Non-Adaptive Methods for Fetal ECG Signal Processing: A Review and Appraisal
Abstract
1. Introduction
1.1. Fetal Electrocardiography
1.2. Basic Distribution of Signal Processing Methods
2. Single Channel Signal Sources
2.1. Wavelet Transform
2.2. Correlation Technique
2.3. Subtraction Technique
2.4. Averaging Technique
2.5. Filtering Methodologies
2.6. De-Shape Short Time Fourier Transform and Nonlocal Median
2.7. Single Channel Blind Source Separation
2.8. Template Subtraction
2.9. Sequential Total Variation Denoising
2.10. Empirical Mode Decomposition
2.11. Summary of Single Channel Methods
- Overall performance—this parameter reflects the robustness of the method used, and it can be divided into three groups:
- -
- Low—methods suitable primarily for NI-fECG preprocessing, these methods are not able to extract fECG, but only remove some specific types of interference, e.g., baseline wandering, power line interference, and so on (improvement ≤5 dB; based on fECG extraction from synthetic records).
- -
- Medium—methods suitable for advanced preprocessing eliminating most of the interference in NI-fECG (e.g., power-line interference, myopotentials, and electromyographic interference, isoelectric line fluctuations, motion artifacts, etc.). These methods partly suppress the maternal component allowing detection of the fQRS complex and thus fHR determination; further morphological analysis is not possible (improvement ≤20 dB; based on fECG extraction from synthetic records).
- -
- High—the most powerful comprehensive NI-fECG processing methods that provide information on fHR, and fECG morphology—PR, QT, ST intervals and so on (improvement ≥20 dB; based on fECG extraction from synthetic records).
- SNR improvement—this parameter takes into account the improvement of SNR, can be divided into three categories: low, medium, and high. It should be noted that the SNR parameter objectively determines the efficacy of the method with regards to the reference; however, in terms of the clinical use, the used SNR as a parameter may be very misleading. The methods that show excellent SNR improvement can be very inaccurate in fQRS complex detection.
- Computational cost—this parameter evaluates the demands of the methods in terms of computational complexity; the categories are low, medium, and high.
- Real-time—parameter defining whether the method can be used in online mode (real-time) from the point of view of its feasibility using currently available hardware devices in clinical practice.
- Implementation complexity—this parameter, divided into three categories low, medium, and high, evaluates the overall complexity in terms of its deployment in clinical practice. The complexity of hardware and software must be economically viable for the public health system to be available to all pregnant women.
3. Multichannel Signal Sources
3.1. Independent Component Analysis
3.2. Singular Value Decomposition
3.3. Principal Component Analysis
3.4. Period Component Analysis
3.5. Sequential Analysis
3.6. Barros’s Algorithm
3.7. Zhang’s Algorithm
3.8. Skewness Method
3.9. Quality Index Optimization
3.10. Polynomial Matrix Eigenvalue Decomposition
3.11. Fuzzy C-Means Clustering Method
3.12. Compressed Sensed
3.13. Maternal Component Suppression Method
3.14. Tucker
3.15. Multivariate Empirical Mode Decomposition
3.16. Summary of Multichannel Methods
4. Hybrid Methods
4.1. ICA-EEMD-WS
4.2. ICA and AF
4.3. ICA and PF
4.4. -ICA
4.5. ICA and PCA
4.6. ICA and SVD
4.7. BA and ZA
4.8. SVD and PC
4.9. PN and SGSF
4.10. Summary of Hybrid Methods
5. Discussion
- fHR (R-R)—this evaluation parameter classifies the effectiveness of the investigated methods from in terms of the fHR determination based on the fetal R-R interval. There are four categories for the assessment:
- -
- Inaccurate—the methods are not sufficient to remove artifacts and noise sufficiently to enable the R–R interval detection; the NI-fECG processed by these methods cannot be used for fHR monitoring.
- -
- Moderately accurate—these methods sufficiently suppress most common interference and thus make RR interval detection possible. However, the noise is not completely eliminated and thus there are many false-detected and undetected significant complexes, i.e., sensitivity (Se) ≤ 80%, positive predictive value (PPV) ≤ 90%, accuracy (ACC) ≤ 80%, total probability of correct detection of beats (F1) ≤ 85%.
- -
- Accurate—these methods allow accurate detection of fHR, i.e., Se ≤ 85%, PPV ≤ 95%, ACC ≤ 85%, F1 ≤ 90%.
- -
- Morphological analysis (T/QRS; QT)—this parameter classifies the efficacy of the investigated methods from a deeper morphological analysis of fECG. The following categories were created for the evaluation:
- -
- Insufficient—these methods cannot estimate fECG in a sufficient quality for morphological analysis.
- -
- Moderately accurate—these methods enable morphological analysis; however, only in the case of some tested real data, the efficacy is significantly affected by gestational age, fetal position, SNR, and so on. Therefore, these methods could not be used for long-term monitoring of T/QRS ratio or QT interval.
- -
- Promising—these mostly hybrid methods have great potential to be used for fECG morphological analysis.
- Dataset—in this column, we provide the source of the data that was used in the studies. Databases with fECG recordings are an important part of this research. A major problem is their insufficient quantity that would be available to the scientific community. Without the databases, i.e., real recordings, the scientists can hardly verify the methods for extracting fECG and thus improve diagnostic quality contrary to the standard ECG of adults.
- Technical aspects—the last column includes the technical details of each study, e.g., number of electrodes, heterogeneity of the patient population, conditions and gestational ages, as well as data quality (duration (T), sampling frequency (Fs), amplitude resolution (res), gestational age (GA), number of electrodes (channels), type of records, and number of records).
