The Identification of Seven Chemical Warfare Mimics Using a Colorimetric Array
Abstract
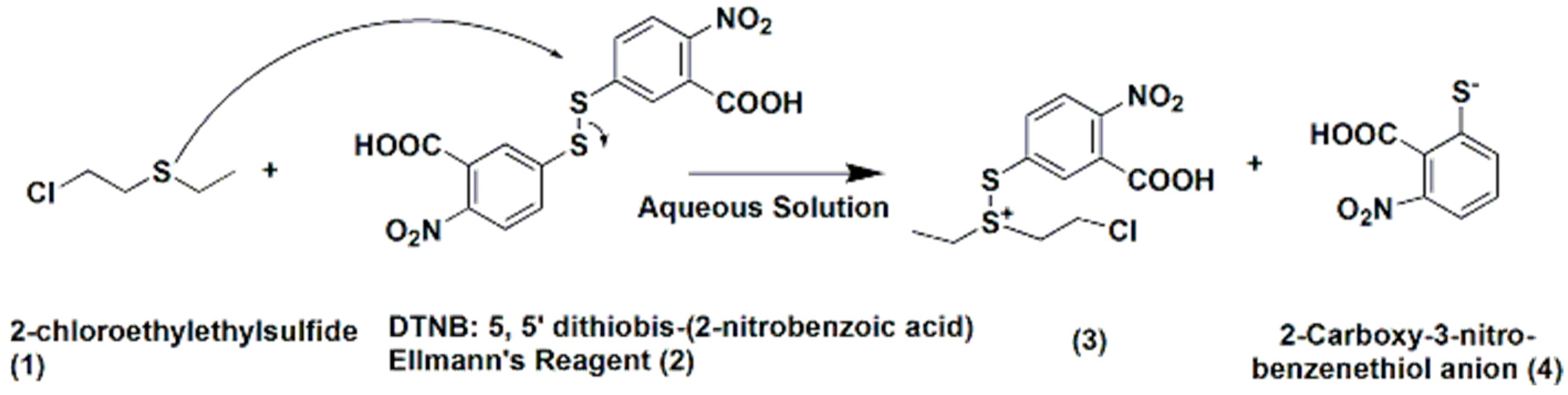
1. Introduction
2. Materials and Methods
3. Results
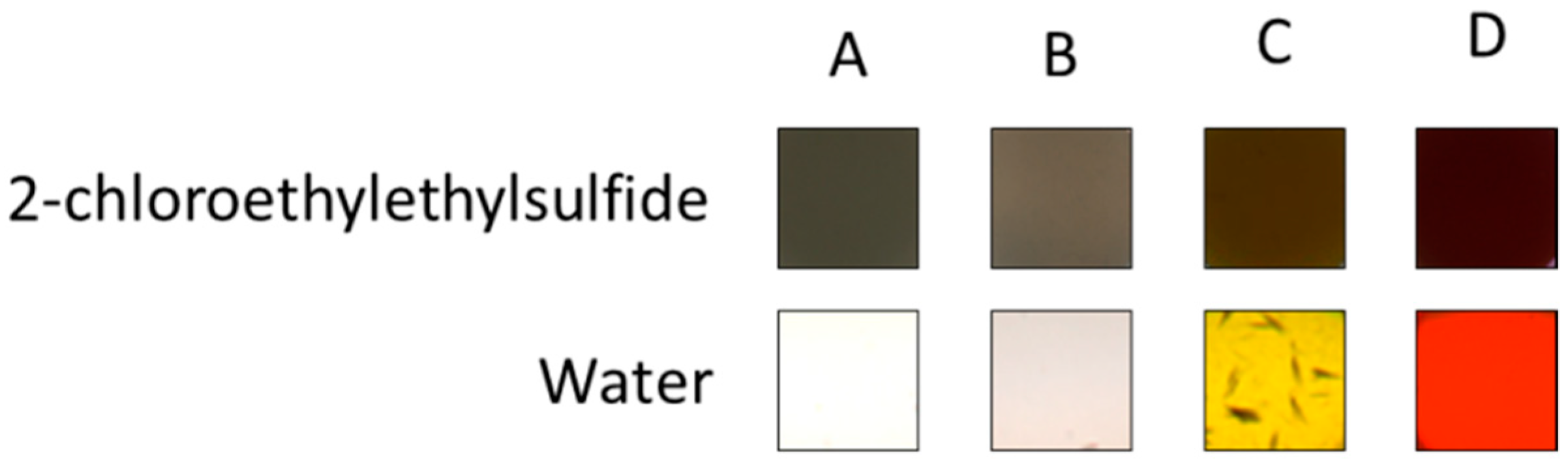
3.1. Visible Colorimetric Changes
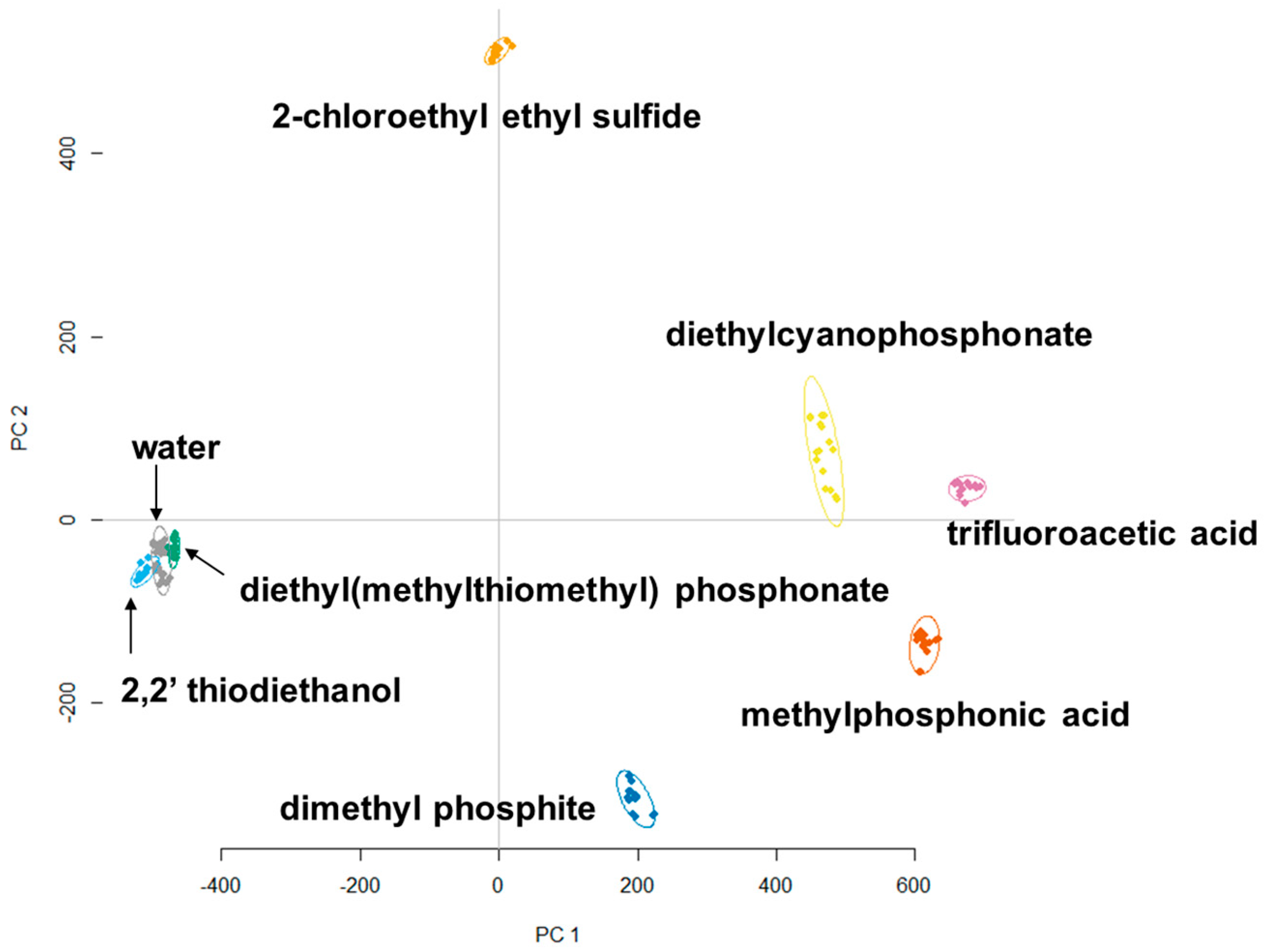
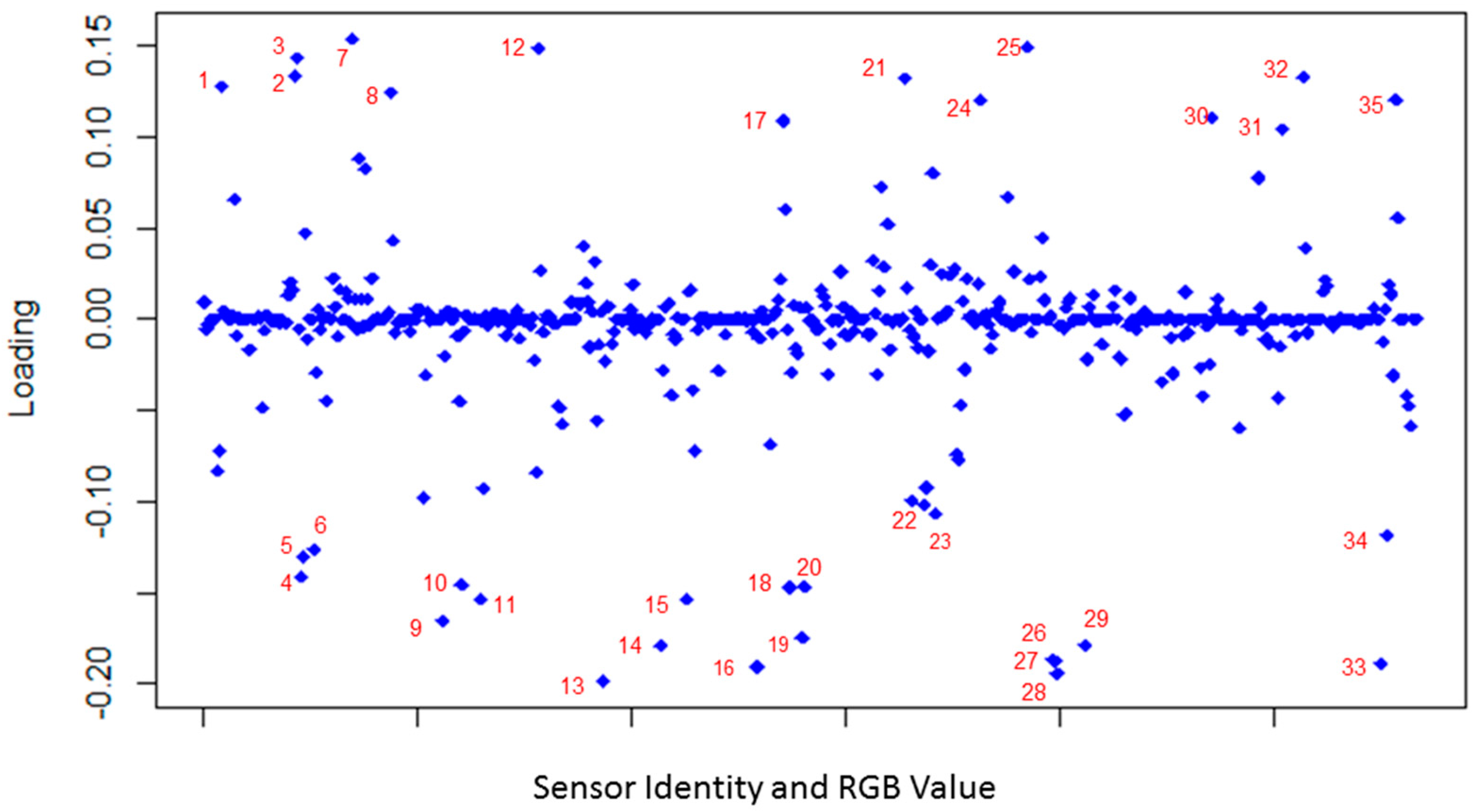
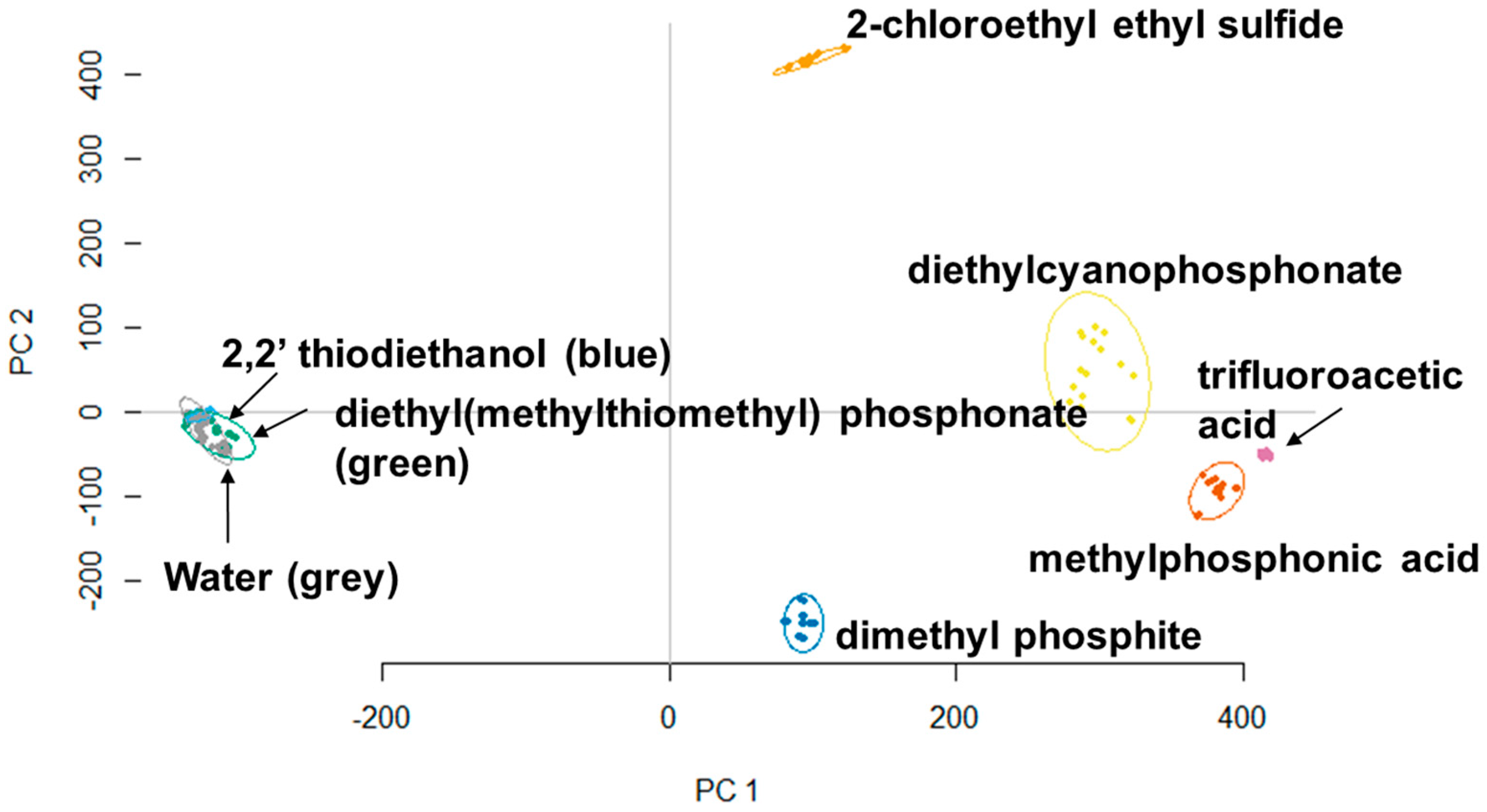
3.2. Principle Component Analysis (PCA)
4. Discussions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kangas, M.J.; Burks, R.M.; Atwater, J.; Lukowicz, R.M.; Williams, P.; Holmes, A.E. Colorimetric Sensor Arrays for the Detection and Identification of Chemical Weapons and Explosives. Crit. Rev. Anal. Chem. 2017, 47, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Sidell, F.R. Chemical Warfare Agents. In Military Preventive Medicine: Mobilization and Deployment; Kelley, P.W., Ed.; Textbooks of Military Medicine; Borden Institute, Walter Reed Army Medical Center: Washington, DC, USA, 2003; Volume 1, pp. 611–625. ISBN 0-16-050500-3. [Google Scholar]
- Witkiewicz, Z.; Neffe, S.; Sliwka, E.; Quagliano, J. Analysis of the Precursors, Simulants and Degradation Products of Chemical Warfare Agents. Crit. Rev. Anal. Chem. 2018, 48, 337–371. [Google Scholar] [CrossRef] [PubMed]
