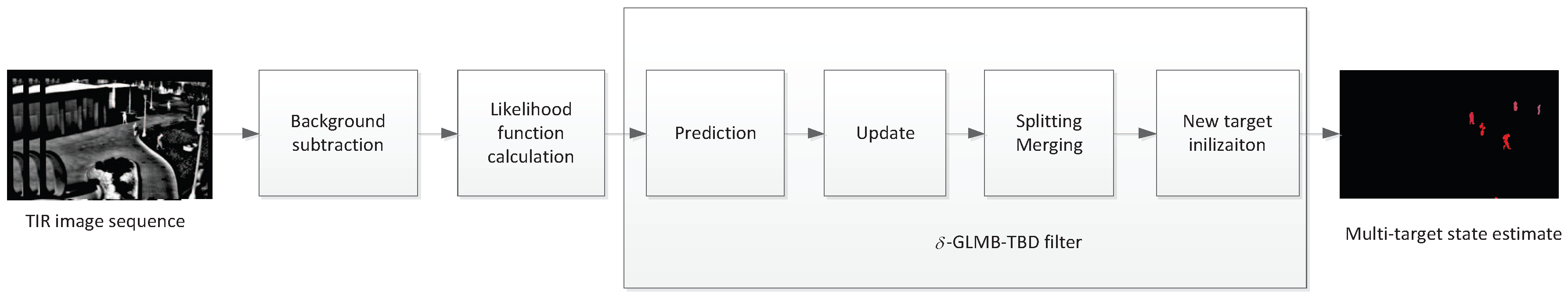
2.3.1. Basic Theory
In this subsection, the standard
-GLMB-TBD filter, consisting of two steps, prediction and update, with a separable likelihood function, is briefly discussed [
23]. Before introducing the recursion of the standard
-GLMB-TBD filter, some notations are shown for convenience.
(1) Notation
For the remainder of the paper, let lowercase letters (e.g.,
x) denote single-target state and uppercase letters (e.g.,
X) denote multi-target states. The labeled target states are indicated by boldface letters (e.g.,
,
). Space is represented by a letter with a tilde (e.g.,
denotes the state space,
denotes the discrete label space). A labeled target can be written as
, where
and
,
k means the target birth time, and
i is a unique index to distinguish targets born at the same time. A labeled multi-Bernoulli (LMB) RFS with state space
and label space
can be written as
, where
and
mean the existence probability and the probability density of a target with label
l.
denotes the standard inner product, and
denotes the multi-target exponential, where
by convention and
,
, and
are real-valued functions. The generalized Kronecker delta function and inclusion function are defined as
(2) Standard -GLMB-TBD Filter
From [
23], the multi-target posterior at time
k has the following
-GLMB form:
where
is the current multi-target state;
denotes the distinct label indicator, which means that the cardinalities of the set of labels and the set of state vectors are identical;
denotes the label space of targets born between time 0 and time
k, where the subscript 0:k means time interval
;
denotes collections of all finite subsets of a given space;
is a projection from space
to
and hence
is the set of labels of
; and
denotes the joint existence probability of the label set
I, while the multi-target exponential
denotes the joint probability density of
, conditional on their corresponding label set
I.
The new birth model covering labeled Poisson, labeled identically and independently distributed cluster and labeled multi-Bernoulli filter can be given by [
20]
where
is the state of new birth targets, and
and
are the joint existence probability and probability density. This model can also be written as the LMB birth model [
23,
31] as follows:
where
and
mean the existence probability and the probability density of a birth target with label
l.
Proposition 1. If the multi-target state posterior with δ-GLMB form is given as (15) at time k, with the new birth model (16), the multi-target prediction density also has a δ-GLMB form [23]:where denotes a single-target transition function. denotes the survival probability of target x. is the label space of a target born at time . Proposition 2. If the multi-target prediction density has the form of δ-GLMB as (19), then, with the measurement set S and separable likelihood function , the multi-target posterior density also has the same form [23]:where Proposition 3. If the multi-target state posterior with the δ-GLMB form is given as (15) at time k, the cardinality distribution can be given by [20]The cardinality estimates can be obtained by The multi-target state estimate is the mean estimate of the multi-target state conditioned on the estimated cardinality
as in [
20].
2.3.2. Recursion
Particle implementation is based on the standard
-GLMB filter implementation [
20,
21]. Each target density
with label l in
is modeled as a set of weighted samples
, where
is the number of particles, and
denotes the state
and label
l. Besides the prediction and update steps, we add four other parts: pruning, splitting, merging, and new birth target initialization to form the whole solution. The details are as follows.