6. Conclusions
7. Ethics Statement
Funding
Conflicts of Interest
References
- Martinek, R.; Kahankova, R.; Jezewski, J.; Jaros, R.; Mohylova, J.; Fajkus, M.; Nedoma, J.; Janku, P.; Nazeran, H. Comparative Effectiveness of ICA and PCA in Extraction of Fetal ECG From Abdominal Signals: Toward Non-invasive Fetal Monitoring. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Jagannath, D.; Selvakumar, A.I. Issues and research on foetal electrocardiogram signal elicitation. Biomed. Signal Process. Control 2014, 10, 224–244. [Google Scholar] [CrossRef]
- Sameni, R.; Clifford, G.D. A review of fetal ECG signal processing; issues and promising directions. Open Pac. Electrophysiol. Ther. J. 2010, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- KováCs, F.; HorváTh, C.; Balogh, Á.T.; Hosszú, G. Fetal phonocardiography—Past and future possibilities. Comput. Methods Progr. Biomed. 2011, 104, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Adithya, P.C.; Sankar, R.; Moreno, W.A.; Hart, S. Trends in fetal monitoring through phonocardiography: Challenges and future directions. Biomed. Signal Process. Control 2017, 33, 289–305. [Google Scholar] [CrossRef]
- Kahánková, R.; Jaroš, R.; Martinek, R.; Jezewski, J.; He, W.; Jezewski, M.; Kawala-Janik, A. Non-Adaptive Methods of Fetal ECG Signal Processing. Adv. Electr. Electron. Eng. 2017, 15, 476–490. [Google Scholar] [CrossRef]
- Verdurmen, K.M.; Lempersz, C.; Vullings, R.; Schroer, C.; Delhaas, T.; van Laar, J.O.; Oei, S.G. Normal ranges for fetal electrocardiogram values for the healthy fetus of 18–24 weeks of gestation: A prospective cohort study. BMC Preg. Childbirth 2016, 16, 227. [Google Scholar] [CrossRef] [PubMed]
- Karvounis, E.; Tsipouras, M.; Papaloukas, C.; Tsalikakis, D.; Naka, K.; Fotiadis, D. A non-invasive methodology for fetal monitoring during pregnancy. Methods Inf. Med. 2010, 49, 238–253. [Google Scholar] [CrossRef] [PubMed]
- Jezewski, J.; Wrobel, J.; Horoba, K. Comparison of Doppler ultrasound and direct electrocardiography acquisition techniques for quantification of fetal heart rate variability. IEEE Trans. Biomed. Eng. 2006, 53, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Jezewski, J.; Matonia, A.; Kupka, T.; Roj, D.; Czabanski, R. Determination of fetal heart rate from abdominal signals: Evaluation of beat-to-beat accuracy in relation to the direct fetal electrocardiogram. Biomed. Tech. 2012, 57, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Matonia, A.; Kupka, T.; Jezewski, J.; Momot, A.; Jeżewski, M.; Bernys, M. Comparison of instantaneous fetal heart rate extracted from abdominal and direct fetal electrocardiograms. J. Med. Inf. Technol. 2012, 19, 101–107. [Google Scholar]
- Martinek, R.; Nedoma, J.; Fajkus, M.; Kahankova, R.; Konecny, J.; Janku, P.; Kepak, S.; Bilik, P.; Nazeran, H. A phonocardiographic-based fiber-optic sensor and adaptive filtering system for noninvasive continuous fetal heart rate monitoring. Sensors 2017, 17, 890. [Google Scholar] [CrossRef] [PubMed]
- Jaros, R.; Kahankova, R.; Martinek, R.; Nedoma, J.; Fajkus, M.; Slanina, Z. Fetal phonocardiography signal processing from abdominal records by non-adaptive methods. In Photonics Applications in Astronomy, Communications, Industry, and High-Energy Physics Experiments 2018; International Society for Optics and Photonics: Wilga, Poland, 2018; Volume 10808, p. 108083E. [Google Scholar] [CrossRef]
- Skutova, H.; Martinek, R.; Jaros, R.; Kahankova, R. A Noise Suppression Technique for Fetal Phonocardiogram Monitoring Using Adaptive Neuro-Fuzzy Interference System. IFAC Pap. Online 2018, 51, 456–461. [Google Scholar] [CrossRef]
- Kahankova, R.; Martinek, R.; Jaros, R.; Nedoma, J.; Fajkus, M.; Vanus, J. Least Mean Squares Adaptive Algorithms Optimization for Fetal Phonocardiogram Extraction. IFAC Pap. Online 2018, 51, 60–65. [Google Scholar] [CrossRef]
- Varady, P.; Wildt, L.; Benyó, Z.; Hein, A. An advanced method in fetal phonocardiography. Comput. Methods Progr. Biomed. 2003, 71, 283–296. [Google Scholar] [CrossRef]
- Yang, W.; Yang, K.; Jiang, H.; Wang, Z.; Lin, Q.; Jia, W. Fetal heart rate monitoring system with mobile internet. In Proceedings of the 2014 IEEE International Symposium on Circuits and Systems (ISCAS), Melbourne, Australia, 1–5 June 2014; pp. 443–446. [Google Scholar] [CrossRef]
- Kovacs, F.; Horváth, C.; Balogh, Á.; Hosszú, G. Extended noninvasive fetal monitoring by detailed analysis of data measured with phonocardiography. IEEE Trans. Biomed. Eng. 2011, 58, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Khandoker, A.; Ibrahim, E.; Oshio, S.; Kimura, Y. Validation of beat by beat fetal heart signals acquired from four-channel fetal phonocardiogram with fetal electrocardiogram in healthy late pregnancy. Sci. Rep. 2018, 8, 13635. [Google Scholar] [CrossRef] [PubMed]
- Popov, B.; Lanzo, V.F.; Agarwal, R. Non-Invasive Measurement of Second Heart Sound Components. U.S. Patent 7,909,772, 22 March 2011. [Google Scholar]
- Xie, K.; Zhang, H.; Xie, S.; Kun, C. Method and Apparatus for Detecting Instantaneous Fetal Heart Rate of Doppler Fetal Heart Sound Based on Time-Frequency Analysis. U.S. Patent 15/335,564, 3 May 2018. [Google Scholar]
- Zhang, D.; Zhang, Y.; Ren, W.; Sun, F.; Guo, Y.; Sun, W.; Wang, Y.; Huang, L.; Cai, A. Prenatal diagnosis of fetal interrupted aortic arch type A by two-dimensional echocardiography and four-dimensional echocardiography with B-flow imaging and spatiotemporal image correlation. Echocardiography 2016, 33, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Fetal, Echocardiography Task Force; American Institute of Ultrasound in Medicine Clinical Standards Committee. AIUM practice guideline for the performance of fetal echocardiography. J. Ultrasound Med. 2011, 30, 127. [Google Scholar] [CrossRef]
- Persico, N.; Moratalla, J.; Lombardi, C.; Zidere, V.; Allan, L.; Nicolaides, K. Fetal echocardiography at 11–13 weeks by transabdominal high-frequency ultrasound. Ultrasound Obstet. Gynecol. 2011, 37, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Bowman, A.W. A Practical Guide to Fetal Echocardiography: Normal and Abnormal Hearts; Radcliffe Cardiology: Buckinghamshire, UK, 2010. [Google Scholar]
- Quartero, H.; Stinstra, J.; Golbach, E.; Meijboom, E.; Peters, M. Clinical implications of fetal magnetocardiography. Ultrasound Obstet. Gynecol. 2002, 20, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Grimm, B.; Haueisen, J.; Huotilainen, M.; Lange, S.; Leeuwen, P.V.; Menendez, T.; Peters, M.J.; Schleussner, E.; Schneider, U. Recommended standards for fetal magnetocardiography. Pac. Clin. Electrophysiol. 2003, 26, 2121–2126. [Google Scholar] [CrossRef]
- Van Leeuwen, P.; Halier, B.; Bader, W.; Geissler, J.; Trowitzsch, E.; Grönemeyer, D. Magnetocardiography in the diagnosis of fetal arrhythmia. BJOG 1999, 106, 1200–1208. [Google Scholar] [CrossRef]
- Hamada, H.; Horigome, H.; Asaka, M.; Shigemitsu, S.; Mitsui, T.; Kubo, T.; Kandori, A.; Tsukada, K. Prenatal diagnosis of long QT syndrome using fetal magnetocardiography. Prenat. Diagn. 1999, 19, 677–680. [Google Scholar] [CrossRef]
- Kähler, C.; Grimm, B.; Schleussner, E.; Schneider, A.; Schneider, U.; Nowak, H.; Vogt, L.; Seewald, H.J. The application of fetal magnetocardiography (FMCG) to investigate fetal arrhythmias and congenital heart defects (CHD). Prenat. Diagn. 2001, 21, 176–182. [Google Scholar] [CrossRef]
- Jezewski, J.; Wrobel, J.; Matonia, A.; Horoba, K.; Martinek, R.; Kupka, T.; Jezewski, M. Is abdominal fetal electrocardiography an alternative to doppler ultrasound for FHR variability evaluation? Front. Physiol. 2017, 8, 305. [Google Scholar] [CrossRef] [PubMed]
- Jezewski, J.; Roj, D.; Wrobel, J.; Horoba, K. A novel technique for fetal heart rate estimation from Doppler ultrasound signal. Biomed. Eng. Online 2011, 10, 92. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, J.; Kupka, T.; Horoba, K.; Matonia, A.; Roj, D.; Jezewski, J. Recognition of Fetal Movements–Automated Detection from Doppler Ultrasound Signals Compared to Maternal Perception. J. Med. Imaging Health Inform. 2015, 5, 1319–1326. [Google Scholar] [CrossRef]
- Monica Healthcare. Available online: http://www.monicahealthcare.com/ (accessed on 17 October 2018).
