An In-Vitro Optical Sensor Designed to Estimate Glycated Hemoglobin Levels
Abstract
:1. Introduction
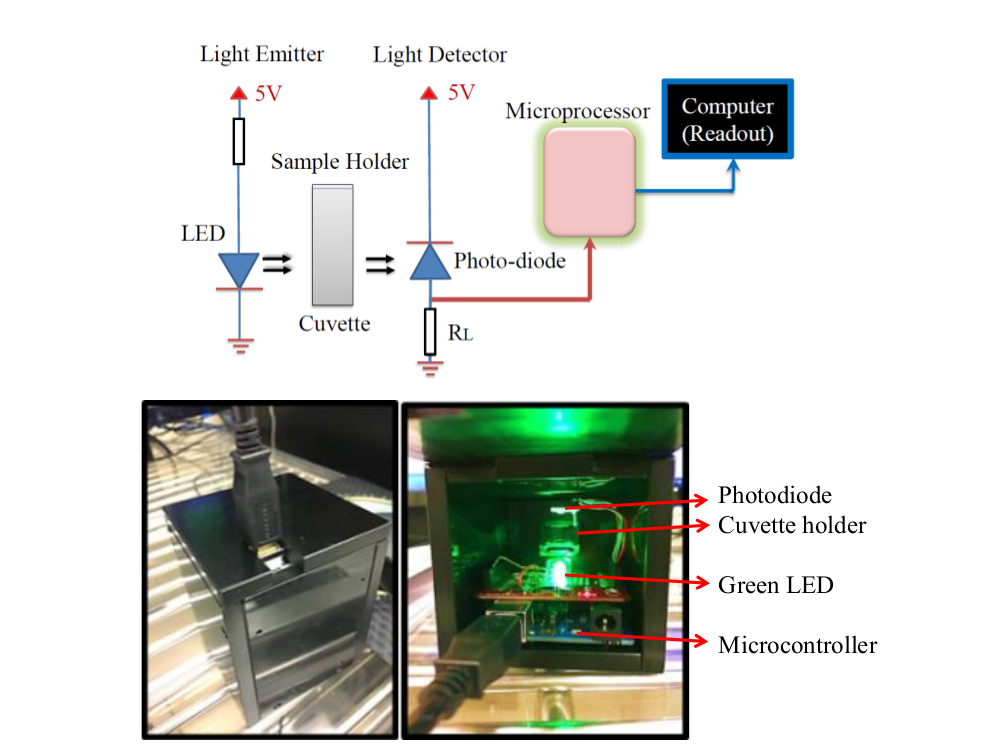
2. Methodology
3. Results and Discussion
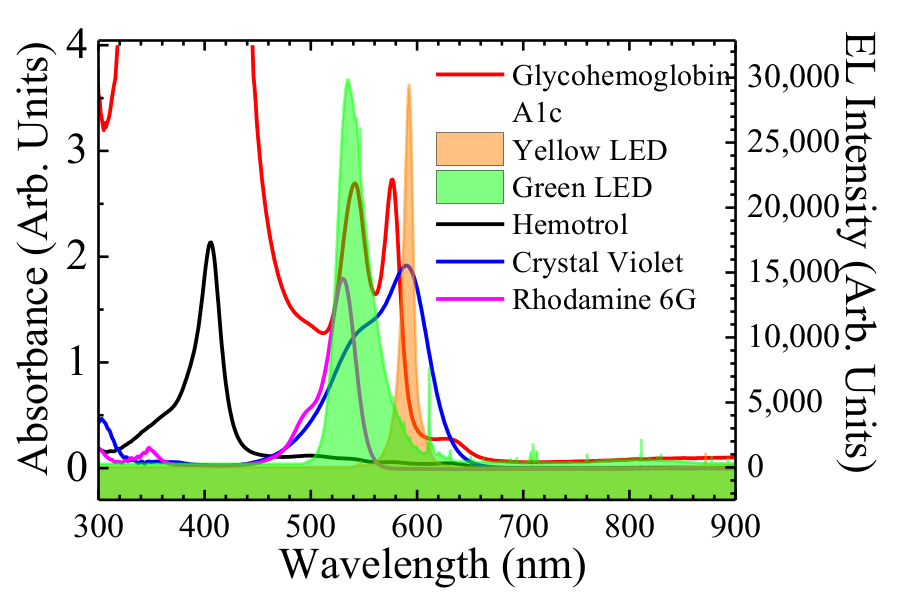
3.1. Molar Absorption Coefficient Calculation
3.2. Estimation of HbA1c Percentage in a Solution
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
#include <EEPROM.h>
#include <Math.h>
// Sensor input variables
int sensorPin = A0; // select the input pin for the photodiode
int ledPin2 = 7; // select the pin for the LED
int sensorValue = 0; // variable to store the value coming from the sensor
int eeAddress = 0; // Location we want the data to be put in EPROM.
