A Novel Spatial Feature for the Identification of Motor Tasks Using High-Density Electromyography
Abstract
:1. Introduction
2. Materials and Methods
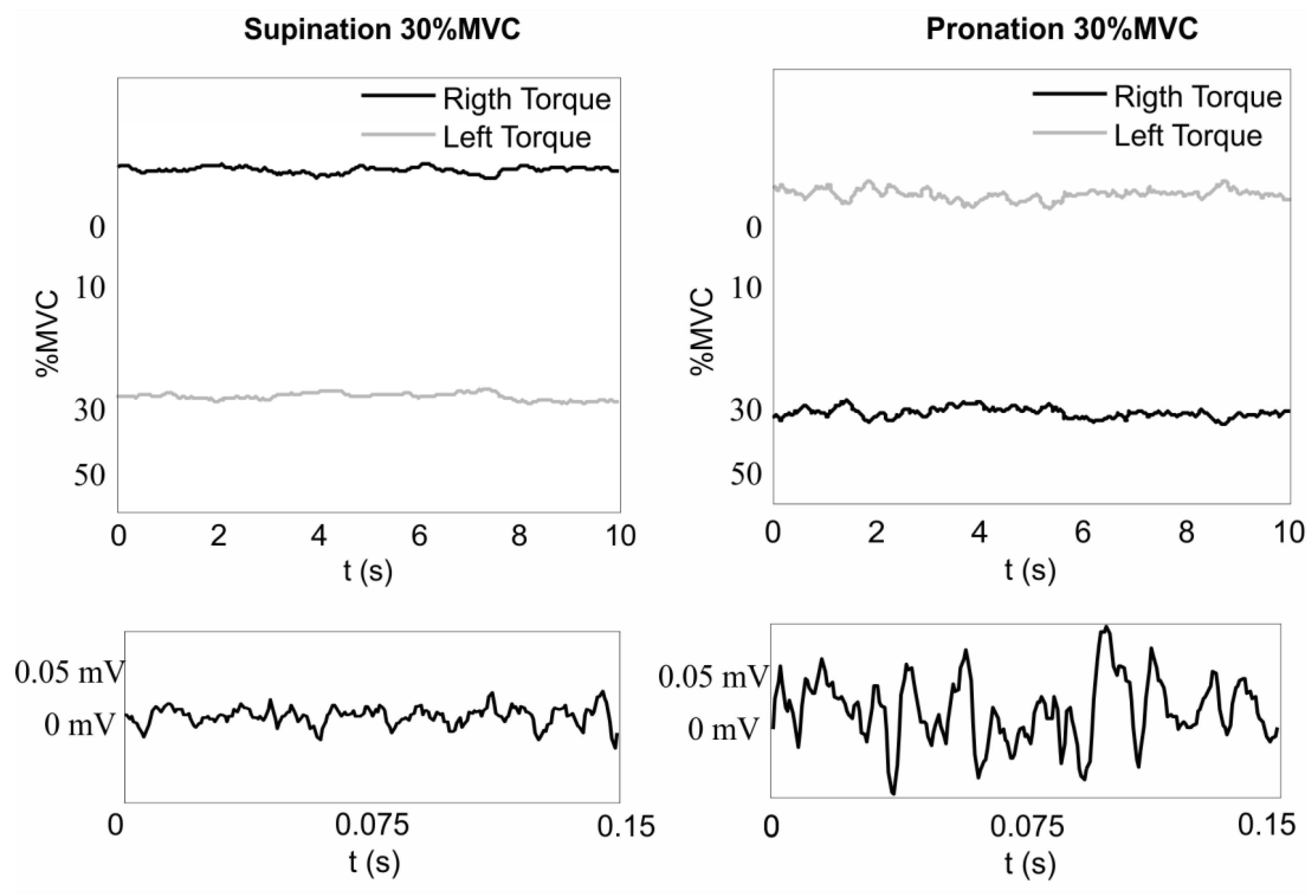