- Selby, R.C. DRSKO Project Ramping Up Quickly to Production. Available online: https://www.army.mil/article/131773/DRSKO_project_ramping_up_quickly_to_production/ (accessed on 17 June 2016).
- Veerabuthiran, S.; Razdan, A.K. LIDAR for detection of chemical and biological warfare agents. Def. Sci. J. 2011, 61, 241–250. [Google Scholar] [CrossRef]
- Hill, H.H., Jr.; Martin, S.J. Conventional analytical methods for chemical warfare agents. Pure Appl. Chem. 2002, 74, 2281–2291. [Google Scholar] [CrossRef]
- Zhang, S.-W.; Swager, T.M. Fluorescent Detection of Chemical Warfare Agents: Functional Group Specific Ratiometric Chemosensors. J. Am. Chem. Soc. 2003, 125, 3420–3421. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, D.; Yoon, J. Recent Advances in the Development of Chromophore-Based Chemosensors for Nerve Agents and Phosgene. ACS Sens. 2018, 3, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Suslick, K.S. An Optoelectronic Nose: “Seeing” Smells by Means of Colorimetric Sensor Arrays. MRS Bull. 2004, 29, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Musto, C.J.; Suslick, K.S. Differential sensing of sugars by colorimetric arrays. Curr. Opin. Chem. Biol. 2010, 14, 758–766. [Google Scholar] [CrossRef]