//sequentially stores 10 readings from your analog sensor into an array, one by one.
//With each new value, the sum of all the numbers is generated and divided
//producing an average value
const int numReadings = 500; // Number of sensor readings you want to take at one concentration
int total = 0; // sensor reading:
double volts = 0;
double current = 0;
double totalV = 0; // the running total Voltage
double totalI = 0; // the running total Current
double averageV = 0; // the average Voltage
double averageI = 0; // the average Current
double concentration = 0; // sample concentration
int maxcount = 11; // No. of sample concentrations
//variables for exponential regression
double x[11]; // always set the size of array equal to maxcount
double y[11]; // always set the size of array equal to maxcount
double dx[10]; // always set the size of array equal to (maxcount-1)
double dy[10]; // always set the size of array equal to (maxcount-1)
double cx[10]; // always set the size of array equal to (maxcount-1)
double dq[10]; // always set the size of array equal to (maxcount-1)
double b_initial[9]; // always set the size of array equal to (maxcount-2)
double a_initial[10]; // always set the size of array equal to (maxcount-1)
double b_int_avg=0;
double a_avg=0;
double c_initial[11]; // always set the size of array equal to maxcount
double lnyc[11]; // always set the size of array equal to maxcount
double xbar=0;
double ybar=0;
double xybar=0;
double xsqbar=0;
double slope=0;
double intercept=0;
double syx = 0;
double std_err = 0;
double stderr_Slope = 0;
double stderr_Intercept = 0;
double a=0;
double b=0;
double c=0;
double a_err=0;
double b_err=0;
double c_err=0;
int abor(double y[11],double cc) // always set the size of array equal to maxcount
{
int returnpara=0;
double e=0;
for (int i=0; i<maxcount-1; i++)
{
e = (y[i+1]-cc)/(y[i]-cc);
if(e<=0)
{
returnpara=1;
break;
}
}
return returnpara;
}
// always set the size of array y and x equal to maxcount, dx equal to maxcount-1
double Devi(double cc, double y[11], double x[11],double dx[10])
{
double avG=0;
double pre_ee=0;
double ee=0;
double growthfactor[maxcount-1];
double Deviation = 0;
for (int i=0; i<maxcount-1; i++)
{
pre_ee = (y[i+1]-cc)/(y[i]-cc);
ee = log(pre_ee);//denominator
avG = avG + ee;
growthfactor[i] = exp(ee/dx[i]); //save
}
avG = exp(avG/(x[maxcount-1]-x[0])); //exp added
ee = 0;
for (int i=0; i<maxcount-1; i++)
{
Deviation = Deviation + abs(avG-growthfactor[i]);
}
return Deviation;
}
void setup()
{ // initialize serial communication with computer:
Serial.begin(9600); // Set data rate to 9600 bps
pinMode(ledPin2, OUTPUT);
}
void loop()
{
digitalWrite(ledPin2, HIGH);
delay (3000); // delay to start recording values or LED stabilizing delay
for (int j=0; j<maxcount; j++)
{
totalV = 0;
total = 0;
totalI = 0;
for (int i = 0; i<numReadings; i++)
{
sensorValue = analogRead(sensorPin); // read the value from the sensor:
volts = double (sensorValue) * (5.0 / 1023.0); // map it to the range of the analog out:
Serial.print(“\t Sensor = ”); // print the results to the serial monitor:
Serial.println(sensorValue);
Serial.print(“ Volts = ”);
Serial.println(volts,4);
delay(10); // wait 10 milisecond before the next loop
// for the analog-to-digital converter to settle
// after the last reading:
total = total+ sensorValue;
totalV = totalV+volts;
}
averageV = totalV / numReadings; // calculate the average Voltage:
if (j==1)
concentration=concentration + 4;
else if (j>1)
concentration = concentration + 2;
Serial.print(“ \t Average Voltage(Volts) = ”); // print the result to the serial monitor
Serial.println(averageV,4); // send it to the computer as ASCII digits
Serial.print(“ Concentration (mM) = ”); // print the result to the serial monitor
Serial.println(concentration,3); // send it to the computer as ASCII digits
x[j] = concentration;
y[j] = averageV;
delay(14000); // delay to change sample concentration
}
// Exponetial curve fitting t equation y = A+B*exp(C*x)
// Estimation of c using a and b.
// initial variables
for (int j=0; j<maxcount-1; j++)
{
dx[j] = x[j+1]-x[j];
dy[j] = y[j+1]-y[j];
cx[j] = (x[j]+x[j+1])/2;
dq[j] = dy[j]/dx[j];
}
// pre-estimation of b
b_int_avg =0;
for (int jj=0; jj<maxcount-2; jj++)
{
b_initial[jj] = (log(dq[jj+1]/dq[jj]))/(cx[jj+1]-cx[jj]);
}
for (int jj=0; jj<maxcount-2; jj++)
{
b_int_avg = b_int_avg + b_initial[jj];
}
b_int_avg = b_int_avg/(maxcount-2);
Serial.print(“ b-initial = ”);
Serial.println(b_int_avg,4);
// pre-estimaton of a
for (int j=0; j<maxcount-1; j++)
{
a_initial[j]= dy[j]/(exp(b_int_avg*x[j+1])-exp(b_int_avg*x[j]));
}
a_avg =0;
for (int j=0; j<maxcount-1; j++)
{
a_avg += a_initial[j];
}
a_avg = a_avg/(maxcount-1);
Serial.print(“ a-initial = ”);
Serial.println(a_avg,4);
// estimation of c and stanndard error in c;
for (int j=0; j<maxcount; j++)
{
c_initial[j] = y[j] - a_avg*exp(b_int_avg*x[j]);
}
for (int j=0; j<maxcount; j++)
{
c += c_initial[j];
}
c=c/maxcount;
double scaledY = 0.001;
c=y[maxcount-1]-scaledY;
double deltaC =0;
if (a_avg>0)
deltaC = -scaledY; //move away from asymptote first
else
deltaC = scaledY;
double abortt=0;
abortt = abor(y,c);
while (abortt>0)
{
c=c+deltaC;
abortt = abor(y,c);
}
Serial.print(“ c-initial = ”);
Serial.println(c,4);
double Bvar1=0;
double Bvar2=0;
Bvar1=Devi(c,y,x,dx);
deltaC=-deltaC;
Serial.print(“ delta C = ”);
Serial.println(deltaC,4);
double w = 0;
while(abs(deltaC)>0.00001 && w<100)
{ w=w+1;
Serial.print(“ w = ”);
Serial.print(w);
Serial.print(“ deltaC = ”);
Serial.println(deltaC,6);
Serial.print(“ c = ”);
Serial.println(c,6);
Serial.print(“ Bvar1 = ”);
Serial.println(Bvar1,4);
abortt = abor(y,c+deltaC);
Serial.print(“ abort = ”);
Serial.println(abortt,4);
if (abortt>0)
{
deltaC=deltaC/2;
}
else
{
c=c+deltaC;
Bvar2 = Devi(c,y,x,dx);
Serial.print(“ Bvar2 = ”);
Serial.println(Bvar2,4);
if (Bvar2<Bvar1)
{
Bvar1=Bvar2;
}
else
