Optical DNA Biosensor Based on Square-Planar Ethyl Piperidine Substituted Nickel(II) Salphen Complex for Dengue Virus Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Instrumentation
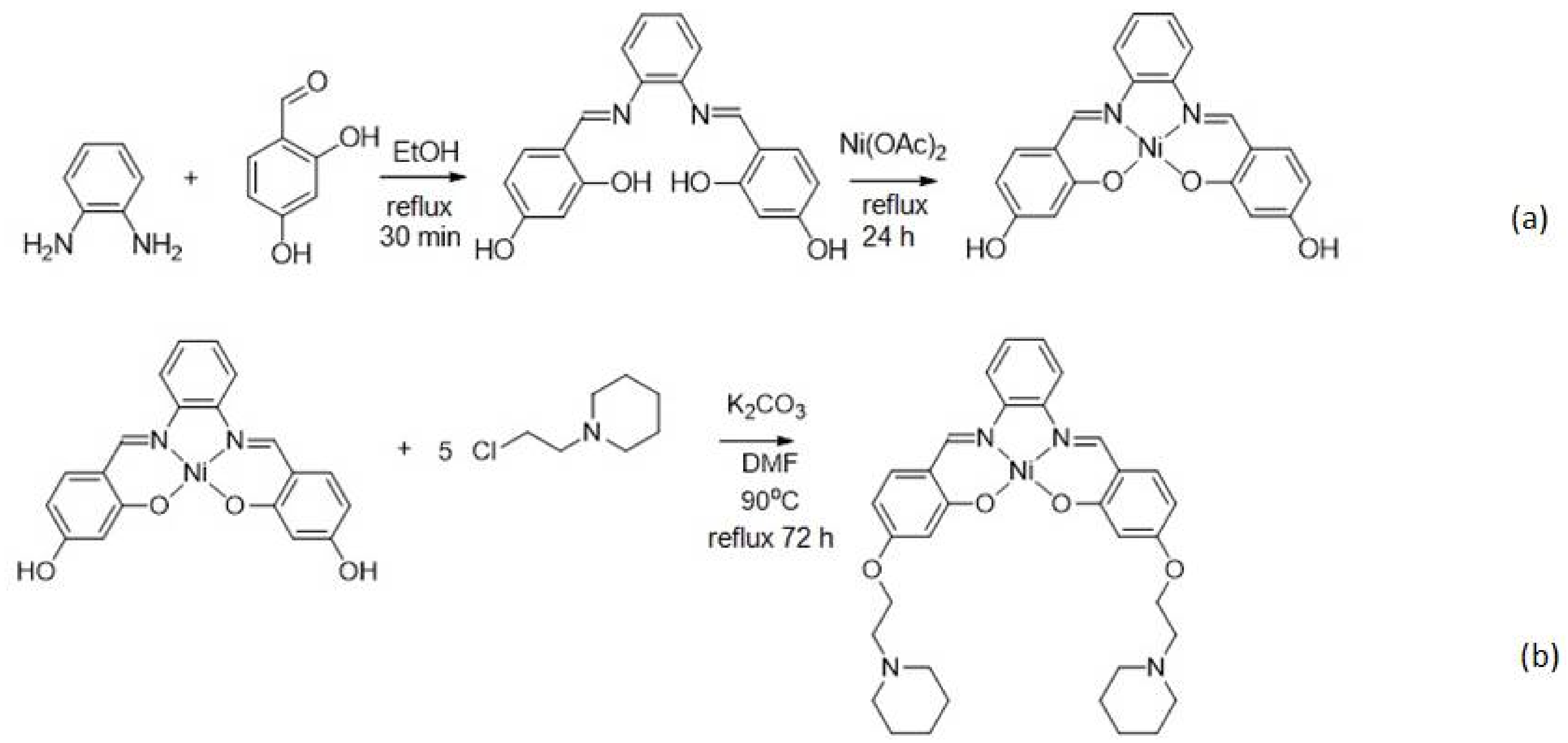
2.3. Synthesis of Nickel(II) Salphen Complex with Piperidine Side Chain
2.4. Synthesis of Aminated Porous Silica Particles
2.5. Fabrication of PSiNs-Based Optical DNA Biosensor
2.6. Characterization and Performance Evaluation of Reflectance Dengue Virus DNA Biosensor
3. Results and Discussion
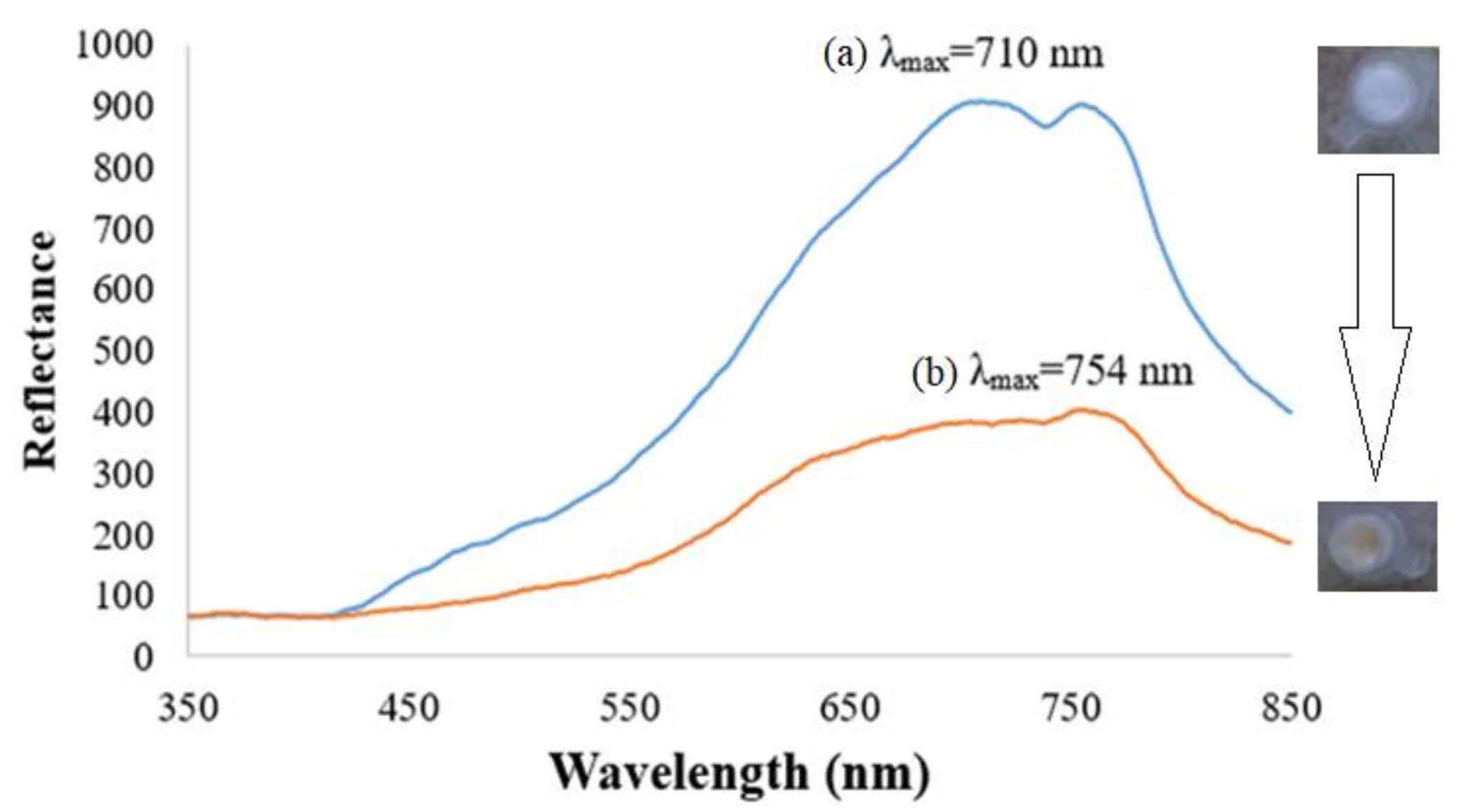
3.1. Characterization of Piperidine Side Chain-Modified Nickel(II) Salphen Complex
3.2. UV-Vis Titration Studies to Evaluate the Interaction between Nickel(II) Salphen Complex and DNA
3.3. DNA Biosensor Based on APTS-Modified Porous Silica Nanospheres
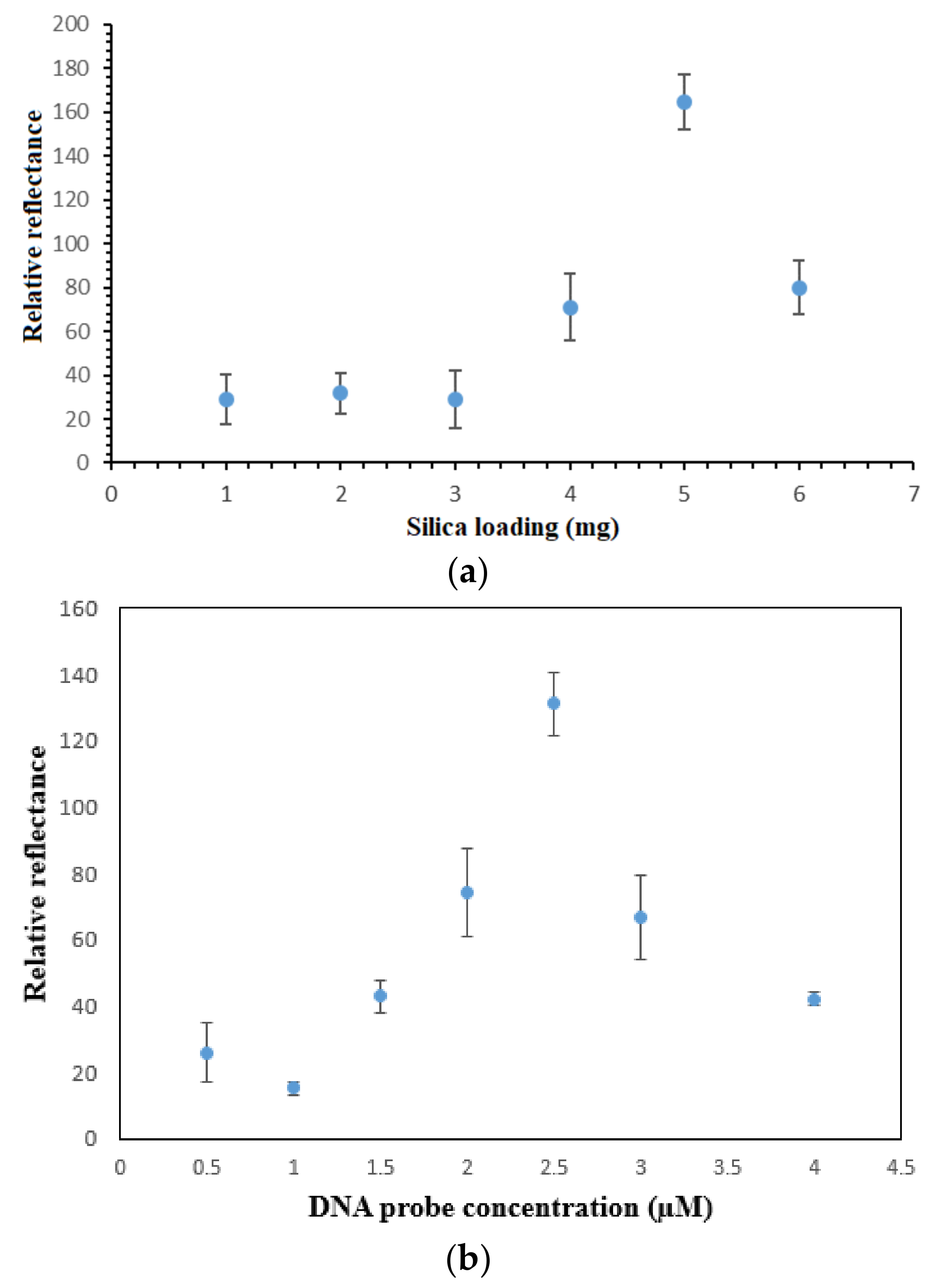
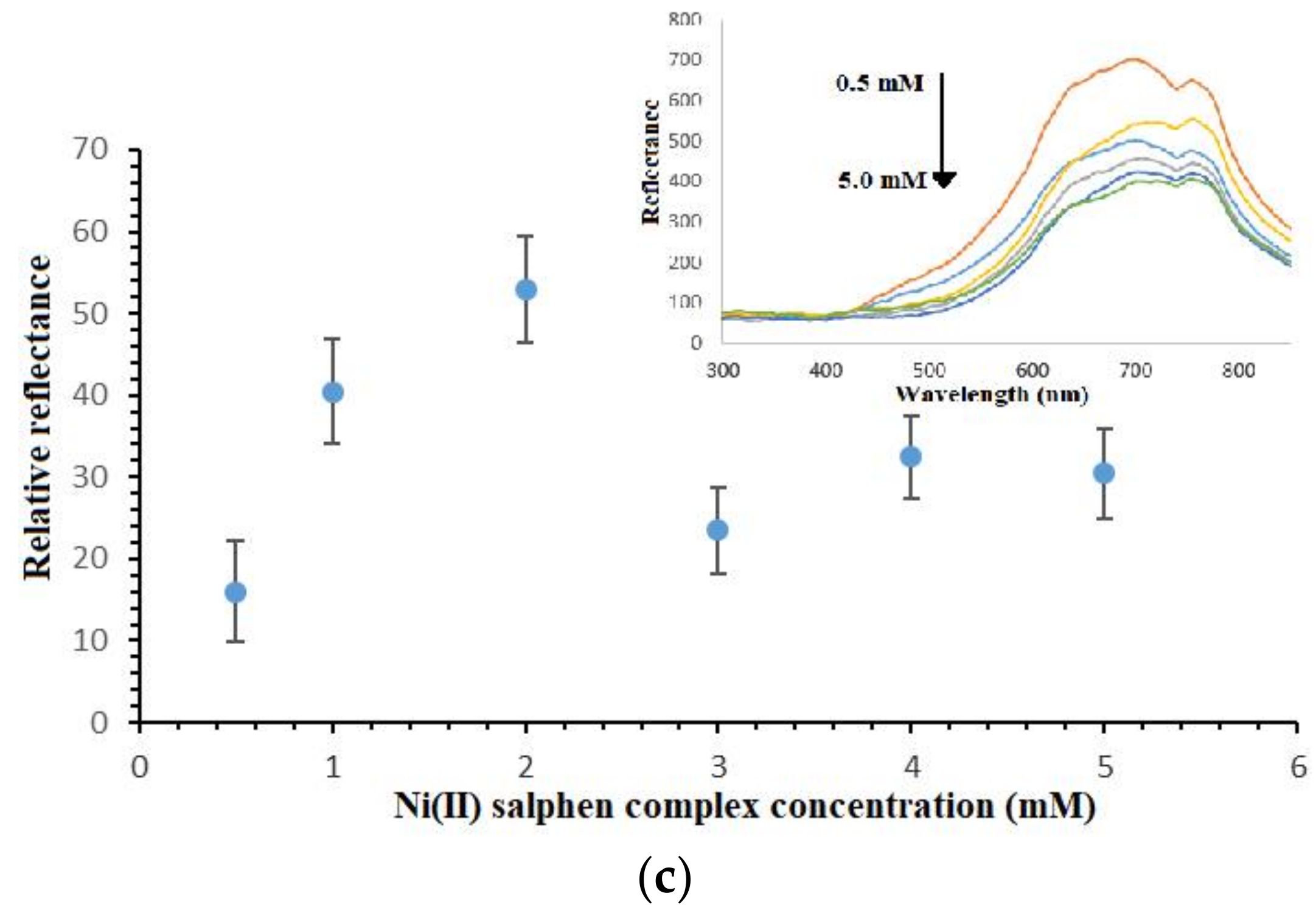
3.4. PSiNs, DNA Probe and Metal Complex Loading Optimizations
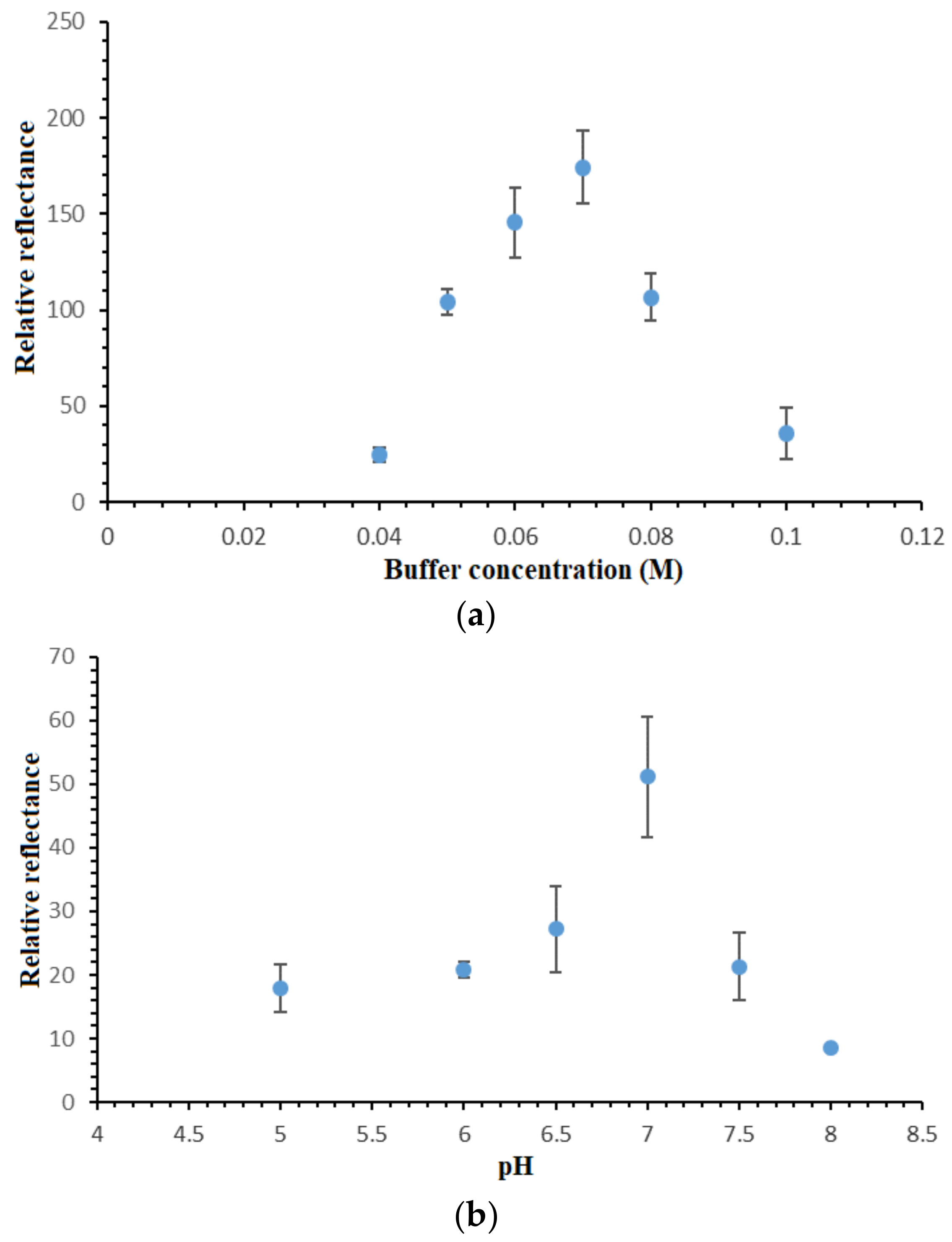
3.5. Effect of Buffer Capacity and pH on the DNA Hybridization Reaction
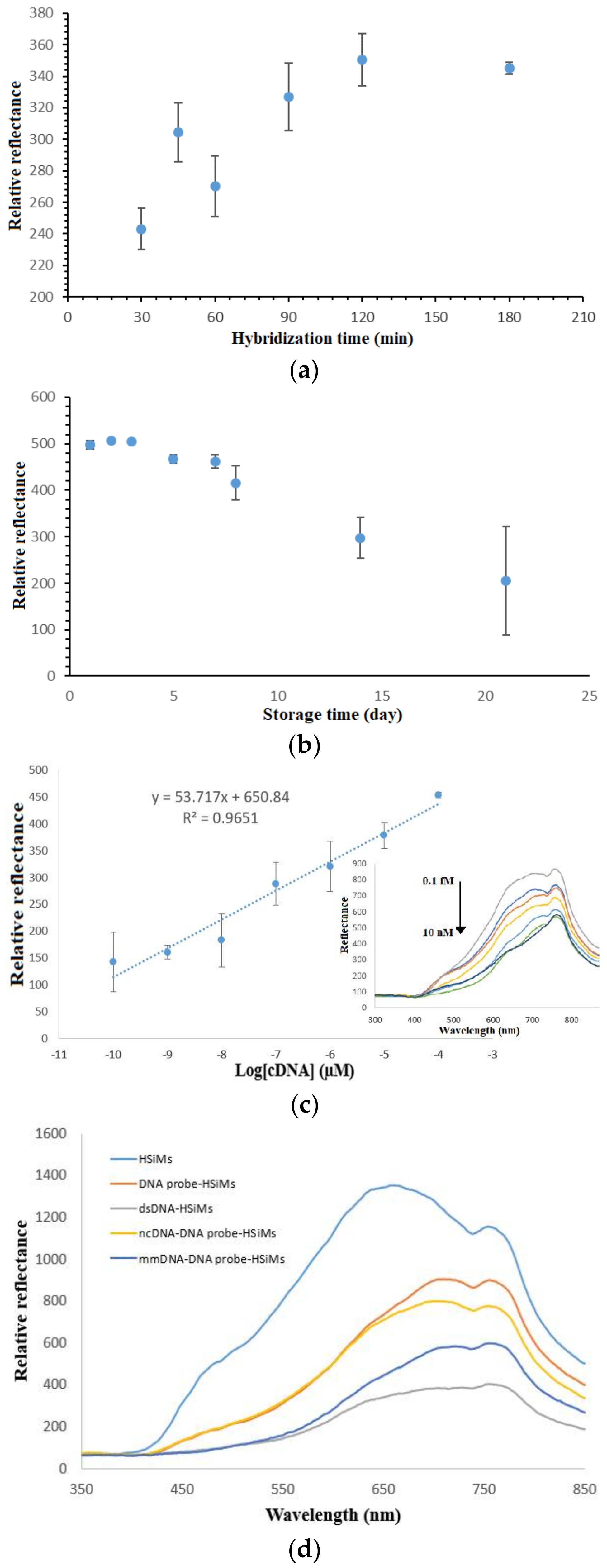
3.6. Determination of DNA Biosensor’s Hybridization Duration, Lifetime, Linear Range and Selectivity
3.7. Recovery Test