- Feng, L.; Musto, C.J.; Kemling, J.W.; Lim, S.H.; Suslick, K.S. A colorimetric sensor array for identification of toxic gases below permissible exposure limits. Chem. Commun. 2010, 46, 2037–2039. [Google Scholar] [CrossRef]
- Kangas, M.J.; Burks, R.M.; Atwater, J.; Lukowicz, R.M.; Garver, B.; Holmes, A.E. Comparative chemometric analysis for classification of acids and bases via a colorimetric sensor array. J. Chemom. 2018, 32, e2961. [Google Scholar] [CrossRef]
- Royo, S.; Martínez-Máñez, R.; Sancenón, F.; Costero, A.M.; Parra, M.; Gil, S. Chromogenic and fluorogenic reagents for chemical warfare nerve agents’ detection. Chem. Commun. 2007, 46, 4839–4847. [Google Scholar] [CrossRef]
- Chulvi, K.; Gaviña, P.; Costero, A.M.; Gil, S.; Parra, M.; Gotor, R.; Royo, S.; Martínez-Máñez, R.; Sancenóna, F.; Vivancos, J.-L. Discrimination of nerve gases mimics and other organophosphorous derivatives in gas phase using a colorimetric probe array. Chem. Commun. 2012, 48, 10105–10107. [Google Scholar] [CrossRef] [PubMed]
- Costero, A.M.; Gil, S.; Parra, M.; Mancini, P.M.E.; Martínez-Máñez, R.; Sancenón, F.; Royo, S. Chromogenic detection of nerve agent mimics. Chem. Commun. 2008, 45, 6002–6004. [Google Scholar] [CrossRef] [PubMed]
