A Label-Free Fluorescent Array Sensor Utilizing Liposome Encapsulating Calcein for Discriminating Target Proteins by Principal Component Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Liposomes
2.3. Fabricated Photometric System
3. Results and Discussions
3.1. Optimization of Fluorescence of Calcein in Arrayed Microwells
3.2. Effect of Cholesterol on Liposome-Protein Interaction
3.3. Effect of Phospholipid Species on Liposome-Protein Interaction
3.4. Evaluation by Principal Component Analysis
3.5. Performance Analysis of the Array Sensor
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tachi, T.; Kaji, M.; Tokeshi, M.; Baba, Y. Microchip-based immunoassay. Anal. Sci. 2007, 56, 521–534. [Google Scholar] [CrossRef][Green Version]
- Nyberg, L.; Persson, F.; Åkerman, B.; Westerlund, F. Heterogeneous staining: A tool for studies of how fluorescent dyes affect the physical properties of DNA. Nucleic Acids Res. 2013, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.; Min, K.; Jung, J.; Jang, W.; Kwon, Y. Chemical biology-based approaches on fluorescent labeling of proteins in live cells. Mol. BioSyst. 2013, 9, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Sapsford, K.E.; Sun, S.; Francis, J.; Sharma, S.; Kostov, Y.; Rasooly, A. A fluorescence detection platform using spatial electroluminescent excitation for measuring botulinum neurotoxin A activity. Biosens. Bioelectron. 