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nascimento, H.P.O.; Oliveira, M.D.L.; Melo, C.P.D.; Silva, G.J.L.; Cordeiro, M.T.; Andrade, C.A.S. An impedimetric biosensor for detection of dengue serotype at picomolar concentration based on gold nanoparticle-polyaniline hybrid composites. Colloids Surf. B Biointerfaces 2011, 86, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Toh, C.S. Impedimetric DNA biosensor based on a nanoporous alumina membrane for the detection of the specific oligonucleotide sequence of dengue virus. Sensors 2013, 13, 7774–7785. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Xu, Y.; Lu, P.; Xie, Y.; Xi, Z. The endosymbiotic bacterium wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Zhang, L.; Huang, M.J.; Luo, Z.H.H.; Tay, G.K.I.; Lim, E.A.; Kang, T.G.; Chen, Y. Silicon nanowire biosensor for highly sensitive and rapid detection of Dengue virus. Sens. Actuators B Chem. 2010, 146, 138–144. [Google Scholar] [CrossRef]
- Mizuno, Y.; Kotaki, A.; Harda, F.; Tajima, S.; Kurane, I.; Takasaki, T. Confirmation of dengue virus infection by detection of dengue virus type 1 genome in urine and saliva but not in plasma. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 738–739. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Dean-Nystrom, E.A.; Casey, T.A. Semi-automated flurogenic PCR assays (TaqMan) for rapid detection of Escherichia coli O157:H7 and other Shiga toxigenic E. coli. Mol. Cell Probes 1999, 13, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.K.; Devine, P.L. Evaluation of capture ELISA and rapid immunochromatographic test for the determination of Ig M dan Ig G antibodies produced during Dengue infection. Clin. Diagn. Virol. 