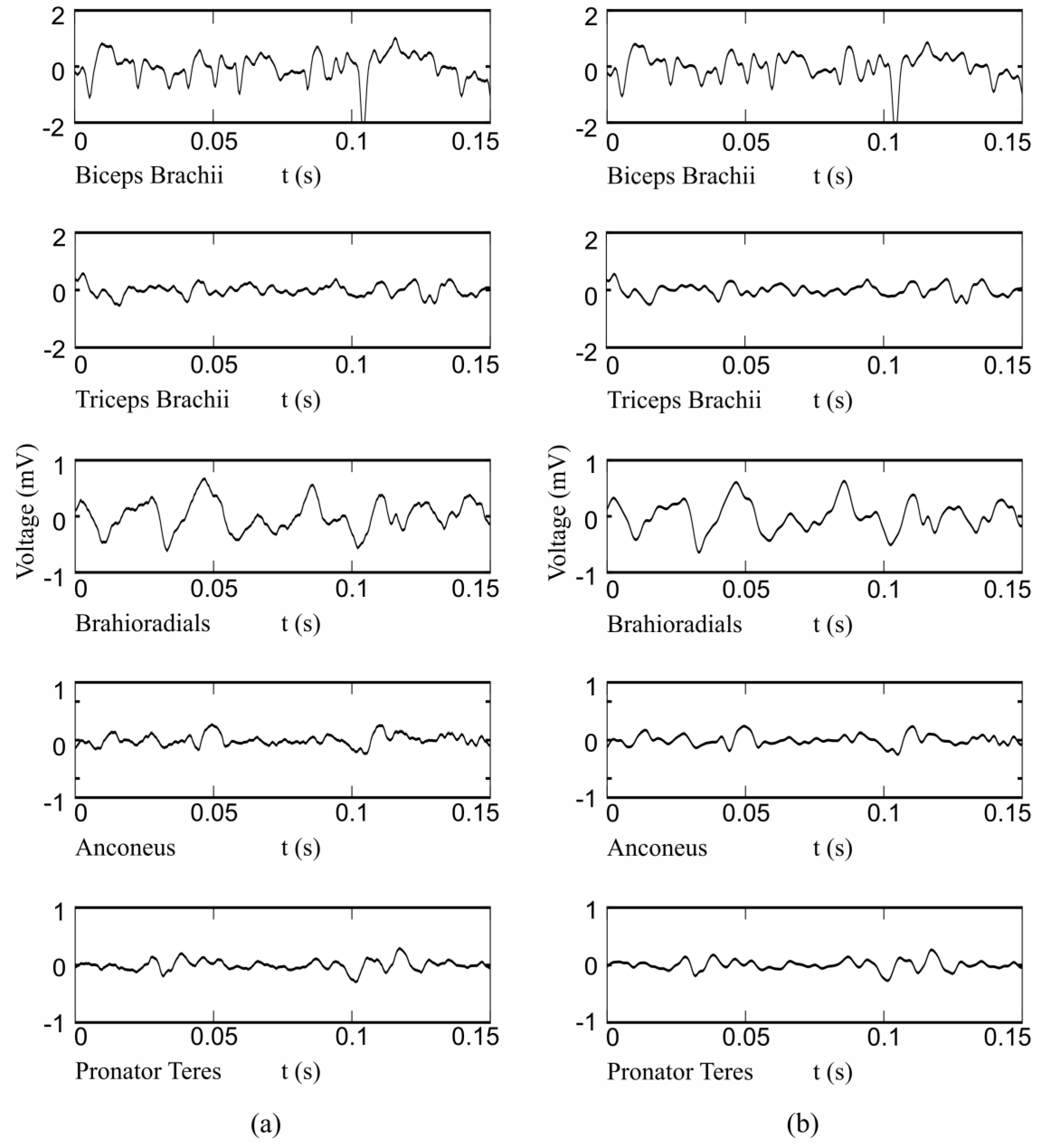
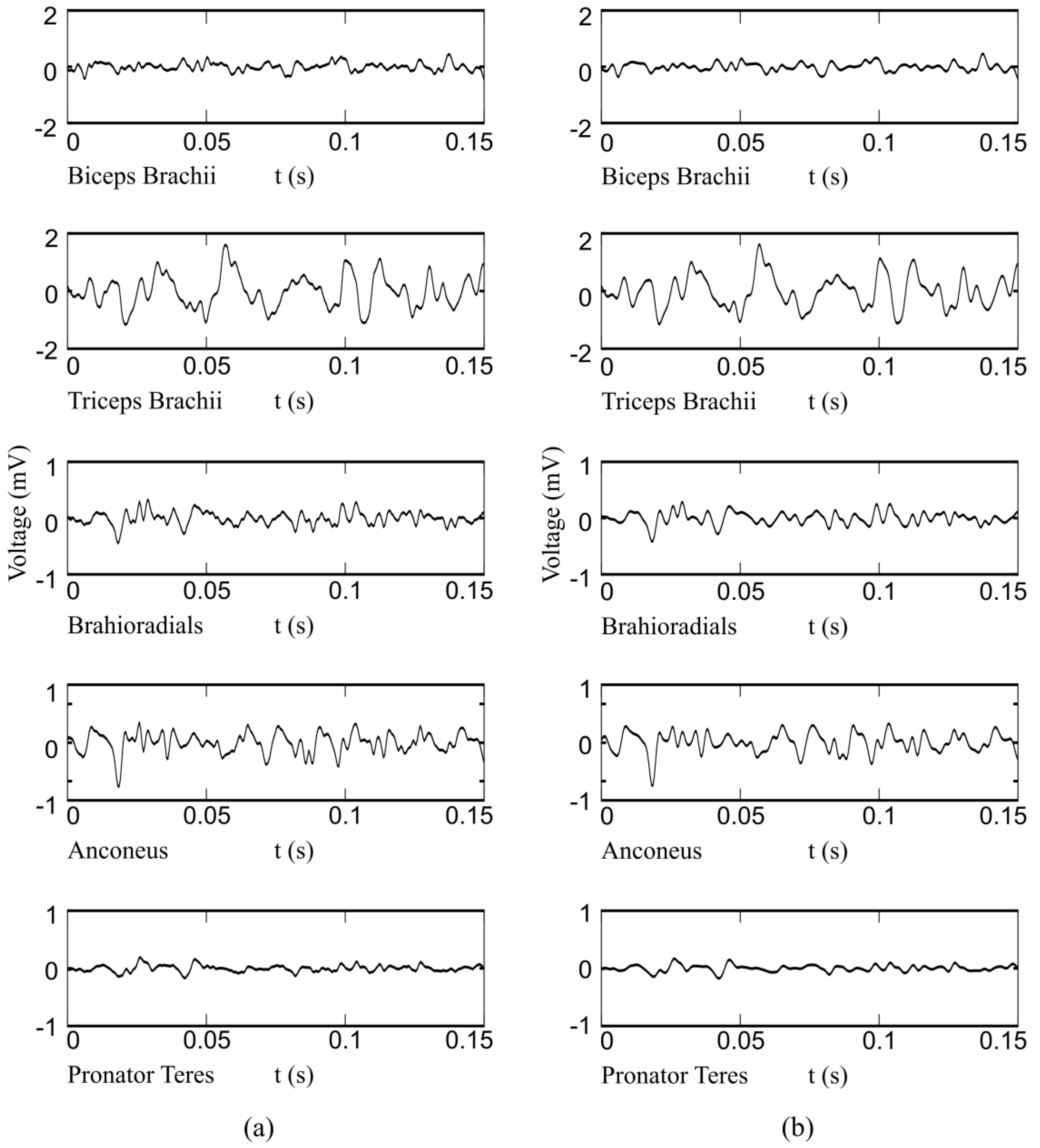