2008, 24, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Van Weemen, B.K.; Schuurs, A.H. Immunoassay using antigen-enzyme conjugates. FEBS Lett. 1971, 15, 232–236. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Skopinskaya, S.N.; Yarkov, S.P.; Zherdev, A.V.; Dzantiev, B.B. Development of liposome immune lysis assay for the herbicide atrazine. J. Immunoass. Immunochem. 2004, 25, 279–294. [Google Scholar] [CrossRef]
- Tomioka, K.; Nakayama, T.; Kumada, Y.; Katoh, S. Characterization of competitive measurement of antigens by use of antigen-coupled liposomes. Kagaku Kogaku Ronbunnshu 2005, 31, 346–351. [Google Scholar] [CrossRef]
- Kumada, Y.; Tomioka, K.; Katoh, S. Characteristics of liposome immunosorbent assay (LISA) using liposomes encapsulating cpenzyme β–NAD+. J. Chem. Eng. Jpn. 2001, 34, 943–947. [Google Scholar] [CrossRef]
- Takada, K.; Fujimoto, T.; Shimanouchi, T.; Fukuzawa, M.; Yamashita, K.; Noda, M. A new discrimination method of target biomolecules with miniaturized sensor array utilizing liposome encapsulating fluorescent molecules with time course analysis. In Proceedings of the 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences, MicroTAS, Freiburg, Germany, 27–31 October 2013; pp. 194–196. [Google Scholar]