1998, 10, 75–81. [Google Scholar] [CrossRef]
- Subramanian, A.; Irudayaraj, J.; Ryan, T. A mixed self-assembled monolayer-based surface plasmon immunosensor for detection of E. coli O157:H7. Biosens. Bioelectron. 2006, 21, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.S.; Ho, J.S.; Tan, C.H.; Wong, J.P.S.; Ng, L.C.; Toh, C.S. Development of an electrochemical membrane-based nanobiosensor for ultrasensitive detection of dengue virus. Anal. Chim. Acta 2012, 725, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.; Souza, E.; Ferreira, D.; Bezerra, W.; Borba, M.A.; Arruda, M.; Lopes, K.; Nascimento, G.; Martins, D.; Cordeiro, M.; et al. A sensitive and selective label-free electrochemical DNA biosensor for the detection of specific dengue virus serotypes 3 sequences. Sensors 2015, 15, 15562–15577. [Google Scholar] [CrossRef] [PubMed]
- Bouffier, L.; Wang, B.S.; Roget, A.; Livache, T.; Demeunynck, M.; Mailley, P. Electrochemical transduction of DNA hybridization at modified electrodes by using an electroactive pyridoacridone intercalator. Anal. Bioanal. Chem. 2014, 406, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Shoora, S.K.; Kumawat, L.K.; Jain, A.K. A