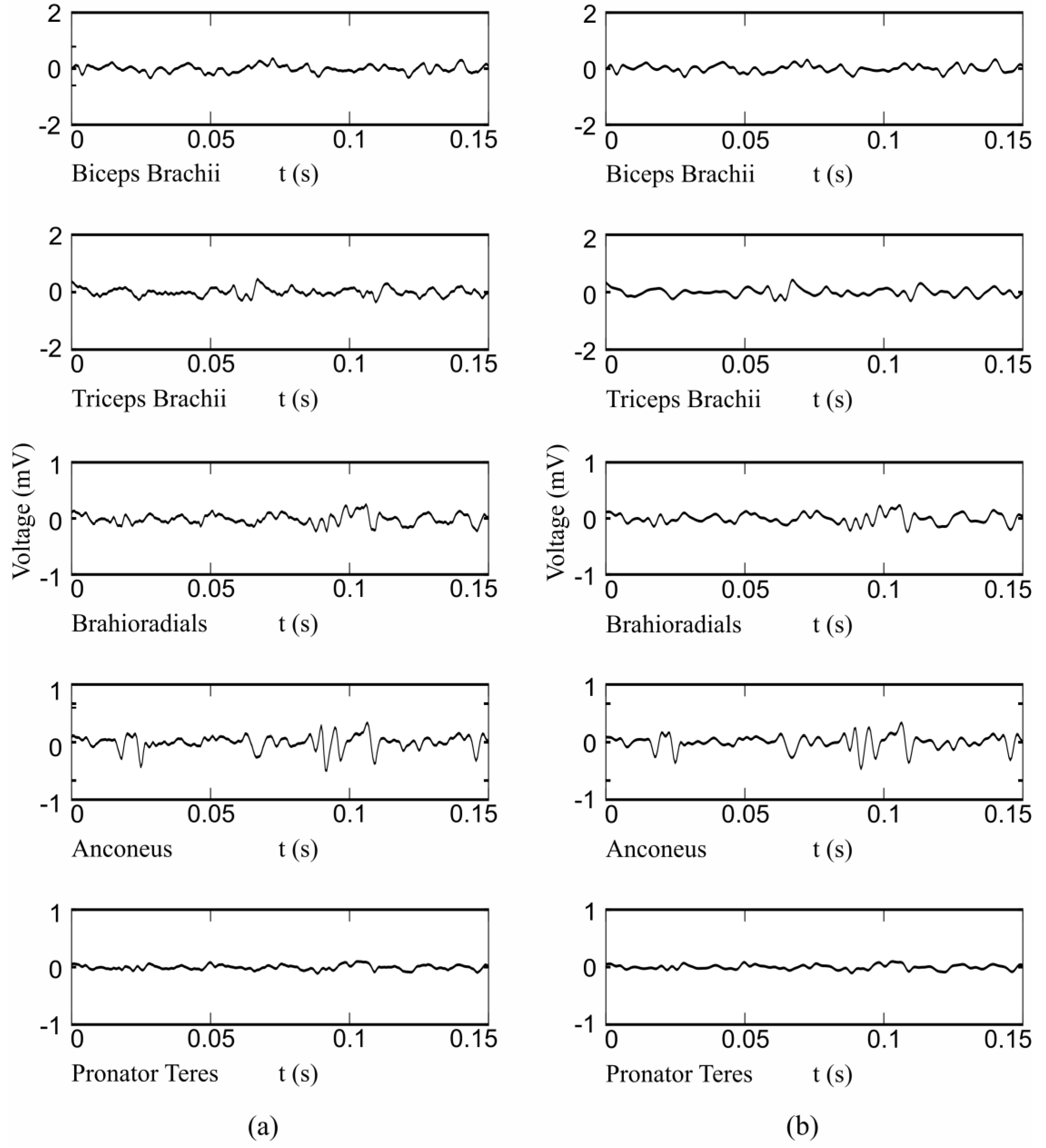
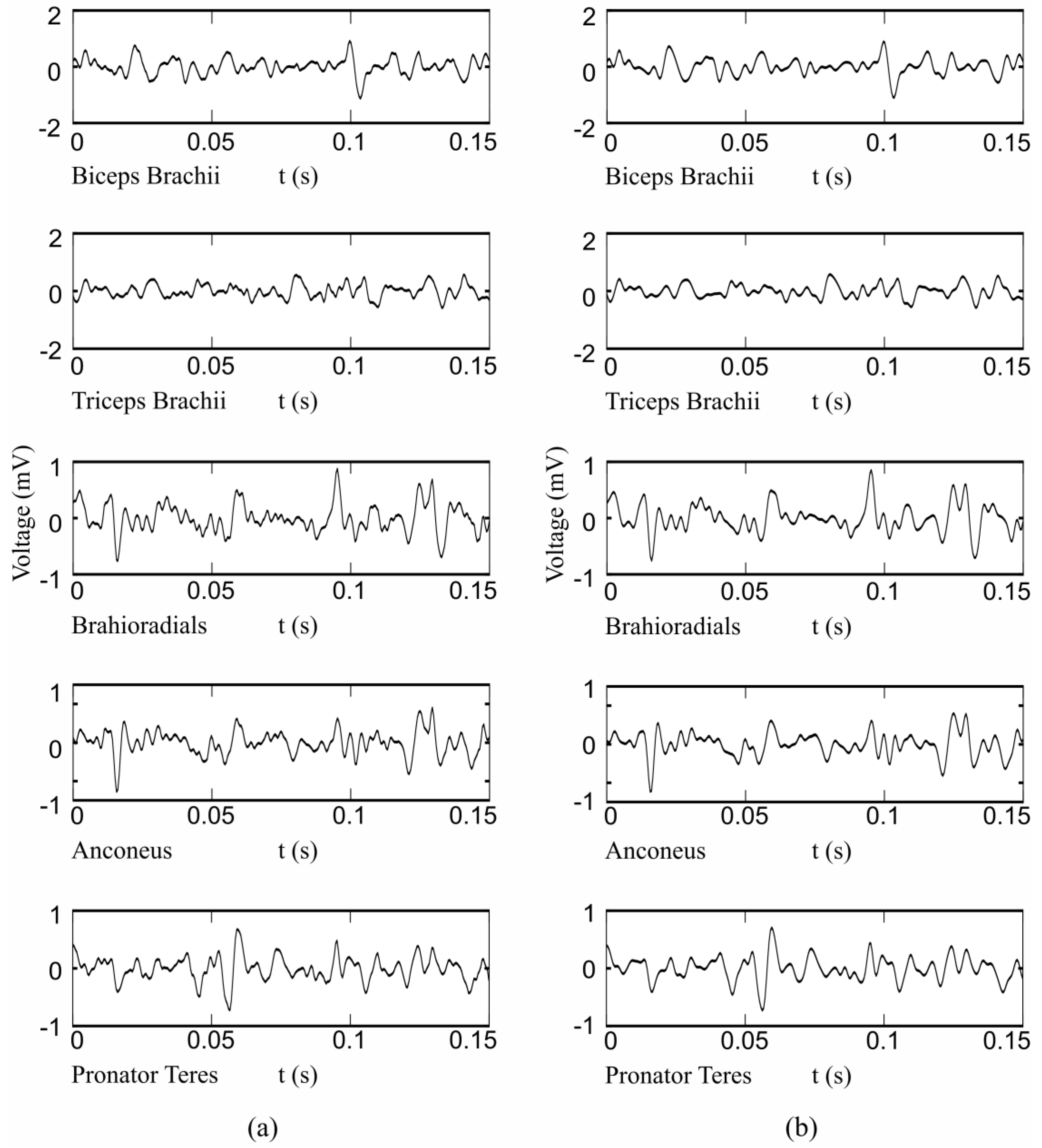
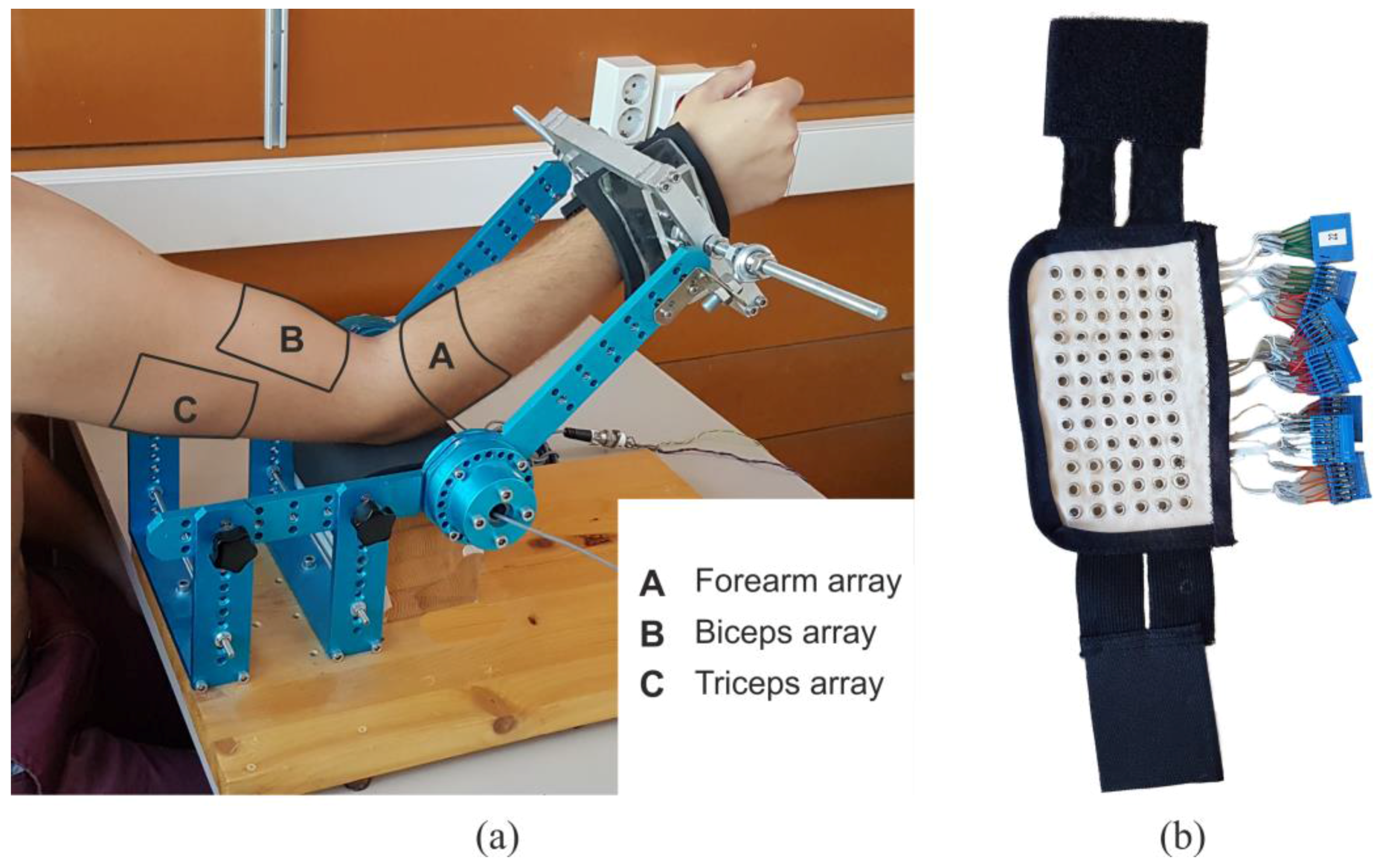