- Mindchild. Available online: http://www.mindchild.com/ (accessed on 17 October 2018).
- Community Research and Development Information Service. Available online: https://cordis.europa.eu/home_en.html/ (accessed on 17 October 2018).
- Clifford, G.D.; Silva, I.; Behar, J.; Moody, G.B. Non-invasive fetal ECG analysis. Physiol. Meas. 2014, 35, 1521. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Li, T.; Qiu, T.; Park, Y. Fetal Heart Rate Monitoring from Phonocardiograph Signal Using Repetition Frequency of Heart Sounds. J. Electr. Comput. Eng. 2016, 2016. [Google Scholar] [CrossRef]
- Vaisman, S.; Salem, S.Y.; Holcberg, G.; Geva, A.B. Passive fetal monitoring by adaptive wavelet denoising method. Comput. Biol. Med. 2012, 42, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Sänger, N.; Hayes-Gill, B.; Schiermeier, S.; Hatzmann, W.; Yuan, J.; Herrmann, E.; Louwen, F.; Reinhard, J. Prenatal Foetal Non-invasive ECG instead of Doppler CTG—A Better Alternative? Geburtshilfe Frauenheilkund 2012, 72, 630. [Google Scholar] [CrossRef] [PubMed]
- Cohen, W.R.; Hayes-Gill, B. Influence of maternal body mass index on accuracy and reliability of external fetal monitoring techniques. Acta Obstet. Gynecol. Scand. 2014, 93, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Agostinelli, A.; Di Cosmo, M.; Sbrollini, A.; Burattini, L.; Morettini, M.; Di Nardo, F.; Fioretti, S.; Burattini, L. Quantification of Fetal ST-Segment Deviations. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017. [Google Scholar] [CrossRef]
- Marcantoni, I.; Vagni, M.; Agostinelli, A.; Sbrollini, A.; Morettini, M.; Burattini, L.; Di Nardo, F.; Fioretti, S.; Burattini, L. T-Wave Alternans Identification in Direct Fetal Electrocardiography. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017. [Google Scholar] [CrossRef]
- Joachim, B.; Fernando, A.; Sebastian, Z.; Julien, O.; Gari, D. A practical guide to non-invasive foetal electrocardiogram extraction and analysis. Physiol. Meas. 37 R1–R35. [CrossRef]
- Agostinelli, A.; Grillo, M.; Biagini, A.; Giuliani, C.; Burattini, L.; Fioretti, S.; Di Nardo, F.; Giannubilo, S.R.; Ciavattini, A.; Burattini, L. Noninvasive fetal electrocardiography: An overview of the signal electrophysiological meaning, recording procedures, and processing techniques. Ann. Noninvasive Electrocardiol. 2015, 20, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Neoventa: It’s All in the Beat. Available online: https://www.neoventa.com/products/stan/ (accessed on 17 October 2018).
- Matonia, A.; Jezewski, J.; Kupka, T.; Horoba, K.; Wrobel, J.; Gacek, A. The influence of coincidence of fetal and maternal QRS complexes on fetal heart rate reliability. Med. Biol. Eng. Comput. 2006, 44, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Kotas, M.; Jezewski, J.; Horoba, K.; Matonia, A. Application of spatio-temporal filtering to fetal electrocardiogram enhancement. Comput. Methods Progr. Biomed. 2011, 104, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kotas, M.; Jezewski, J.; Matonia, A.; Kupka, T. Towards noise immune detection of fetal QRS complexes. Comput. Methods Progr. Biomed. 2010, 97, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. Physiobank, physiotoolkit, and physionet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour, H.; Parsaei, A. Fetal ECG extraction using wavelet transform. In Proceedings of the 2006 International Conference on Computational Inteligence for Modelling Control and Automation and International Conference on Intelligent Agents Web Technologies and International Commerce (CIMCA’06) Vienna, Austria, 22 May 2006; p. 179. [Google Scholar] [CrossRef]
- Bhoker, R.; Gawande, J. Fetal ECG extraction using wavelet transform. ITSI Trans. Electr. Electron. Eng. 2013, 1, 2320–8945. [Google Scholar]
- De Moor, B.; De Gersem, P.; De Schutter, B.; Favoreel, W. DAISY: A database for identification of systems. J. A 1997, 38, 4–5. [Google Scholar]
- Silva, I.; Behar, J.; Sameni, R.; Zhu, T.; Oster, J.; Clifford, G.D.; Moody, G.B. Noninvasive fetal ecg: The physionet/computing in cardiology challenge 2013. In Proceedings of the IEEE Computing in Cardiology Conference (CinC), Zaragoza, Spain, 22–25 September 2013; pp. 149–152. [Google Scholar]
- Sober, M.M.; Marco, J.G. Non-Invasive Fetal Electrocardiogram Database. PhysioNet 1997. [Google Scholar] [CrossRef]
- Karvounis, E.; Papaloukas, C.; Fotiadis, D.; Michalis, L. Fetal heart rate extraction from composite maternal ECG using complex continuous wavelet transform. In Proceedings of the IEEE Computers in Cardiology, Chicago, IL, USA, 19–22 September 2004; pp. 737–740. [Google Scholar] [CrossRef]
- Ravindrakumar, S.; Raja, K.B. Fetal ECG extraction and enhancement in prenatal monitoring—Review and implementation issues. In Proceedings of the IEEE Trendz in Information Sciences &Computing (TISC), Chennai, India, 17–19 December 2010; pp. 16–20. [Google Scholar] [CrossRef]
- Kumar, P.; Sharma, S.K.; Prasad, S. CAD for Detection of Fetal Electrocardiogram by using Wavelets and. Int. J. Appl. Eng. Res. 2016, 11, 2321–2326. [Google Scholar]
- Van Bemmel, J. Detection of weak foetal electrocardiograms by autocorrelation and crosscorrelation of envelopes. IEEE Trans. Biomed. 1968, 17–23. [Google Scholar] [CrossRef]
- Bergveld, P.; Meijer, W.J. A new technique for the suppression of the MECG. IEEE Trans. Biomed. 1981, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Levkov, C.; Mihov, G.; Ivanov, R.; Daskalov, I.; Christov, I.; Dotsinsky, I. Removal of power-line interference from the ECG: A review of the subtraction procedure. Biomed. Eng. Online 2005, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Hon, E.; Lee, S. Averaging techniques in fetal electrocardiography. Med. Electron. Biol. Eng. 1964, 2, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Varady, P. Wavelet-based adaptive denoising of phonocardiographic records. In Proceedings of the 23rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Istanbul, Turkey, 25–28 October 2001; Volume 2, pp. 1846–1849. [Google Scholar] [CrossRef]
- Alcaraz, R.; Rieta, J. Adaptive singular value QRST cancellation for the analysis of short single lead atrial fibrillation electrocardiograms. In Proceedings of the IEEE Computers in Cardiology, Durham, NC, USA, 30 September–3 October 2007; pp. 513–516. [Google Scholar] [CrossRef]
- Chmelka, L.; Kozumplik, J. Wavelet-basedwiener filter for electrocardiogram signal denoising. In Proceedings of the IEEE Computers in Cardiology, Lyon, France, 25–28 September 2005; pp. 771–774. [Google Scholar] [CrossRef]
- Sun, Y.; Chan, K.L.; Krishnan, S.M. ECG signal conditioning by morphological filtering. Comput. Biol. Med. 2002, 32, 465–479. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Wu, H.T. Extract fetal ECG from single-lead abdominal ECG by de-shape short time Fourier transform and nonlocal median. Front. Appl. Math. Stat. 2017, 3, 2. [Google Scholar] [CrossRef]
- Andreotti, F.; Behar, J.; Zaunseder, S.; Oster, J.; Clifford, G.D. An open-source framework for stress-testing non-invasive foetal ECG extraction algorithms. Physiol. Meas. 2016, 37, 627–648. [Google Scholar] [CrossRef] [PubMed]