highly selective colorimetric and turn-on fluorescent chemosensor based on 1-(2-pyridylazo)-2-naphthol for the detection of aluminium (III) ions. Sens. Actuators B Chem. 2015, 209, 15–24. [Google Scholar] [CrossRef]
- Shamsipur, M.; Memari, M.; Ganjali, M.R.; Norouzi, P.; Faridbod, F. Highly sensitive gold nanoparticles-based optical sensing of DNA hybridization using bis(8-hydroxyquinoline-5-solphonate)cerium(III) chloride as a novel fluorescence probe. J. Pharm. Biomed. Anal. 2016, 118, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Ghauch, A.; Rima, J.; Charef, A.; Suptil, J.; Fachinger, C.; Martin-Bouyer, M. Quantitative measurements of ammonium, hydrogenophosphate and Cu(II) by diffuse reflectance spectrometry. Talanta 1999, 48, 385–392. [Google Scholar] [CrossRef]
- Mariappan, M.; Suenaga, M.; Mukhopadhyay, A.; Maiya, B.G. Synthesis, structure, DNA binding and photonuclease activity of a nickel(II) complex with a N,N′-Bis(salicylidene)-9-(3,4-diaminophenyl)acridine ligand. Inorg. Chim. Acta 2012, 390, 95–104. [Google Scholar] [CrossRef]
- Vilar, R.; Arnal, A.A.; Buchholz, J.B.; Neidle, S. Effects of metal coordination geometry on stabilization of human telomeric quadruplex DNA by square-planar and square-pyramidal metal complexes. Inorg. Chem. 2008, 47, 11910–11919. [Google Scholar]