{
deltaC=-deltaC/2;
}
}
}
// linear regression on ln(y-c) = ln(a) + b*x using estimated c value
// consider a linear plot for ln(y-c) vs. x; slope=b and intercept = ln (a) => a = exp (intercept)
for (int j=0; j<maxcount; j++)
{
lnyc[j] = log(y[j]-c);
}
for (int j=0; j<maxcount; j++)
{
xbar+=x[j];
ybar+=lnyc[j];
xybar+=x[j]*lnyc[j];
xsqbar+=x[j]*x[j];
}
xbar=xbar/maxcount;
ybar=ybar/maxcount;
xybar=xybar/maxcount;
xsqbar=xsqbar/maxcount;
slope=(xybar-(xbar*ybar))/(xsqbar-(xbar*xbar));
intercept=ybar-(slope*xbar);
double x_dev =0;
for (int j=0; j<maxcount; j++)
{
x_dev += sq(x[j] - xbar);
}
double y_cap[maxcount];
for (int j=0; j<maxcount; j++)
{
y_cap[j] = slope*x[j] + intercept;
syx += sq(y[j] - y_cap[j]);
}
std_err = sqrt(syx/(maxcount-2) );
stderr_Slope = std_err / sqrt(x_dev) ;
stderr_Intercept = stderr_Slope * sqrt(xsqbar);
b = slope;
b_err = stderr_Slope;
a = exp(intercept);
a_err = exp(stderr_Intercept);
Serial.print(“ Exponential fit to the equation: Y = B*exp(C*X)+ A. \n B = ”);
Serial.println(a,4);
Serial.print(“ C = ”);
Serial.println(b,4);
Serial.print(“ A = ”);
Serial.println(c,6);
//save the slope and intercept data in EPR0M
EEPROM.put(eeAddress, a);
eeAddress += sizeof(a);
EEPROM.put(eeAddress, a_err);
eeAddress += sizeof(a_err);
EEPROM.put(eeAddress, b);
eeAddress += sizeof(b);
EEPROM.put(eeAddress, b_err);
eeAddress += sizeof(b_err);
EEPROM.put(eeAddress, c);
eeAddress += sizeof(c);
EEPROM.put(eeAddress, c_err);
eeAddress += sizeof(c_err);
delay (1000);
exit(0);
}
Appendix B
Appendix C
References
- World Health Organization (WHO). Global Report on Diabetes; WHO: Geneva, Switzerland, 2016; p. 88. ISBN 9789241565257. [Google Scholar]
- Chen, C.; Xie, Q.; Yang, D.; Xiao, H.; Fu, Y.; Tan, Y.; Yao, S. Recent advances in electrochemical glucose biosensors: A review. RCS Adv. 2013, 3, 4473–4491. [Google Scholar] [CrossRef]
- Bandodkar, A.J.; Wang, J. Non-invasive wearable electrochemical sensors: A review. Trends Biotechnol. 2014, 32, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Huang, Z.; Rogers, M.; Boutelle, M.; Cass, A.E. Evaluation of a minimally invasive glucose biosensor for continuous tissue monitoring. Anal. Bioanal. Chem. 2016, 408, 8427–8435. [Google Scholar] [CrossRef] [PubMed]
- Kagie, A.; Bishop, D.K.; Burdick, J.; La Belle, J.T.; Dymond, R.; Felder, R.; Wang, J. Flexible Rolled Thick-Film Miniaturized Flow-Cell for Minimally Invasive Amperometric Sensing. Electroanalysis 2008, 20, 1610–1614. [Google Scholar] [CrossRef]
- Li, M.; Bo, X.; Mu, Z.; Zhang, Y.; Guo, L. Electrodeposition of nickel oxide and platinum nanoparticles on electrochemically reduced graphene oxide film as a nonenzymatic glucose sensor. Sens. Actuators B Chem. 2014, 192, 261–268. [Google Scholar] [CrossRef]
- Mandal, S.; Marie, M.; Kuchuk, A.; Manasreh, M.O.; Benamara, M. Sensitivity enhancement in an in-vitro glucose sensor using gold nanoelectrode ensembles. J. Mater. Sci. Mater. Electron. 2017, 28, 5452–5459. [Google Scholar] [CrossRef]