2.1. Instrumentation and Measurement Protocol
2.2. HD-EMG Processing
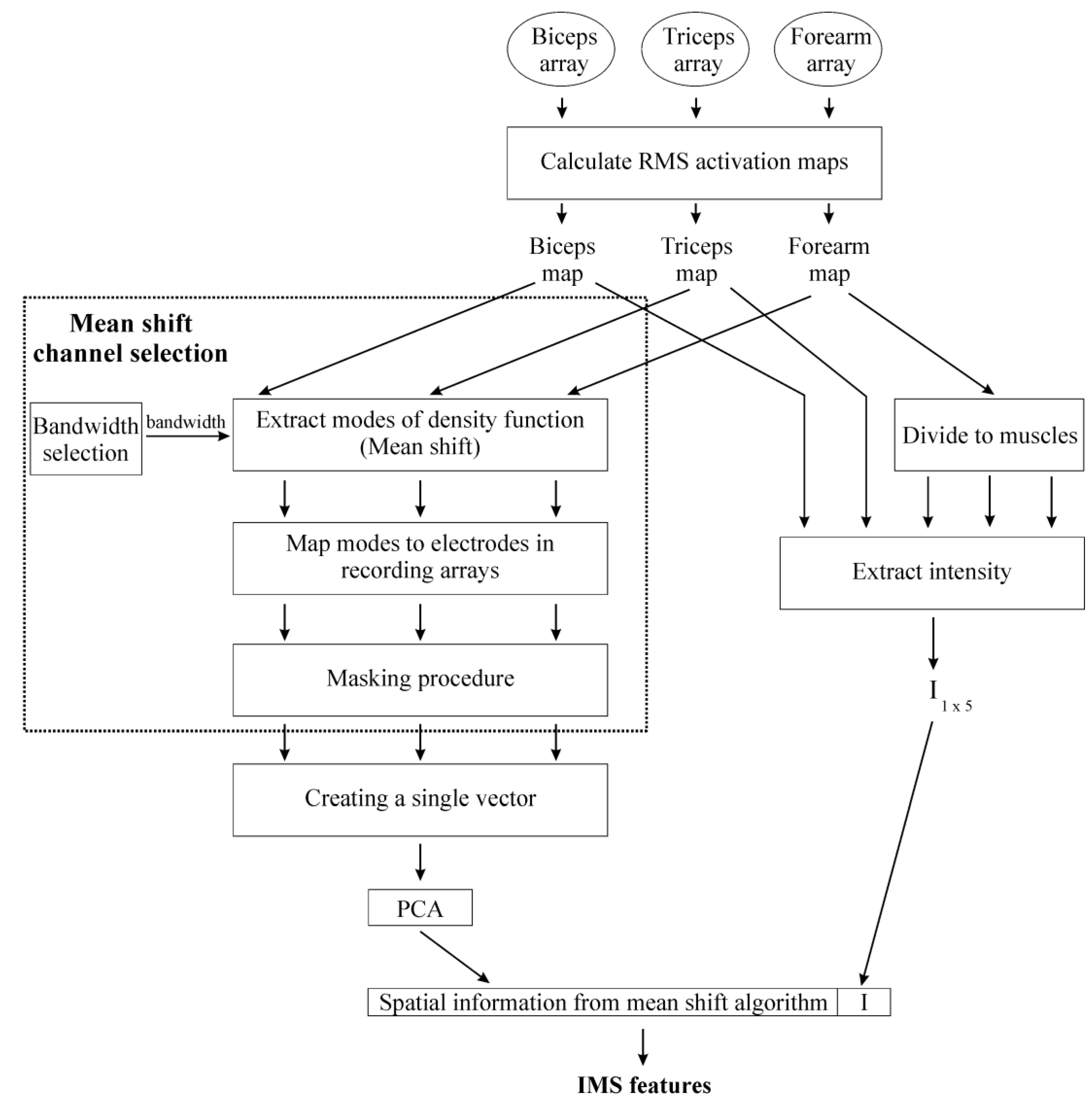
2.3. Feature Extraction
2.4. Task Identification
- Short-term identification