- Nur-Fadhilah, M.; Tan, L.L.; Nurul Huda, A.K.; Lee, Y.H.; Reza, M.I.H. Optical biosensing using newly synthesized metal salphen complexes: A potential DNA diagnostic tool. Sens. Actuators B Chem. 2017, 242, 176–188. [Google Scholar]
- Campbell, N.H.; Nurul Huda, A.K.; Parkinson, G.N.; Gunaratnam, M.; Petrucci, V.; Todd, A.K.; Vilar, R.; Neidle, S. Molecular Basis of Structure−Activity Relationships between Salphen Metal Complexes and Human Telomeric DNA Quadruplexes. J. Med. Chem. 2012, 55, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Sukri, S.A.M.; Lee, Y.H.; Nurul Huda, A.K. Synthesis and characterization of platinum(II) salphen complex and its interaction with calf tymus DNA. AIP Conf. Proc. 2014, 1614, 419–426. [Google Scholar]
- Kashanian, S.; Gholivand, M.B.; Ahmadi, F.; Taravati, A.; Colagar, A.H. DNA interaction with Al-N, N′-Bis(Salicylidene) 2,2′-Phenylendiamine Complex. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2007, 67, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Castor, K.J. Transition Metal Complexes as G-quadruplex DNA Binders. Ph.D. Thesis, McGill University, Montreal, QC, Canada, February 2014. [Google Scholar]
- Zhao, J.; Li, W.; Ma, R.; Chen, S.; Ren, S.; Jiang, T. Design, synthesis and DNA interaction study of new potential DNA bis-intercalators based on glucuronic acid. Int. J. Mol. Sci. 2013, 14, 16851–16865. [Google Scholar] [CrossRef] [PubMed]