- Dale, T.J.; Rebek, J., Jr. Fluorescent Sensors for Organophosphorus Nerve Agent Mimics. J. Am. Chem. Soc. 2006, 128, 4500–4501. [Google Scholar] [CrossRef] [PubMed]
- Foster, L.S.; Gruntfest, I.J. Demonstration experiments using universal indicators. J. Chem. Educ. 1937, 14, 274–276. [Google Scholar] [CrossRef]
- Nomeir, A.A.; Burka, L.T.; Matthews, H.B. Analysis of Dimethyl Hydrogen Phosphite and Its Stability under Simulated Physiological Conditions. J. Anal. Toxicol. 1988, 12, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Abramoff, M.D.; Magalhaes, P.J.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Soldat, D.J.; Barak, P.; Lepore, B.J. Microscale Colorimetric Analysis Using a Desktop Scanner and Automated Digital Image Analysis Douglas. J. Chem. Educ. 2009, 86, 617–620. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Available online: http://github.com/vqv/ggbiplot (accessed on 5 December 2011).
- Salles, M.O.; Meloni, G.N.; de Aaujo, W.R.; Paixão, T.R.L.C. Explosive colorimetric discrimination using a smartphone, paper device and chemometrical approach. Anal. Methods 2014, 6, 2047–2052. [Google Scholar] [CrossRef]
- Bang, J.H.; Lim, S.H.; Park, E.; Suslick, K.S. Chemically Responsive Nanoporous Pigments: Colorimetric Sensor Arrays and the Identification of Aliphatic Amines. Langmuir 2008, 24, 13168–13172. [Google Scholar] [CrossRef]