- Shimanouchi, T.; Oyama, E.; Thi Vu, H.; Ishii, H.; Umakoshi, H.; Kuboi, R. Membranomics research on interactions between liposome membranes with membrane chip analysis. Membrane 2009, 34, 1–9. [Google Scholar]
- Shimanouchi, T.; Oyama, E.; Thi Vu, H.; Ishii, H.; Umakoshi, H.; Kuboi, R. Monitoring of membrane damages by dialysis treatment: Study with membrane chip analysis. Desalination Water Treat. 2010, 17, 45–51. [Google Scholar] [CrossRef]
- Shimanouchi, T.; Ishii, H.; Yoshimoto, N.; Umakoshi, H.; Kuboi, R. Calcein Permeation across phosphatidylcholine bilayer membrane: Effects of membrane fluidity, liposome size, and immobilization. Colloids Surf. B 2009, 73, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Kuboi, R.; Shimanouchi, T.; Yoshimoto, M.; Umakoshi, H. Detection of protein conformation under stress conditions using liposomes as sensor materials. Sens. Mater. 2004, 16, 241–254. [Google Scholar]
- Hertog, A.L.D.; Sang, H.W.F.; Kraayenhof, R.; Bolscher, J.G.M.; Hof, W.V.; Veerman, E.C.I.; Amerongen, A.V.N. Interactions of histatin 5 and histatin 5-derived peptides with liposome membranes: Surface effects, translocation and permeabilization. Biochem. J. 2004, 379, 556–672. [Google Scholar] [CrossRef] [PubMed]