- Topală, T.; Bodoki, A.; Oprean, L.; Oprean, R. Experimental Techniques Employed in the Study of Metal Complexes-DNA-Interactions. Farmacia 2014, 62, 1049–1061. [Google Scholar]
- Li, F.; Chen, W.; Zhang, S. Development of DNA electrochemical biosensor based on covalent immobilization of probe DNA by direct coupling of sol–gel and self-assembly technologies. Biosens. Bioelectron. 2008, 24, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Keighley, S.D.; Li, P.; Estrela, P.; Migliorato, P. Optimization of DNA immobilization on gold electrodes for label-free detection by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2008, 23, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Watterson, J.H.; Piunno, P.A.E.; Krull, U.J. Towards the optimization of an optical DNA sensor: Control of selectivity coefficients and relative surface affinities. Anal. Chim. Acta 2002, 457, 29–38. [Google Scholar] [CrossRef]
- Wong, E.L.S.; Gooding, J.J. Charge transfer through DNA: A selective electrochemical DNA biosensor. Anal. Chem. 2006, 78, 2138–2144. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tan, W.; Wang, K.; Xiao, D.; Yang, X.; He, X.; Tang, Z. Ultrasensitive Optical DNA Biosensor Based on Surface Immobilization of Molecular Beacon by a Bridge Structure. Anal. Sci. 2001, 17, 1149–1153. [Google Scholar] [CrossRef] [PubMed]