- Kangas, M.J.; Wilson, C.L.; Burks, R.M.; Atwater, J.; Lukowicz, R.M.; Garver, B.; Mayer, M.; Havenridge, S.; Holmes, A.E. An Improved Comparison of Chemometric Analyses for the Identification of Acids and Bases with Colorimetric Sensor Arrays. Int. J. Chem. 2018, 10, 36–55. [Google Scholar] [CrossRef]
- Kangas, M.J.; Lukowicz, R.; Atwater, J.; Pliego, A.; Al-Shdifat, Y.; Havenridge, S.; Burks, R.; Garver, B.; Mayer, M.; Holmes, A.E. Printed Colorimetric Arrays for the Identification and Quantification of Acids and Bases. Anal. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Askim, J.R.; Mahmoudi, M.; Suslick, K.S. Optical sensor arrays for chemical sensing: The optoelectronic nose. Chem. Soc. Rev. 2013, 42, 8649–8682. [Google Scholar] [CrossRef] [PubMed]
- Okuom, M.O.; Wilson, M.V.; Jackson, A.; Holmes, A.E. Intermolecular interactions between eosin Y and caffeine using 1H-NMR spectroscopy. Int. J. Spectrosc. 2013, 2013, 245376. [Google Scholar] [CrossRef] [PubMed]
- Okuom, M.O.; Holmes, A.E. Developing a Color-Based Molecular Sensing Device: DETECHIP®. Sens. Transducers 2014, 183, 30–33. [Google Scholar]
- Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]