- Davidsen, J.; Jørgensen, K.; Andresen, T.L.; Mouritsen, O.G. Secreted phospholipase A2 as a new enzymatic trigger mechanism for localised liposomal drug release and absorption in diseased tissue. Biochim. Biophys. Acta Biomembr. 2003, 1609, 95–101. [Google Scholar] [CrossRef]
- Katsu, T.; Imamura, T.; Komagoe, K.; Masuda, K.; Mizushima, T. Simultaneous measurements of K+ and calcein release from liposomes and the determination of pore size formed in a membrane. Anal. Sci. 2007, 23, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Wang, C.; Ma, L.; Sun, Y.; Shang, D. Investigation of the role of tryptophan residues in cationic antimicrobial peptides to determine the mechanism of antimicrobial action. J. Appl. Microbiol. 2013, 115, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.; Lee, J.K.; Lee, J.H.; Kim, Y.G.; Oh, G.C.; Seo, C.H.; Park, Y. Effect of repetitive lysine-tryptophan motifs on the eukaryotic membrane. Int. J. Mol. Sci. 2013, 14, 2190–2202. [Google Scholar] [CrossRef] [PubMed]
- Komura, S.; Imai, M. Physics of Heterogeneous structures in biomembranes. J. Phys. Soc. Jpn. 2013, 68, 1–10. [Google Scholar]