- Alizar, U.; Lee, Y.H.; Sharina, A.H.; Tan, L.L. An electrochemical DNA microbiosensor based on succinimide-modified acrylic microspheres. Sensors 2012, 12, 5445–5460. [Google Scholar]
- Metzenberg, S. Working with DNA: The Basics; Taylor & Francis Group: Florence, KY, USA, 2007; p. 56. [Google Scholar]
- Teles, F.R.R.; Foncessca, L.P. Trends in DNA biosensors. Talanta 2008, 77, 606–623. [Google Scholar] [CrossRef]
- Hames, B.B.; Higgins, S.J. Nucleic Acid Hybridisation—A Practical Approach; Oxford University Press: New York, NY, USA, 1985; p. 78. [Google Scholar]
- Chan, C.P.; Choi, J.W.; Cao, K.Y.; Wang, M.; Gao, Y.; Zhuo, D.H.; Di, B.; Xu, H.F.; Leung, M.F.; Bergmann, A.; et al. Detection of serum neopterin for early assessment of dengue virus infection. J. Infect. 2006, 53, 152–158. [Google Scholar] [CrossRef] [PubMed]


| DNA | Base Sequences |
|---|---|
| DNA probe | 5′-NH2-TTTTGTCCTGCTCTTCATTTAGGCTGGGTT-3′ |
| cDNA | 5′-AACCCAGCCTAAATGAAGAGCAGGACAAAA-3′ |
| ncDNA | 5′-AACGCCGATACCATTACTTATACCGCGACG-3′ |
| 1-base mismatched DNA | 5′-AACCCATCCTAAATGAAGAGCAGGACAAAA-3′ |
| Samples | Spiked cDNA Concentration (µM) 1 | Reflectance Response (a.u.) | cDNA Concentration Determined by DNA Biosensor (µM) 2 | Recovery (%) 3 |
|---|---|---|---|---|
| Saliva | 0.00 | 1111.22 ± 83.32 | 7.65 × 10−13 | - |
| 1.00 × 10−4 | 678.12 ± 14.26 | 9.80 × 10−5 | 98.0 | |
| 1.00 × 10−5 | 727.67 ± 12.37 | 1.13 × 10−5 | 113.0 | |