| Sensor [Channel] | PC1 Loading | Sensor [Channel] | PC2 Loading |
|---|---|---|---|
| Methyl Orange [Red] | −0.198 | Eosin Y [Red] | −0.274 |
| Ellman’s Reagent [Blue] | −0.194 | Phloxine B [Red] | −0.259 |
| Thymol Blue [Red] | −0.191 | Methyl Orange [Red] | 0.223 |
| Xylenol Blue [Red] | −0.188 | Morin [Red] | −0.203 |
| Ellman’s Reagent [Green] | −0.187 | o-Dianisidine [Blue] | −0.202 |
| Ellman’s Reagent [Blue] | −0.186 | Bromocresol Purple [Red] | −0.201 |
| Orange IV [Red] | −0.179 | Bromothymol Blue [Red] | −0.199 |
| Metanil Yellow [Red] | −0.179 | Thymol Blue [Red] | −0.196 |
| Alizarin Yellow GG [Red] | −0.174 | o-Dianisidine [Green] | −0.195 |
| Eosin Y [Red] | −0.165 | o-Dianisidine [Red] | −0.194 |
| Bromocresol Green [Red] | 0.154 | Morin [Green] | −0.187 |
| Phloxine B [Red] | −0.153 | Bromophenol blue [Red] | −0.180 |
| Fluorescein [Red] | −0.153 | Alizarin Red S [Green] | 0.174 |
| Nitrazine Yellow [Red] | 0.149 | Orange IV [Red] | 0.170 |
| Hexammine Cobalt (III) Chloride [Red] | 0.148 | Alizarin Red S [Red] | 0.163 |
| Yellow A2 [Red] | −0.147 | Ferric Chloride [Blue] | −0.154 |
| Alizarin Yellow GG[Green] | −0.146 | Alizarin Yellow R [Red] | 0.153 |