- Komura, S.; Shirotori, H. Phase transition and phase separation in biomembranes. J. Phys. Soc. Jpn. 2005, 60, 128–132. [Google Scholar]
- Sharma, N.; Litoriya, R. Incorporating Data Mining Techniques on Software Cost Estimation: Validation and Improvement. IJETAE 2012, 2, 301–309. [Google Scholar]
- Noda, M.; Shimanouchi, T.; Okuyama, M.; Kuboi, R. A bio-thermochemical microbolometer with immobilized intact liposome on sensor solid surface. Sens. Actuators B Chem. 2008, 135, 40–45. [Google Scholar] [CrossRef]
- Murata, N.; Imamura, R.; Du, W.; Fukuzawa, M.; Noda, M. An image processing platform for expendable bio-sensing system with arrayed biosensor and embedded imager. In Proceedings of the 3rd International Conference on Applied Computing and Information Technology/2nd International Conference on Computational Science and Intelligence, Okayama, Japan, 12–16 July 2015; pp. 239–242. [Google Scholar]
- Hamann, S.; Kiilgaard, J.F.; Litman, T.; Alvarez-Leefmans, F.J.; Winther, B.R.; Zeuthen, T. Measurement of cell volume changes by fluorescence self-quenching. J. Fluoresc. 2002, 12, 139–145. [Google Scholar] [CrossRef]
- Roberts, K.E.; O’Keeffe, A.K.; Lloyd, C.J.; Clarke, D.J. Selective dequenching by photobleaching increases fluorescence probe visibility. J. Fluoresc. 2003, 13, 513–517. [Google Scholar] [CrossRef]