- He, P.J.; Chen, X.M.; Liang, Y.; Zeng, H.Z. Extraction for fetal ECG using single channel blind source separation algorithm based on multi-algorithm fusion. In Proceedings of the MATEC Web of Conferences, EDP Sciences, Hong Kong, China, 26–27 April 2016; Volume 44. [Google Scholar]
- Agostinelli, A.; Sbrollini, A.; Burattini, L.; Fioretti, S.; Di Nardo, F.; Burattini, L. Noninvasive fetal electrocardiography part II: Segmented-Beat Modulation Method for signal denoising. Open Biomed. Eng. J. 2017, 11, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.J.; Lee, B. Sequential total variation denoising for the extraction of fetal ECG from single-channel maternal abdominal ECG. Sensors 2016, 16, 1020. [Google Scholar] [CrossRef] [PubMed]
- Warbhe, A.D.; Dharaskar, R.V.; Kalambhe, B. A single channel phonocardiograph processing using EMD, SVD, and EFICA. In Proceedings of the 2010 3rd International Conference on Emerging Trends in Engineering and Technology (ICETET), Goa, India, 19–21 November 2010; pp. 578–581. [Google Scholar] [CrossRef]
- Ahuja, E.; Shaikh, F. A Novel Approach to FEG Extraction Based on Fast ICA. Int. Res. J. Eng. Technol. 2016, 3, 2450–2453. [Google Scholar]
- Pani, D.; Argiolas, S.; Raffo, L. A dsp algorithm and system for real-time fetal ecg extraction. In Proceedings of the IEEE Computers in Cardiology, Bologna, Italy, 14–17 September 2008; pp. 1065–1068. [Google Scholar] [CrossRef]
- BIOMED. Biomed Database; Katholieke Universiteit Leuven: Leuven, Belgium, 2005. [Google Scholar]
- De Lathauwer, L. Private Communication; Katholieke Universiteit Leuven: Leuven, Belgium, 2010. [Google Scholar]
- Ananthanag, K.; Sahambi, J. Investigation of blind source separation methods for extraction of fetal ECG. In Proceedings of the IEEE Canadian Conference on Electrical and Computer Engineering (CCECE), Montreal, QC, Canada, 4–7 May 2003; Volume 3, pp. 2021–2024. [Google Scholar] [CrossRef]
- Marossero, D.E.; Erdogmus, D.; Euliano, N.; Principe, J.C.; Hild, K. Independent components analysis for fetal electrocardiogram extraction: A case for the data efficient mermaid algorithm. In Proceedings of the IEEE 13th Workshop on Neural Networks for Signal Processing (NNSP’03), Toulouse, France, 17–19 September 2003; pp. 399–408. [Google Scholar] [CrossRef]
- Camargo-Olivares, J.L.; Martín-Clemente, R.; Hornillo-Mellado, S.; Elena, M.; Román, I. The maternal abdominal ECG as input to MICA in the fetal ECG extraction problem. IEEE Signal Process. Lett. 2011, 18, 161–164. [Google Scholar] [CrossRef]
- Sevim, Y.; Atasoy, A. Performance evaluation of nonparametric ICA algorithm for fetal ECG extraction. Turk. J. Electr. Eng. Comput. Sci. 2011, 19, 657–666. [Google Scholar]
- Ye, Y.; Zhang, Z.L.; Zeng, J.; Peng, L. A fast and adaptive ICA algorithm with its application to fetal electrocardiogram extraction. Appl. Math. Comput. 2008, 205, 799–806. [Google Scholar] [CrossRef]
- Cichocki, A.; Amari, S.I.; Siwek, K.; Tanaka, T.; Phan, A.H.; Zdunek, R.; Cruces, S.; Georgiev, P.; Washizawa, Y.; Leonowicz, Z.; et al. ICALAB Toolboxes. Available online: http://www.bsp.brain.riken.jp/ICALAB (accessed on 17 October 2018).
- Leach, S. Singular Value Decomposition—A Primer. Available online: http://www.citeulike.org/group/474/article/493979 (accessed on 17 October 2018).
- De Lathauwer, L.; De Moor, B.; Vandewalle, J. SVD-based methodologies for fetal electrocardiogram extraction. In Proceedings of the IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP’00), Istanbul, Turkey, 5–9 June 2000; Volume 6, pp. 3771–3774. [Google Scholar] [CrossRef]
- Romero, I. PCA-based noise reduction in ambulatory ECGs. In Proceedings of the IEEE Computing in Cardiology, Belfast, UK, 26–29 September 2010; pp. 677–680. [Google Scholar]
- Bacharakis, E.; Nandi, A.K.; Zarzoso, V. Foetal ECG extraction using blind source separation methods. In Proceedings of the 8th IEEE European Signal Processing Conference (EUSIPCO), Trieste, Italy, 10–13 September 1996; pp. 1–4. [Google Scholar]
- Kharabian, S.; Shamsollahi, M.B.; Sameni, R. Fetal R-wave detection from multichannel abdominal ECG recordings in low SNR. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Hilton Minneapolis, MN, USA, 3–6 September 2009; pp. 344–347. [Google Scholar] [CrossRef]
- Martens, S.M.; Rabotti, C.; Mischi, M.; Sluijter, R.J. A robust fetal ECG detection method for abdominal recordings. Physiol. Meas. 2007, 28, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Barros, A.K.; Cichocki, A. Extraction of specific signals with temporal structure. Neural Comput. 2001, 13, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Ye, Y. Extended Barros’s extraction algorithm with its application in fetal ECG extraction. In Proceedings of the IEEE International Conference on Neural Networks and Brain (ICNN&B’05), Beijing, China, 13–15 October 2005; Volume 2, pp. 1077–1080. [Google Scholar] [CrossRef]
- Zhang, Z.L.; Yi, Z. Extraction of a source signal whose kurtosis value lies in a specific range. Neurocomputing 2006, 69, 900–904. [Google Scholar] [CrossRef]
- Jafari, F.; Tinati, M.A.; Mozaffari, B. A new fetal ECG extraction method using its skewness value which lies in specific range. In Proceedings of the 18th Iranian Conference on Electrical Engineering (ICEE), Isfahan, Iran, 11–13 May 2010; pp. 30–34. [Google Scholar] [CrossRef]
- Varanini, M.; Tartarisco, G.; Balocchi, R.; Macerata, A.; Pioggia, G.; Billeci, L. A new method for QRS complex detection in multichannel ECG: Application to self-monitoring of fetal health. Comput. Biol. Med. 2017, 85, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Redif, S. Fetal electrocardiogram estimation using polynomial eigenvalue decomposition. Turk. J. Electr. Eng. Comput. Sci. 2016, 24, 2483–2497. [Google Scholar] [CrossRef]
- Tan, B.; Peng, Q.; Lin, J.; Li, M. A novel method for estimating source number of fetal ECG. In Proceedings of the 2015 International Conference on Wireless Communications & Signal Processing (WCSP), Nanjing, China, 15–17 October 2015; pp. 1–6. [Google Scholar] [CrossRef]
- Da Poian, G.; Bernardini, R.; Rinaldo, R. Separation and analysis of fetal-ecg signals from compressed sensed abdominal ecg recordings. IEEE Trans. Biomed. Eng. 2016, 63, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Matonia, A.; Jezewski, J.; Horoba, K.; Gacek, A.; Labaj, P. The maternal ECG suppression algorithm for efficient extraction of the fetal ECG from abdominal signal. In Proceedings of the 28th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS’06), New York, NY, USA, 30 August–3 September 2006; pp. 3106–3109. [Google Scholar] [CrossRef]
- Akbari, H.; Shamsollahi, M.B.; Phlypo, R. Fetal ECG extraction using πTucker decomposition. In Proceedings of the International Conference on Systems, Signals and Image Processing (IWSSIP), London, UK, 10–12 September 2015; pp. 174–178. [Google Scholar] [CrossRef]
- Gupta, P.; Sharma, K.; Joshi, S. Fetal heart rate extraction from abdominal electrocardiograms through multivariate empirical mode decomposition. Comput. Biol. Med. 2016, 68, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Radek Martinek. Available online: www.sites.google.com/site/martinekradek/scientific-activity (accessed on 17 October 2018).