- Rama, E.C.; Costa-Garcia, A.; Fernandez-Abedul, M.T. Pin-based electrochemical glucose sensor with multiplexing possibilities. Biosens. Bioelectron. 2017, 88, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.G.; Jung, D.; Kong, S.H. A Lab-on-a-Chip-Based Non-Invasive Optical Sensor for Measuring Glucose in Saliva. Sensors 2017, 17, 2607. [Google Scholar] [CrossRef] [PubMed]
- Bruen, D.; Delaney, C.; Florea, L.; Diamond, D. Glucose sensing for diabetes monitoring: Recent developments. Sensors 2017, 17, 1866. [Google Scholar] [CrossRef] [PubMed]
- Tierney, M.J.; Tamada, J.A.; Potts, R.O.; Jovanovic, L.; Garg, S. Clinical evaluation of the GlucoWatch® biographer: A continual, non-invasive glucose monitor for patients with diabetes. Biosens. Bioelectron. 2001, 16, 621–629. [Google Scholar] [CrossRef]
- Lane, J.D.; Krumholz, D.M.; Sack, R.A.; Morris, C. Tear Glucose Dynamics in Diabetes Mellitus. Curr. Eye Res. 2006, 31, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, M.; Lee, M.S.; Kim, K.; Ji, S.; Kim, Y.T.; Park, J.; Na, K.; Bae, K.H.; Kim, H.K.; et al. Wearable smart sensor systems integrated on soft contact lenses for wireless ocular diagnostics. Nat. Commun. 2017, 8, 14997. [Google Scholar] [CrossRef] [PubMed]
- Talary, M.S.; Dewarrat, F.; Huber, D.; Caduff, A. In vivo life sign application of dielectric spectroscopy and non-invasive glucose monitoring. J. Non-Cryst. Solids 2007, 353, 4515–4517. [Google Scholar] [CrossRef]
- Arnold, M.A.; Small, G.W. Noninvasive Glucose Sensing. Anal. Chem. 2005, 77, 5429–5439. [Google Scholar] [CrossRef] [PubMed]
- Weykamp, C. HbA1c: A Review of Analytical and Clinical Aspects. Ann. Lab. Med. 2013, 33, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Koenig, R.J.; Cerami, A. Hemoglobin AIc and diabetes mellitus. Annu. Rev. Med. 1980, 31, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.P.; Pavlovich, J.G.; Little, R.; England, J.; Peterson, C.M. What is hemoglobin A1c? An analysis of glycated hemoglobins by electrospray ionization mass spectrometry. Clin. Chem. 1998, 44, 1951–1958. [Google Scholar] [PubMed]
- Koenig, R.; Peterson, C.; Jones, R.; Saudek, C.; Lehrman, M.; Cerami, A. Correlation of glucose regulation and hemoglobin A1c in diabetes mellitus. N. Engl. J. Med. 1976, 295, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, E.J.; Roith, D.L.; Bloomgarden, Z. Review of hemoglobin A1c in the management of diabetes. J. Diabetes 2009, 1, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Nathan, D.M.; Kuenen, J.; Borg, R.; Zheng, H.; Schoenfeld, D.; Heine, R.J. Translating the A1C assay into estimated average glucose values. Diabetes Care 2008, 31, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Audit MicroControls. Control FD Glycohemoglobin A1c. Available online: http://www.auditmicro.com/ control-fd-glycohemoglobin-a1c.html (accessed on 13 March 2018).
