Scholar]
- Beal, M.J.; Ghahramani, Z.; Rasmussen, C.E. The Infinite Hidden Markov Model. In Advances in Neural Information Processing Systems 14; Neural Information Processing Systems: Montréal, QC, Canada, 2002; pp. 577–584. [Google Scholar]
- Hand, D.J.; Yu, K. Idiot’s Bayes—Not so stupid after all? Int. Stat. Rev. 2001, 69, 385–398. [Google Scholar]
- Hughes, M.C.; Stephenson, W.T.; Sudderth, E. Scalable Adaptation of State Complexity for Nonparametric Hidden Markov Models. In Advances in Neural Information Processing Systems 28; Neural Information Processing Systems: Montréal, QC, Canada, 2015; pp. 1198–1206. [Google Scholar]
- Raykov, Y.P.; Boukouvalas, A.; Little, M.A. Simple approximate MAP inference for Dirichlet processes mixtures. Electron. J. Stat. 2016, 10, 3548–3578. [Google Scholar] [CrossRef]
- Leech, C.; Raykov, Y.P.; Ozer, E.; Merrett, G.V. Real-time room occupancy estimation with Bayesian machine learning using a single PIR sensor and microcontroller. In Proceedings of the 2017 IEEE Sensors Applications Symposium (SAS), Glassboro, NJ, USA, 13–15 March 2017; pp. 1–6. [Google Scholar]
- Pavel, M.; Hayes, T.; Tsay, I.; Erdogmus, D.; Paul, A.; Larimer, N.; Jimison, H.; Nutt, J. Continuous assessment of gait velocity in Parkinson’s disease from unobtrusive measurements. In Proceedings of the 2007 3rd International IEEE/EMBS Conference on Neural Engineering, Kohala Coast, HI, USA, 2–5 May 2007; pp. 700–703. [Google Scholar]
| Behavior | BA | TP | TN |
|---|---|---|---|
| Walking | 95% | 96% | 93% |
| Standing up straight | 95% | 98% | 91% |
| Phone stationary | 98% | 100% | 95% |
| Sustained phonation | 98% | 99% | 97% |
| Walking Tests | Balance Tests | Voice Tests | |
|---|---|---|---|
| Nonparametric switching AR + naive Bayes | |||
| BA | 85% (11%) | 81% (14%) | 89% (8%) |
| TP | 85% (18%) | 81% (16%) | 88% (9%) |
| TN | 90% (8%) | 88% (9%) | 91% (9%) |
| GMM + running median filtering | |||
| BA | 62% | 24% | 99% |
| TP | 80% | 74% | 86% |
| TN | 89% | 82% | 96% |
| randomised classifier | |||
| BA | 50% (1%) | 50% (0.2%) | 53% (24%) |
| TP | 1% (0.4%) | 0.4% (0.02%) | 99% (1%) |
| TN | 100% | 100% | 6% (23%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badawy, R.; Raykov, Y.P.; Evers, L.J.W.; Bloem, B.R.; Faber, M.J.; Zhan, A.; Claes, K.; Little, M.A. Automated Quality Control for Sensor Based Symptom Measurement Performed Outside the Lab. Sensors 2018, 18, 1215. https://doi.org/10.3390/s18041215
Badawy R, Raykov YP, Evers LJW, Bloem BR, Faber MJ, Zhan A, Claes K, Little MA. Automated Quality Control for Sensor Based Symptom Measurement Performed Outside the Lab. Sensors. 2018; 18(4):1215. https://doi.org/10.3390/s18041215
Chicago/Turabian StyleBadawy, Reham, Yordan P. Raykov, Luc J. W. Evers, Bastiaan R. Bloem, Marjan J. Faber, Andong Zhan, Kasper Claes, and Max A. Little. 2018. "Automated Quality Control for Sensor Based Symptom Measurement Performed Outside the Lab" Sensors 18, no. 4: 1215. https://doi.org/10.3390/s18041215
APA StyleBadawy, R., Raykov, Y. P., Evers, L. J. W., Bloem, B. R., Faber, M. J., Zhan, A., Claes, K., & Little, M. A. (2018). Automated Quality Control for Sensor Based Symptom Measurement Performed Outside the Lab. Sensors, 18(4), 1215. https://doi.org/10.3390/s18041215