- Liu, G.; Luan, Y. An adaptive integrated algorithm for noninvasive fetal ECG separation and noise reduction based on ICA-EEMD-WS. Med. Biol. Eng. Comput. 2015, 53, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Behar, J.; Andreotti, F.; Zaunseder, S.; Li, Q.; Oster, J.; Clifford, G.D. An ECG simulator for generating maternal-foetal activity mixtures on abdominal ECG recordings. Physiol. Meas. 2014, 35, 1537. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Srivastava, M.; Khandelwal, V.; Gupta, A. A Novel approach to fetal ECG extraction and enhancement using blind source separation (BSS-ICA) and adaptive fetal ECG enhancer (AFE). In Proceedings of the IEEE 6th International Conference on Information, Communications & Signal Processing, Singapore, 10–13 December 2007; pp. 1–4. [Google Scholar] [CrossRef]
- Kotas, M. Combined application of independent component analysis and projective filtering to fetal ECG extraction. Biocybern. Biomed. Eng. 2008, 28, 75–93. [Google Scholar]
- Bergveld, P.; Kolling, A.J.; Peuscher, J.H. Real-time fetal ECG recording. IEEE Trans. Biomed. Eng. 1986, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Sameni, R.; Jutten, C.; Shamsollahi, M.B. Multichannel electrocardiogram decomposition using periodic component analysis. IEEE Trans. Biomed. Eng. 2008, 55, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- Martin-Clemente, R.; Camargo-Olivares, J.L.; Hornillo-Mellado, S.; Elena, M.; Roman, I. Fast technique for noninvasive fetal ECG extraction. IEEE Trans. Biomed. Eng. 2011, 58, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Chang, E.C.; Wyse, L. Blind separation of fetal ECG from single mixture using SVD and ICA. In Proceedings of the Joint Conference of the Fourth International Conference on Information, Communications and Signal Processing and Fourth Pacific Rim Conference on Multimedia, Singapore, 15–18 December 2003; Volume 3, pp. 1418–1422. [Google Scholar] [CrossRef]
- Ma, M.; Yang, Y.L.; Lei, S.Y. Blind extraction of fECG combining periodicity and kurtosis. In Proceedings of the 3rd International Conference on Bioinformatics and Biomedical Engineering (ICBBE), Beijing, China, 11–13 June 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Ayat, M.; Assaleh, K.; Al-Nashash, H. Fetal ECG extraction from a single abdominal ECG signal using SVD and polynomial classifiers. In Proceedings of the IEEE Workshop on Machine Learning for Signal Processing (MLSP), Cancun, Mexico, 16–19 October 2008; pp. 250–254. [Google Scholar] [CrossRef]
- Ayat, M.; Assaleh, K.; Al-Nashash, H. Extracting fetal ECG from a single maternal abdominal record. In Proceedings of the IEEE 8th GCC Conference and Exhibition (GCCCE), Muscat, Oman, 1–4 February 2015; pp. 1–4. [Google Scholar] [CrossRef]
- Baska-Vincze, B.; Baska, F.; Szenci, O. Fetal heart rate and fetal heart rate variability in Lipizzaner broodmares. Acta Vet. Hung. 2015, 63, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, P.; Alphonse, J.; Henry, A.; Wang, J.; Redmond, S.; Welsh, A. Beat-to-beat variability of fetal myocardial performance index. Ultrasound Obstet. Gynecol. 2017, 50, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Fruhman, G.; Gavard, J.A.; McCormick, K.; Wilson-Griffin, J.; Amon, E.; Gross, G.A. Standard External Doppler Fetal Heart Tracings (efhr) Versus External Fetal Ecg (fecg) in Premature Gestations [17q]. Obstet. Gynecol. 2016, 127, 143S. [Google Scholar] [CrossRef]
- Becker, J.H.; Krikhaar, A.; Schuit, E.; Mårtendal, A.; Maršál, K.; Kwee, A.; Visser, G.H.; Amer-Wåhlin, I. The added predictive value of biphasic events in ST analysis of the fetal electrocardiogram for intrapartum fetal monitoring. Acta Obstet. Gynecol. Scand. 2015, 94, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Sacco, A.; Muglu, J.; Navaratnarajah, R.; Hogg, M. ST analysis for intrapartum fetal monitoring. Obstet. Gynaecol. 2015, 17, 5–12. [Google Scholar] [CrossRef]