- Eurotrol. HemoTrol. Available online: https://www.eurotrol.com/products/hemocue-controls/hemotrol (accessed on 13 March 2018).
- Ball, D.W. Field Guide to Spectroscopy; SPIE Press: Bellingham, WA, USA, 2006; ISBN 9780819463524. [Google Scholar]
- Chan, E.D.; Chan, M.M.; Chan, M.M. Pulse oximetry: Understanding its basic principles facilitates appreciation of its limitations. Respir. Med. 2013, 107, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Pittman, R.N. Regulation of Tissue Oxygenation. In Colloquium Series on Integrated Systems Physiology: From Molecule to Function; Morgan & Claypool Life Sciences: San Rafael, CA, USA, 2011; ISBN 9781615041770. [Google Scholar]
- Prahl, S. Tabulated Molar Extinction Coefficient for Hemoglobin in Water. 4 March 1998. Available online: https://omlc.org/spectra/hemoglobin/summary.html (accessed on 13 March 2018).
- Zollinger, H. Color Chemistry: Syntheses, Properties and Applications of Organic Dyes and Pigments; VCH Verlagsgesellschaft mbH: Weinheim, Germany, 1987; ISBN 3906390233. [Google Scholar]
- Birge, R. Kodak Laser Dyes; Kodak Publications: Rochester, NY, USA, 1987; ISBN-10 OCLC: 83459316. [Google Scholar]
- Edelhoch, H. Spectroscopic Determination of Tryptophan and Tyrosine in Proteins. Biochemistry 1967, 6, 1948–1954. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.C.; Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Pace, N.C.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [PubMed]
- Burnett, R.W. Accurate Measurement of Molar Absorptivities. J. Res. Natl. Bur. Stand. A Phys. Chem. 1972, 76, 483–489. [Google Scholar] [CrossRef]
- Safar, M.E.; Boudier, H.S. Vascular Development, Pulse Pressure, and the Mechanisms of Hypertension. Hypertension 2005, 46, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Moyle, J.T. Uses and abuses of pulse oximetry. Arch. Dis. Child. 1996, 74, 77–80. [Google Scholar] [CrossRef] [PubMed]



| Sample No. | Molar Concentration of Control FD HbA1c (mmol/L) | R | HbA1c (8%) | HbA1c (13%) |
|---|---|---|---|---|
| 1 | 0.03 | 2.8206 | 10.31% | 16.75% |
| 2 | 0.035 | 2.9710 | 7.99% | 12.98% |
| 3 | 0.04 | 2.8641 | 9.54% | 15.50% |
| 4 | 0.05 | 2.9573 | 8.16% | 13.27% |
| 5 | 0.0525 | 2.9682 | 8.02% | 13.03% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandal, S.; Manasreh, M.O. An In-Vitro Optical Sensor Designed to Estimate Glycated Hemoglobin Levels. Sensors 2018, 18, 1084. https://doi.org/10.3390/s18041084
Mandal S, Manasreh MO. An In-Vitro Optical Sensor Designed to Estimate Glycated Hemoglobin Levels. Sensors. 2018; 18(4):1084. https://doi.org/10.3390/s18041084
Chicago/Turabian StyleMandal, Sanghamitra, and M. O. Manasreh. 2018. "An In-Vitro Optical Sensor Designed to Estimate Glycated Hemoglobin Levels" Sensors 18, no. 4: 1084. https://doi.org/10.3390/s18041084
APA StyleMandal, S., & Manasreh, M. O. (2018). An In-Vitro Optical Sensor Designed to Estimate Glycated Hemoglobin Levels. Sensors, 18(4), 1084. https://doi.org/10.3390/s18041084