| Erythrosin B [Red] | −0.145 | 1,1′-Diethyl-4,4′-Cyanine Iodide [Red] | −0.149 |
| Alizarin Red S [Green] | 0.143 | Alizarin Yellow R [Green] | 0.144 |
| Alizarin Yellow R [Red] | −0.141 | Chlorophenol Red [Red] | −0.135 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kangas, M.J.; Ernest, A.; Lukowicz, R.; Mora, A.V.; Quossi, A.; Perez, M.; Kyes, N.; Holmes, A.E. The Identification of Seven Chemical Warfare Mimics Using a Colorimetric Array. Sensors 2018, 18, 4291. https://doi.org/10.3390/s18124291
Kangas MJ, Ernest A, Lukowicz R, Mora AV, Quossi A, Perez M, Kyes N, Holmes AE. The Identification of Seven Chemical Warfare Mimics Using a Colorimetric Array. Sensors. 2018; 18(12):4291. https://doi.org/10.3390/s18124291
Chicago/Turabian StyleKangas, Michael J., Adreanna Ernest, Rachel Lukowicz, Andres V. Mora, Anais Quossi, Marco Perez, Nathan Kyes, and Andrea E. Holmes. 2018. "The Identification of Seven Chemical Warfare Mimics Using a Colorimetric Array" Sensors 18, no. 12: 4291. https://doi.org/10.3390/s18124291
APA StyleKangas, M. J., Ernest, A., Lukowicz, R., Mora, A. V., Quossi, A., Perez, M., Kyes, N., & Holmes, A. E. (2018). The Identification of Seven Chemical Warfare Mimics Using a Colorimetric Array. Sensors, 18(12), 4291. https://doi.org/10.3390/s18124291