- Rosen, K. Fetal Scalp Electrode. U.S. Patent 10/380,556, 22 January 2004. [Google Scholar]
- Fuchs, T.; Grobelak, K.; Pomorski, M.; Zimmer, M. Fetal Heart Rate Monitoring Using Maternal Abdominal Surface Electrodes in Third Trimester: Can We Obtain Additional Information Other than CTG Trace? Adv. Clin. Exp. Med. 2016, 25, 309–316. [Google Scholar] [CrossRef] [PubMed]
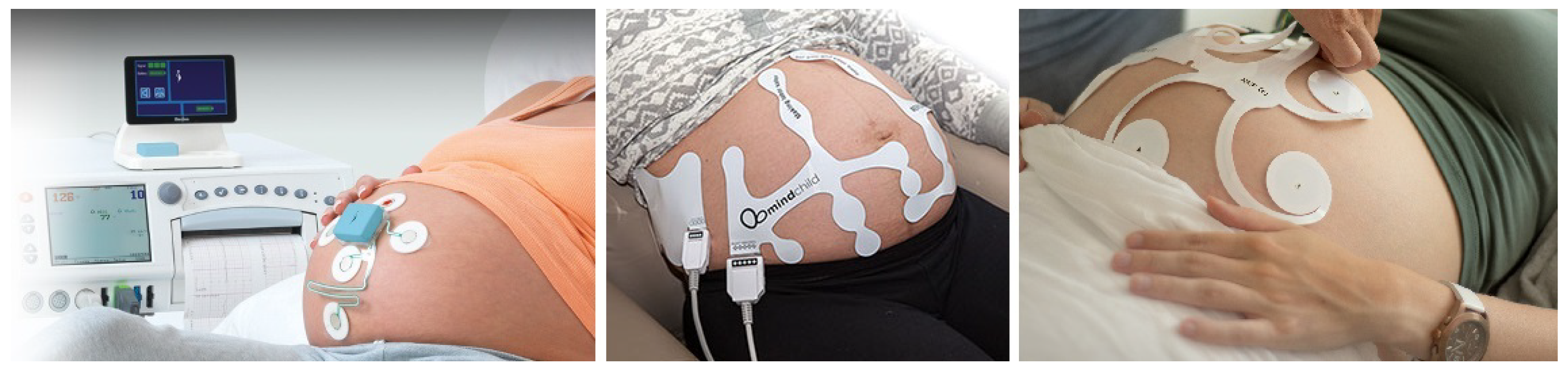

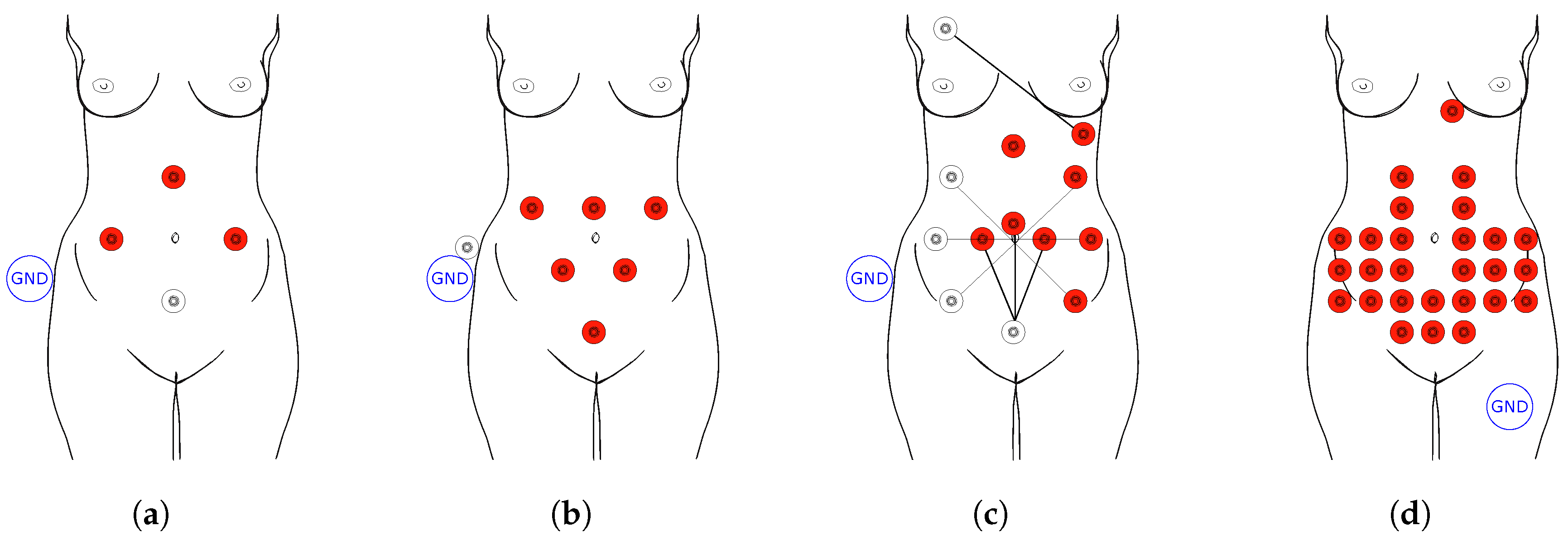
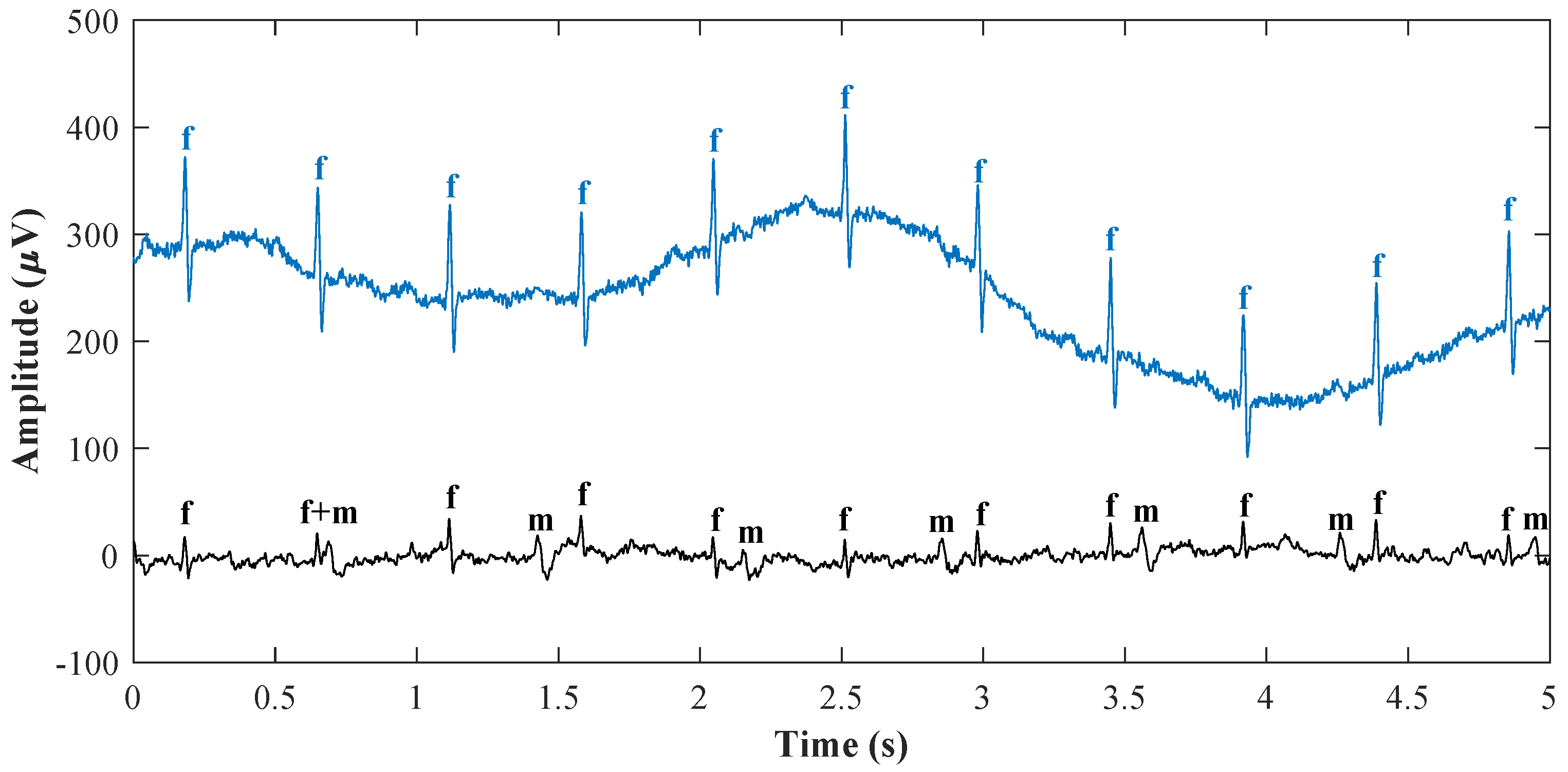
| Method | Gestational Age Restriction | Technical Solution | Advantages | Disadvantages |
|---|---|---|---|---|
| NI-fECG [1,2,3,6,7,8] | ≥20 | Standard ECG electrodes placed on mother’s abdomen (number of electrodes differs) | Cheap Relatively accurate Easy to handle Comfortable (mobility) Continuous monitoring Fetal heart rate monitoring | Low signal-to-noise ratio Significant amount of overlapped undesired signals Without morphological fECG analysis * (ST segment analysis) |
| I-fECG [9,10,11] | Only during labor | Transvaginal scalp electrode (fetal scalp electrode) | fECG morphology analysis [42,43] Continuous monitoring Fetal heart rate monitoring Accurate | Invasive (risk of infection) Expensive Limited movement Uncomfortable Requires skilled personnel |
| CTG [9,31,32,33] | ≥20 Possible to use during labor | One tranducer for fetal heart rate measurement and one tranducer for uterine contractions measurement | Smoothed heart rate time series Rather robust and reliable Most used method in clinical practice Relatively cheap Easy to implement | Not possible to assess beat-to-beat variability Ultrasound irradiation Not passive |
| fECHO [22,23,24,25] | ≥18 | The transducer in the probe serves as a transmitter and receiver for ultrasound signals | Provides reliable data on cardiac morphology as well as deviations and blood flow velocity deviations | Expensive Requires skilled personnel Ultrasound irradiation Not suitable for continual monitoring |
| fPCG [4,5,12,13,14,15,16,17,18,19,20,21] | ≥20 Possible to use | Microphone sensors (or optical ** sensors) attached to the mother’s abdomen | Cheap No energy is transmitted Determination of multiple pregnancies Possibility of home monitoring (cell phone) | Not used in clinical practice Susceptible to movement artifacts |
| fMCG [26,27,28,29,30] | ≥20 | Fetal magnetic field detection by superconducting quantum interferencedevice sensors located near the maternal abdomen | Better morphological analysis due to higher signal-to-noise ratio | Expensive Requires skilled personnel Complexity of measurement No long term monitoring possible to date |
| Method | Overall Performance | SNR Improvement | Computational Cost | Real-Time | Implementation Complexity |
|---|---|---|---|---|---|
| WT | Medium | Medium | Low | Yes | Medium |
| CT | Low | Low | Low | Yes | Simple |
| ST | Low | Low | Low | Yes | Simple |
| AT | Low | Low | Low | Yes | Simple |
| FT | Low | Low | Low | Yes | Simple |
| STFT & NM | Medium | Medium | Medium | No | Medium |
| SCBSS | Medium | Medium | Medium | No | Complex |
| TS | Medium | Low | Low | No | Medium |
| STVD | Medium | Medium | Medium | No | Complex |
| EMD | Medium | Medium | High | No | Medium |
| Method | Overall Performance | SNR Improvement | Computational Cost | Real-Time | Implementation Complexity |
|---|---|---|---|---|---|
| ICA | Medium | Medium | Medium | No | Medium |
| SVD | Low | Low | Low | Yes | Simple |
| PCA | Low | Medium | Low | Yes | Simple |
| CA | Medium | High | Low | No | Simple |
| SA | Medium | Medium | Medium | No | Medium |
| BA | Medium | Medium | Low | No | Simple |
| ZA | Medium | Medium | Low | No | Simple |
| SM | Medium | Medium | Medium | No | Simple |
| QIO | Medium | Medium | Medium | No | Medium |
| PEVD | Medium | Medium | Medium | No | Medium |