- Wang, T.; Smith, E.A.; Chapman, E.R.; Weisshaar, J.C. Lipid mixing and content release in single-vesicle SNARE-driven fusion assay with 1–5 ms resolution. Biophys. J. 2009, 96, 4122–4131. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Y.; Wang, Y.; Ding, H.; Su, Z. Different effects of l-arginine on protein refolding: Suppressing aggregates of hydrophobic interaction, not covalent binding. Biotechnol. Prog. 2008, 24, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Touch, V.; Hayakawa, S.; Fukada, K.; Aratani, Y.; Sun, Y. Preparation of antimicrobial reduced lysozyme compatible in food applications. J. Agric. Food Chem. 2003, 51, 5154–5161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sohgawa, M.; Yamashita, K.; Noda, M. A micromechanical cantilever-based liposome biosensor for characterization of protein-membrane interaction. Electroanalysis 2016, 28, 620–625. [Google Scholar] [CrossRef]
- Oku, N. Novel development of liposome application for development of artificial cell. Anal. NTS Press. 2005, 2005, 7–8. [Google Scholar]
- Yoshimoto, N.; Yoshimoto, M.; Yasuhara, K.; Shimanouchi, T.; Umakoshi, H.; Kuboi, R. Evaluation of temperature and guanidine hydrochloride-induced protein–liposome interactions by using immobilized liposome chromatography. Biochem. Eng. J. 2006, 29, 174–181. [Google Scholar]