| 1.00 × 10−7 | 833.2 ± 3.52 | 1.15 × 10−7 | 115.0 | |
| Urine | 0.00 | 1068.10 ± 89.13 | 7.65 × 10−13 | - |
| 1.00 × 10−4 | 641.80 ± 29.34 | 9.66 × 10−5 | 96.6 | |
| 1.00 × 10−5 | 738.53 ± 63.89 | 9.97 × 10−6 | 99.7 | |
| 1.00 × 10−7 | 820.97 ± 44.61 | 9.16 × 10−8 | 91.6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ariffin, E.Y.; Tan, L.L.; Abd. Karim, N.H.; Yook Heng, L. Optical DNA Biosensor Based on Square-Planar Ethyl Piperidine Substituted Nickel(II) Salphen Complex for Dengue Virus Detection. Sensors 2018, 18, 1173. https://doi.org/10.3390/s18041173
Ariffin EY, Tan LL, Abd. Karim NH, Yook Heng L. Optical DNA Biosensor Based on Square-Planar Ethyl Piperidine Substituted Nickel(II) Salphen Complex for Dengue Virus Detection. Sensors. 2018; 18(4):1173. https://doi.org/10.3390/s18041173
Chicago/Turabian StyleAriffin, Eda Yuhana, Ling Ling Tan, Nurul Huda Abd. Karim, and Lee Yook Heng. 2018. "Optical DNA Biosensor Based on Square-Planar Ethyl Piperidine Substituted Nickel(II) Salphen Complex for Dengue Virus Detection" Sensors 18, no. 4: 1173. https://doi.org/10.3390/s18041173
APA StyleAriffin, E. Y., Tan, L. L., Abd. Karim, N. H., & Yook Heng, L. (2018). Optical DNA Biosensor Based on Square-Planar Ethyl Piperidine Substituted Nickel(II) Salphen Complex for Dengue Virus Detection. Sensors, 18(4), 1173. https://doi.org/10.3390/s18041173