| FCM | Medium | Medium | Medium | Yes | Simple |
| CS | Medium | Medium | Medium | Yes | Medium |
| MCSM | Medium | Medium | Medium | No | Simple |
| Tucker | Medium | Medium | Low | No | Simple |
| MEMD | Medium | Medium | Medium | No | Medium |
| Method | Overall Performance | SNR Improvement | Computational Cost | Real-Time | Implementation Complexity |
|---|---|---|---|---|---|
| ICA-EEMD-WS | High | High | High | Yes | Complex |
| ICA & AF | High | Medium | High | No | Complex |
| ICA & PF | High | Medium | High | No | Complex |
| -ICA | Medium | Medium | Medium | No | Medium |
| ICA & PCA | Medium | Medium | Medium | Yes | Medium |
| ICA & SVD | Medium | Medium | Medium | Yes | Medium |
| BA & ZA | High | Medium | Low | No | Simple |
| SVD & PC | Medium | Medium | Low | No | Medium |
| PN & SGSF | Medium | Medium | Medium | No | Medium |
| Method | fHR (R-R) | Morphology Analysis (T/QRS; QT) | Dataset | Technical Aspects |
|---|---|---|---|---|
| WT [51,52,56,58] Section 2.1 | Moderately accurate | Insufficient | De-Moor [53]; NI-fECG [54,55]; U. Nottingham [57] | 3 synt. records [51]; T = 10 s; Fs = 250 Hz [53]; T = 10 s; Fs = 1 kHz; res = 16 b; GA = 21–40 weeks; 55 real records [54,55]; T = 60 s; Fs = 300 Hz; res = 12 b; data = 15 real records [57] |
| CT [57,59] Section 2.2 | Inaccurate | Insufficient | — | Old method; Without tech. specification [59] |
| ST [60,61] Section 2.3 | Inaccurate | Insufficient | — | Old method; Without tech. specification [60,61] |
| AT [62] Section 2.4 | Inaccurate | Insufficient | — | Old method; Without tech. specification [62] |
| FT [64,65,66] Section 2.5 | Inaccurate | Insufficient | MIT-BIH [67] | T = 15 s; Fs = 1 kHz; 15 real records [64] Fs = 500 Hz [65]; T = 0.5 h; Fs = 360 Hz; res = 11 b; 48 real records [67] |
| STFT & NM [68] Section 2.6 | Accurate | Insufficient | fecgsyndb [69]; adfecgdb [10,47,48,49,50]; ECG physionet challenge 2013 [50] | T = 300 s; Fs = 250 Hz; res = 16 b; 1750 synt. records; 34 channels [69]; T = 300 s; Fs = 1 kHz; res = 16 b; T = 300 s; Fs = 1 kHz; res = 16 b; GA = 38–41 weeks; 5 real records; 5 channels [10,47,48,49,50]; T = 60, 600 and 3600 s; Fs = 1 kHz; res = 12 b; 4 channels; 175 real records [50] |
| SCBSS [70] Section 2.7 | Accurate | Insufficient | MIT-BIH [67] | 1 synt. record [70]; T = 0.5 h; Fs = 360 Hz; res = 11 b; 48 real records [67] |
| TS [45,71] Section 2.8 | Moderately accurate | Insufficient | adfecgdb [10,47,48,49,50] | T = 300 s; Fs = 1 kHz; res = 16 b; GA = 38–41 weeks; 5 real records; 5 channels [10,47,48,49,50] |
| STVD [72] Section 2.9 | Accurate | Insufficient | fecgsyndb [69]; ECG physionet challenge 2013 [50] | T = 300 s; Fs = 250 Hz; res = 16 b; 1750 synt. records; 34 channels [69]; T = 60, 600 and 3600 s; Fs = 1 kHz; res = 12 b; 4 channels; 175 real records [50] |
| EMD [5,73] Section 2.10 | Accurate | Insufficient | — | Without tech. specification [5,73] |
| ICA [74,75] [78,79,80,81,82] Section 3.1 | Accurate | Moderately accurate | Katholieke U. Leuven [76]; de Lathauwer [77]; NI-fECG [54,55]; ICALAB toolbox [83]; De-Moor [53] | T = 10 s; Fs = 250 Hz; res = 12 b; 8 real records [76]; T = 300 s; Fs = 500 Hz; 8 channels[79]; T = 300 s; Fs = 500 Hz; 8 channels[79]; T = 60 s; Fs = 500 Hz; res = 12 b; 8 channels [77]; T = 10 s; Fs = 1 kHz; res = 16 b; GA = 21–40 weeks; 55 real records [54,55]; T = 10 s; Fs = 250 Hz [53] |
| SVD [85] Section 3.2 PCA [88] Section 3.3 | Inaccurate Moderately accurate | Insufficient Insufficient | — — | T = 60 s; Fs = 500 Hz; 8 channels [85] T = 10 s; Fs = 500 Hz; 8 real records [88] |
| CA [88] Section 3.4 | Very accurate | Moderately accurate | — | Fs = 500 Hz; 8 synt. records [88] |
| SA [89] Section 3.5 | Accurate | Moderately accurate | — | 20 real records [89] |
| BA [90,91] Section 3.6 | Moderately accurate | Insufficient | De-Moor [53] | T = 10 s; Fs = 250 Hz [53] |