(1) Prediction
The pixels illuminated by the target can describe the target more effectively than a single point or a rectangle. Let us assume the single-target state x have two fields: a geometric center location and cover area , which can be obtained from . For convenience, let and in this paper. This new approach representing a single-target benefits from the similar target shape in consecutive frames. To accommodate the small amount of deformation, pixel sampling is executed in this prediction step. Each pixel in has the probability of to be selected to survive, the value of which depends on the deformation degree. In general, when the deformation is small, is close to 1. It is obvious that sampling may reduce the covered area in the final estimates. Fortunately, this can be alleviated in the update step.
If at time
k, the multi-target state posterior is given by (
15), when the LMB new birth model
is known (in practice for the unknown birth model, the third part in this subsection describes how to initialize the new birth model), where
is
and
is the number of particles, according to proposition 1, we obtain
and
can be represented as
where
(
),
is generated by a random pixel sample as described above;
;
; and
is a proposal density. The procedure for calculating
in (
20) is bothersome and complex; however, reference [
21] offers a method based on a K-shortest paths algorithm to carry it out. To understand the procedure more clearly, reference [
21] is strongly recommended.
(2) Update
With the assumption that targets are not overlapped, we obtain a separable likelihood function shown as (
12). After adding a distinct label to each target, (
12) can be written as
where
.
According to proposition 2, if each single-target density
is modeled by a particle set
. Then,
and
can be represented as
where
;
; and
. Substituting (
34) into (
26), we obtain
.
Analogous to the standard particle filter, resampling each target density
must be executed to reduce the degeneracy. For simplicity, in this paper, multinomial resampling is used for numerical studies that would otherwise be carried out with other multi-Bernoulli filters [
17].
To eliminate the influence of pixel sampling in the prediction step, a pixel set update procedure is used to correct the shape of the target. Taking target
x as an example, the procedure is described as follows. First, for all
particles representing target
x, count the pixel
i (
) occupied times
. The occupied times are initialized to 0, i.e.,
, and for the jth (
) particle state
,
is given by
Second, compare the with a threshold . When reaches the threshold, the pixel i is classified as an occupied pixel. Third, if the occupied pixel i is greater than an extremely low value in the foreground probability map, then it is considered to be a pixel in the updated pixel set for target x. The updated pixel set is the estimated cover area of target x. The pseudo-code for the pixel set update procedure is presented in Algorithm 2. Algorithm 2 shows us that for each pixel belonging to the covered areas occupied by all particles representing x, the occupied time is calculated (line 3) and then it will be used to determine the true cover area of the target (lines 5–9). The filter will propagate a pixel set as the target state in the recursion and will directly produce the pixel set as its output. This operation can facilitate target recognition and extraction and subsequent processing in computer vision applications.
| Algorithm 2: Pixel set update |
![Sensors 18 03944 i002 Sensors 18 03944 i002]() |
(3) Pruning, Splitting, Merging and Birth Target Initialization
To reduce computational complexity, besides the truncation [
20,
21], the pruning procedure is also needed. The multi-target set
should be discarded if its weight
is less than a threshold
. In practical application, the dynamic processes of multi-targets are complex. To accommodate these processes, we propose splitting and merging, which are described as follows. For each target
, clustering is executed to check whether the target
should be split into several small targets. If yes, the small target with the most pixels inherits the identity (label) of target
, while the others are labeled as new birth targets. For two targets in
with substantial overlap, if the overlap ratio of the pixel intersection area to the smaller target area is higher than a threshold
, these two targets should be merged; the merged label is the same as the earlier born target.
Birth target initialization has two pre-processing steps. Step 1: clustering the foreground probability map to obtain current cluster target (, n denotes the number of all current cluster targets). Step 2: for each cluster target , calculate the overlap ratio between the and all existing targets. If the maximum overlap ratio is higher than a threshold , then remove this . Subsequently, we use the remaining to initiate the new birth target LMB modeled as . For simplicity, the is initialized as a constant and the target density is modeled as , where all have equal weight; and , where denotes the Gaussian distribution, is the geometric center location of one remaining , and is the variance. can be obtained from using random sampling by probability .
These procedures are designed to broaden the applications of the filter. Without splitting and merging, this filter would produce a cardinality estimation error. Missing the new target birth process would invalidate the -GLMB-TBD filter. Using proposition 3 can produce a multi-target estimate.