- Shimanouchi, T.; Yoshimoto, N.; Hiroiwa, A.; Nishiyama, K.; Hayashi, K.; Umakoshi, H. Relationship between the mobility of phosphocholine headgroup and the protein-liposome interacation: A dielectric sperctoscopic study. Colloids Surf. B 2014, 116, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Fujimori, T.; Sato, H.; Ishikawa, G.; Kami, K.; Ohashi, Y. Statistical Hypothesis Testing of Factor Loading in Principal Component Analysis and Its Application to Metabolite Set Enrichment Analysis. BMC Bioinform. 2014, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Dai, Y. Principal component analysis based methods in bioinformatics studies. Brief. Bioinform. 2011, 12, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.C. Multivariate QSAR. J. Braz. Chem. Soc. 2002, 13, 742–753. [Google Scholar] [CrossRef]

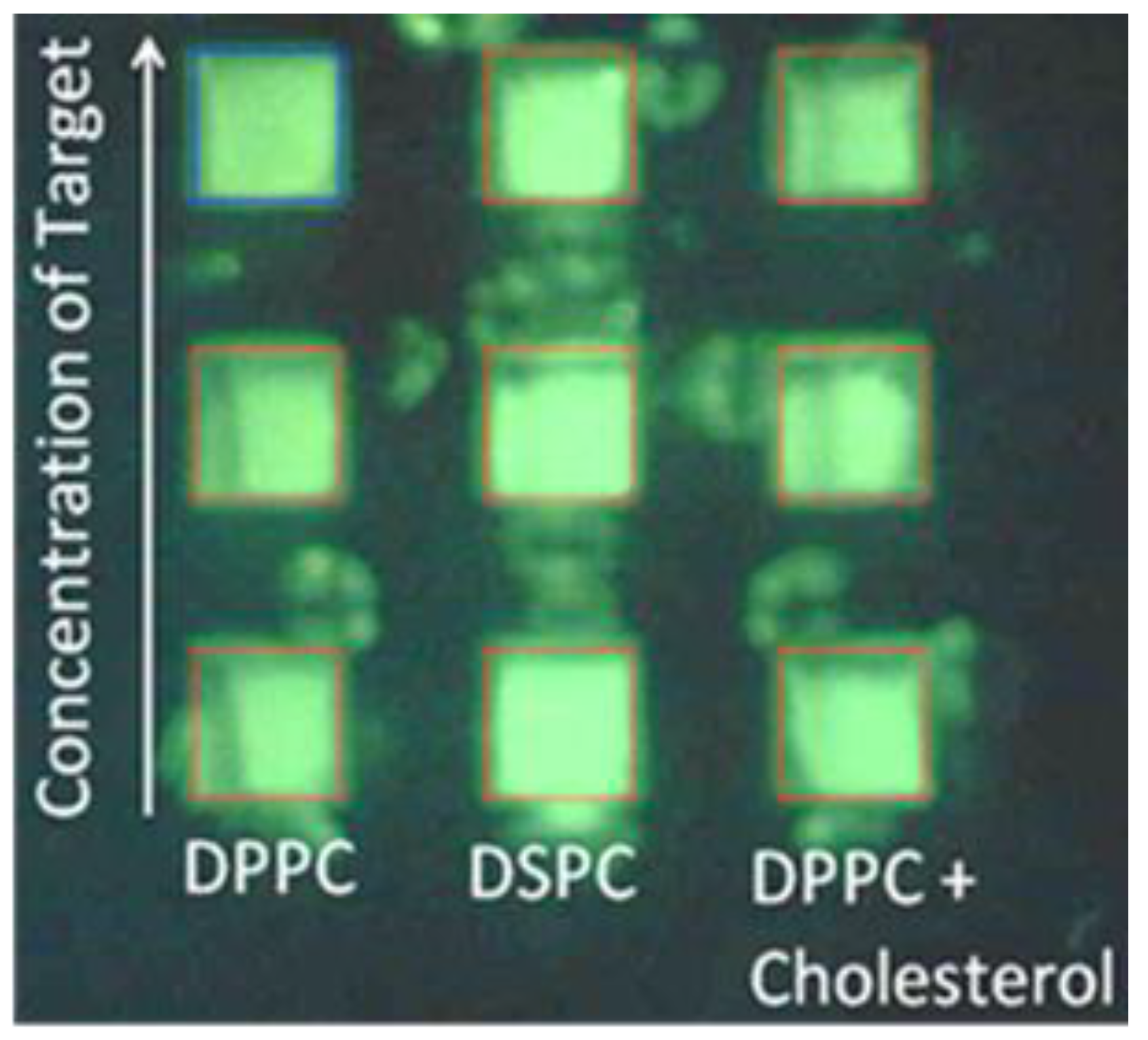
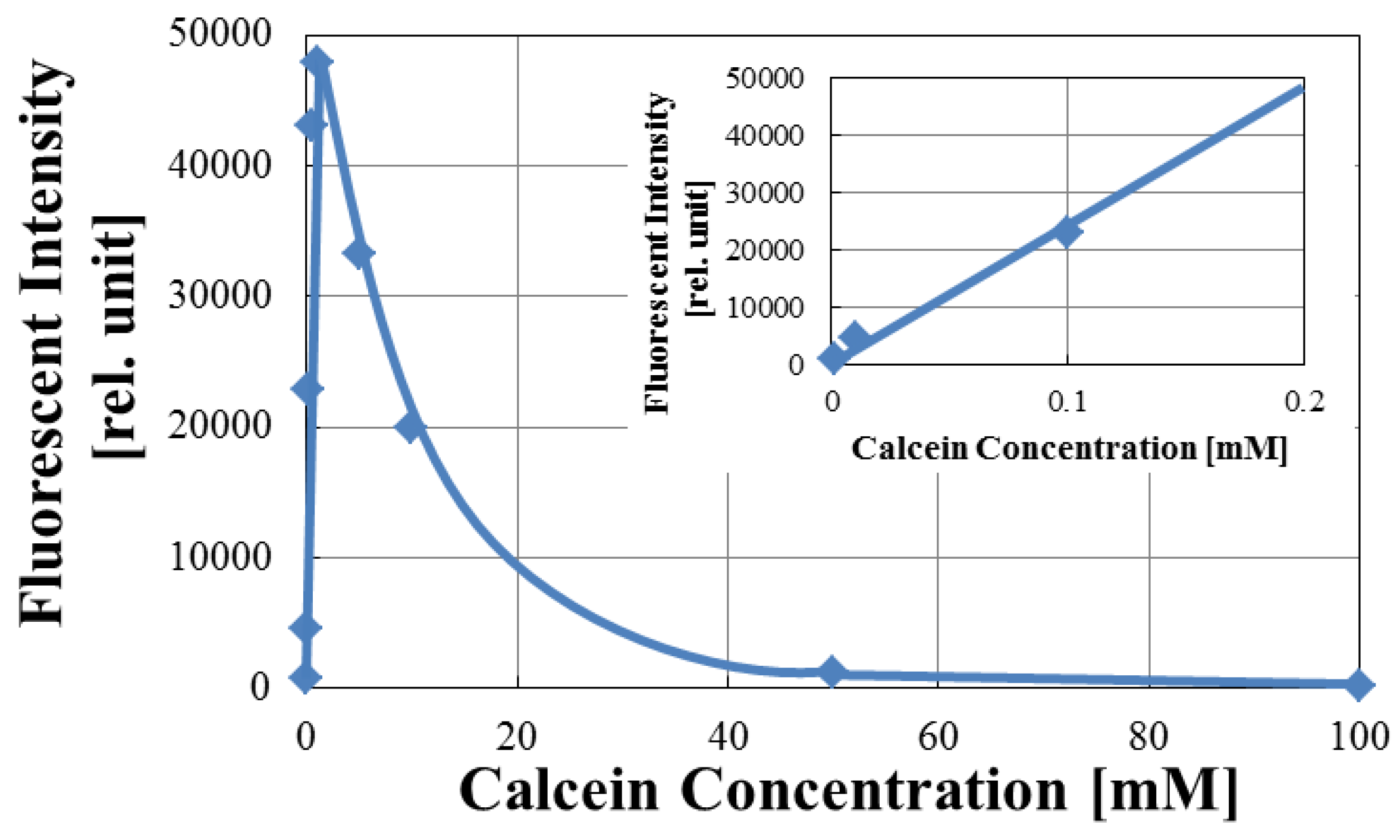
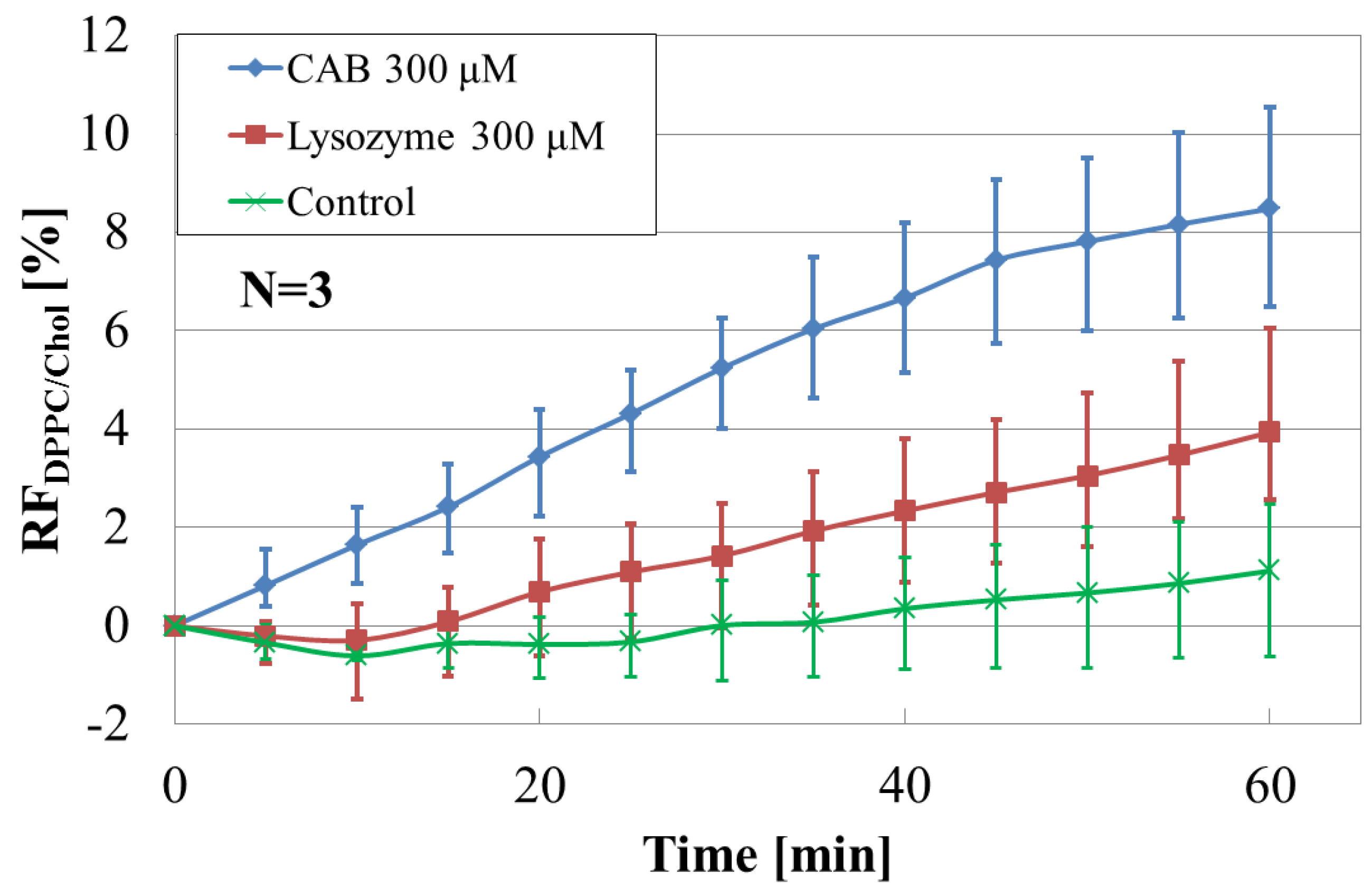
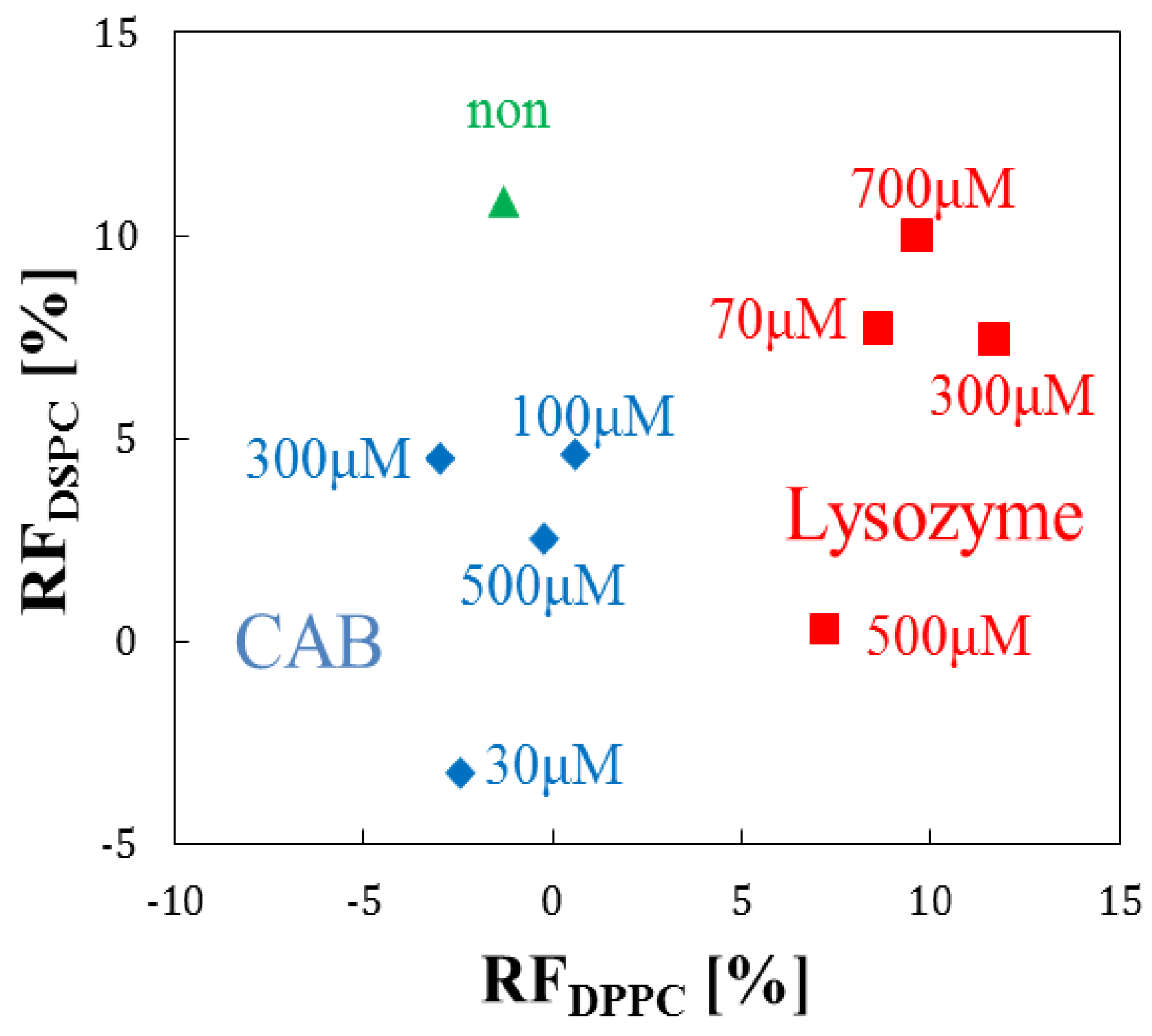
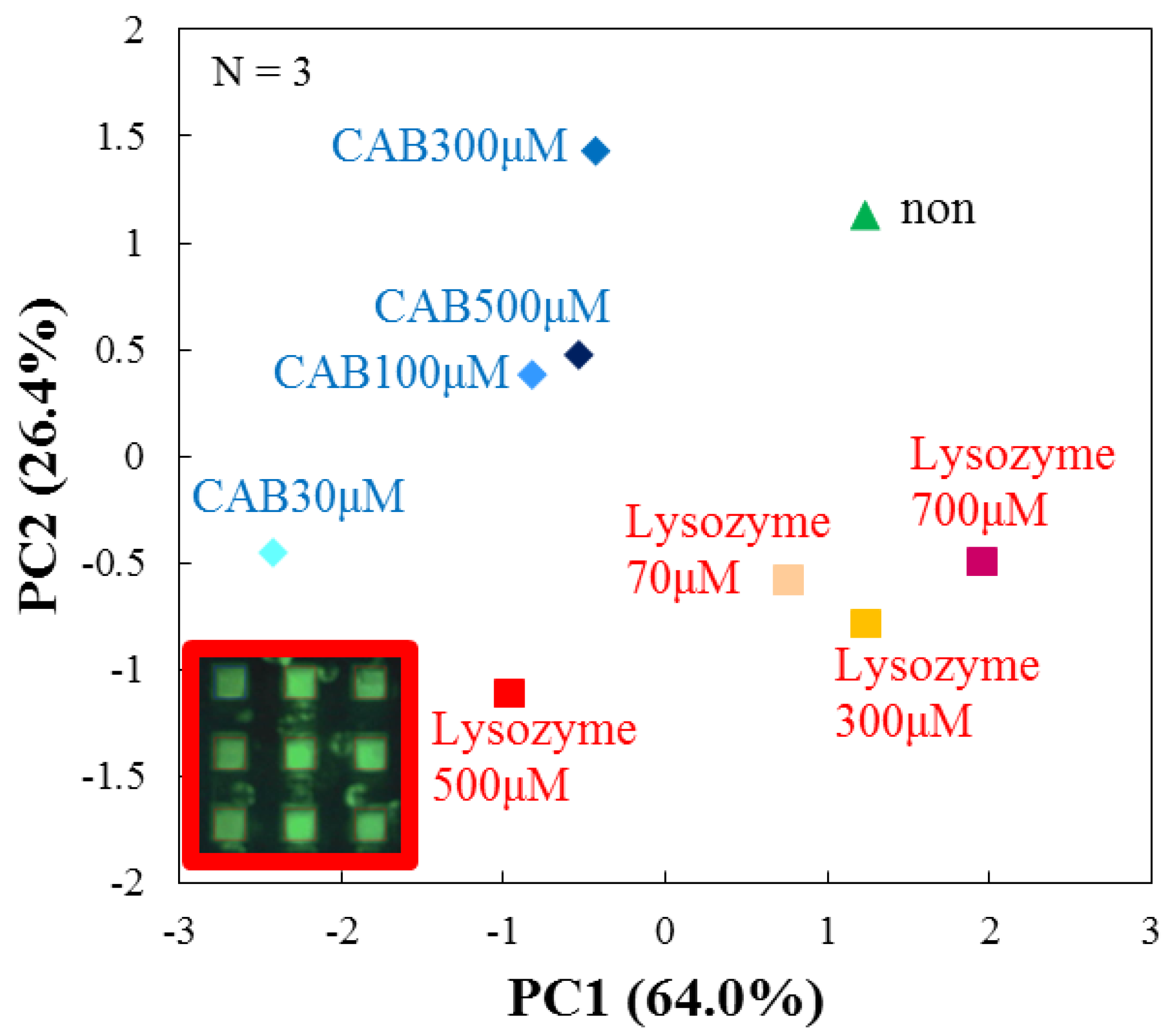
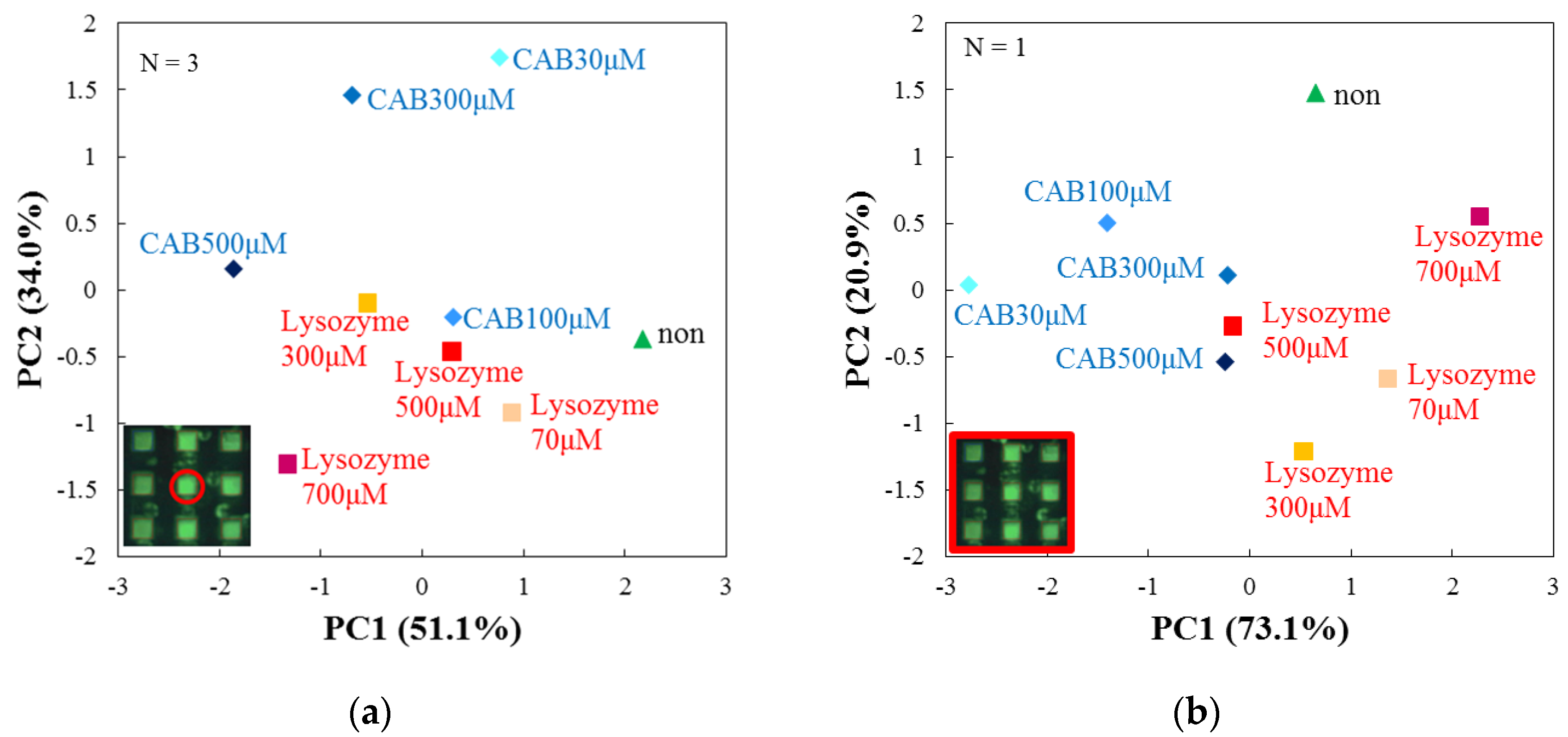
| Target Protein (Conc.) | RF(%) for DPPC | RF(%) for DPPC + Cholesterol |
|---|---|---|
| CAB (100 µM) | −1 ± 2 | −1 ± 2 |