| ZA [92] Section 3.7 | Moderately accurate | Insufficient | De-Moor [53] | 4 synt. records [92]; T = 10 s; Fs = 250 Hz [53] |
| SM [93] Section 3.8 | Accurate | Moderately accurate | De-Moor[53] | T = 10 s; Fs = 250 Hz [53] |
| QIO [94] Section 3.9 | Accurate | Moderately accurate | ECG physionet challenge 2013 [50] | T = 60, 600 and 3600 s; Fs = 1 kHz; res = 12 b; 4 channels; 175 real records [50] |
| PEVD [95] Section 3.10 | Accurate | Moderately accurate | MIT-BIH [67]; ECG physionet challenge 2013 [50] | T = 0.5 h; Fs = 360 Hz; res = 11 b; 48 real records [67]; T = 60, 600 and 3600 s; Fs = 1 kHz; res = 12 b; 4 channels; 175 real records [50] |
| FCM [96] Section 3.11 | Accurate | Moderately accurate | — | T = 7 s; Fs = 500 Hz; 2 real records [96] |
| CS [97] Section 3.12 | Accurate | Moderately accurate | adfecgdb [10,47,48,49,50]; ECG physionet challenge 2013 [50] | T = 300 s; Fs = 1 kHz; res = 16 b; GA = 38–41 weeks; 5 real records; 5 channels [10,47,48,49,50]; T = 60, 600 and 3600 s; Fs = 1 kHz; res = 12 b; 4 channels; 175 real records [50] |
| MCSM [98] Section 3.13 | Accurate | Moderately accurate | — | 3 real records [98] |
| Tucker [99] Section 3.14 | Moderately accurate | Insufficient | adfecgdb [10,47,48,49,50] | T = 300 s; Fs = 1 kHz; res = 16 b; GA = 38–41 weeks; 5 real records; 5 channels [10,47,48,49,50] |
| MEMD [100] Section 3.15 | Accurate | Moderately accurate | adfecgdb [10,47,48,49,50]; | T = 300 s; Fs = 1 kHz; res = 16 b; GA = 38–41 weeks; 5 real records; 5 channels [10,47,48,49,50] 1 real record [101] |
| ICA-EEMD-WS [102] Section 4.1 | Very accurate | Promising | NI-fECG gen. [103]; MIT-BIH [67]; adfecgdb [10,47,48,49,50] | Fs = 1 kHz; 500 synt. records [103]; T = 0.5 h; Fs = 360 Hz; res = 11 b; 48 real records [67]; T = 300 s; Fs = 1 kHz; res = 16 b; GA = 38–41 weeks; 5 real records; 5 channels [10,47,48,49,50] |
| ICA & AF [104] Section 4.2 | Very accurate | Promising | MIT-BIH [67]; | T = 0.5 h; Fs = 360 Hz; res = 11 b; 48 real records [67]; T = 10 s; Fs = 250 Hz [53] |
| ICA & PF [105] Section 4.3 | Very accurate | Promising | — | 4 real records; 4 channels [105] |
| -ICA [107] Section 4.4 | Very accurate | Moderately accurate | De-Moor [53] | T = 10 s; Fs = 250 Hz [53] |
| ICA & PCA [108] Section 4.5 | Accurate | Moderately accurate | ECG toolbox [108]; de Lathauwer [77] | 8 synt. records; 5000 samples [108]; T = 60 s; Fs = 500 Hz; res = 12 b; 8 channels [77]; |
| ICA & SVD [109] Section 4.6 | Accurate | Moderately accurate | — | 2 synt. records [109]; T = 600 s; Fs = 300 Hz; 1 real record [109] |
| BA & ZA [110] Section 4.7 | Very accurate | Promising | De-Moor [110] | T = 10 s; Fs = 250 Hz [110] |
| SVD & PC [111] Section 4.8 | Moderately accurate | Insufficient | de Lathauwer [77] | 1 synt. record [111]; T = 60 s; Fs = 500 Hz; res = 12 b; 8 channels [77]; |
| PN & SGSF [112] Section 4.9 | Accurate | Insufficient | ECG physionet challenge 2013 [50] | 1 synt. record [112]; T = 60, 600 and 3600 s; Fs = 1 kHz; res = 12 b; 4 channels; 175 real records [50] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaros, R.; Martinek, R.; Kahankova, R. Non-Adaptive Methods for Fetal ECG Signal Processing: A Review and Appraisal. Sensors 2018, 18, 3648. https://doi.org/10.3390/s18113648
Jaros R, Martinek R, Kahankova R. Non-Adaptive Methods for Fetal ECG Signal Processing: A Review and Appraisal. Sensors. 2018; 18(11):3648. https://doi.org/10.3390/s18113648
Chicago/Turabian StyleJaros, Rene, Radek Martinek, and Radana Kahankova. 2018. "Non-Adaptive Methods for Fetal ECG Signal Processing: A Review and Appraisal" Sensors 18, no. 11: 3648. https://doi.org/10.3390/s18113648
APA StyleJaros, R., Martinek, R., & Kahankova, R. (2018). Non-Adaptive Methods for Fetal ECG Signal Processing: A Review and Appraisal. Sensors, 18(11), 3648. https://doi.org/10.3390/s18113648