| CAB (300 µM) | 2 ± 1 | 8 ± 2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imamura, R.; Murata, N.; Shimanouchi, T.; Yamashita, K.; Fukuzawa, M.; Noda, M. A Label-Free Fluorescent Array Sensor Utilizing Liposome Encapsulating Calcein for Discriminating Target Proteins by Principal Component Analysis. Sensors 2017, 17, 1630. https://doi.org/10.3390/s17071630
Imamura R, Murata N, Shimanouchi T, Yamashita K, Fukuzawa M, Noda M. A Label-Free Fluorescent Array Sensor Utilizing Liposome Encapsulating Calcein for Discriminating Target Proteins by Principal Component Analysis. Sensors. 2017; 17(7):1630. https://doi.org/10.3390/s17071630
Chicago/Turabian StyleImamura, Ryota, Naoki Murata, Toshinori Shimanouchi, Kaoru Yamashita, Masayuki Fukuzawa, and Minoru Noda. 2017. "A Label-Free Fluorescent Array Sensor Utilizing Liposome Encapsulating Calcein for Discriminating Target Proteins by Principal Component Analysis" Sensors 17, no. 7: 1630. https://doi.org/10.3390/s17071630
APA StyleImamura, R., Murata, N., Shimanouchi, T., Yamashita, K., Fukuzawa, M., & Noda, M. (2017). A Label-Free Fluorescent Array Sensor Utilizing Liposome Encapsulating Calcein for Discriminating Target Proteins by Principal Component Analysis. Sensors, 17(7), 1630. https://doi.org/10.3390/s17071630