- Long term identification
- Identification during fatigue
2.5. Statistical Methods
3. Results
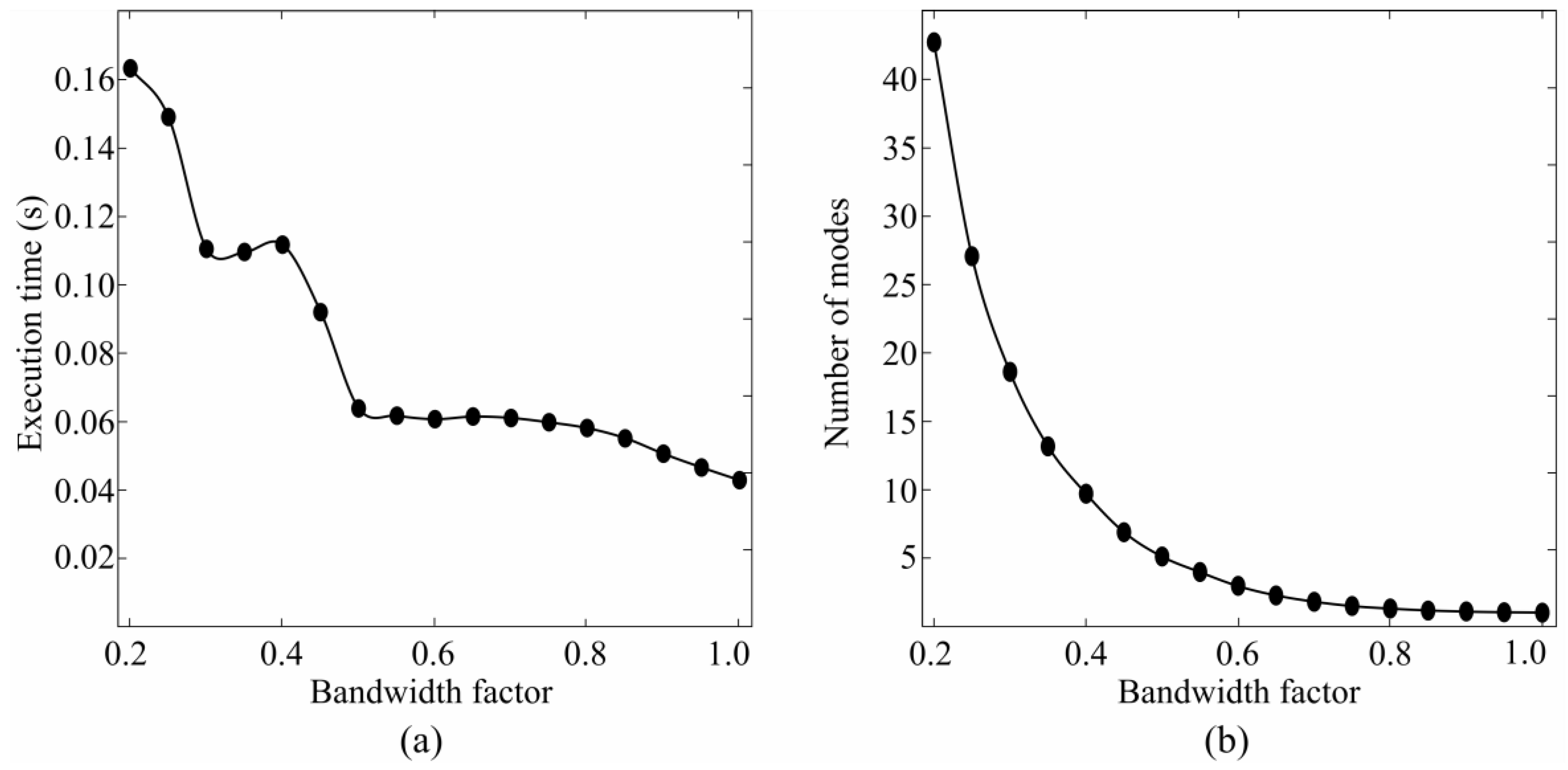
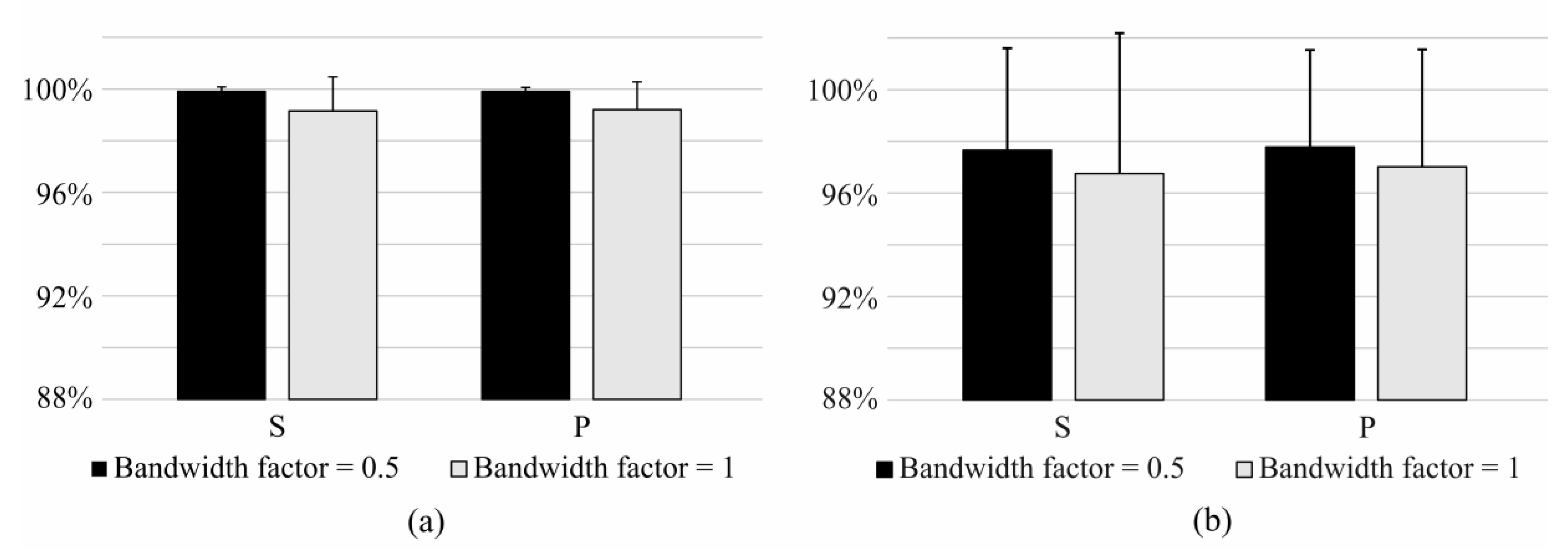
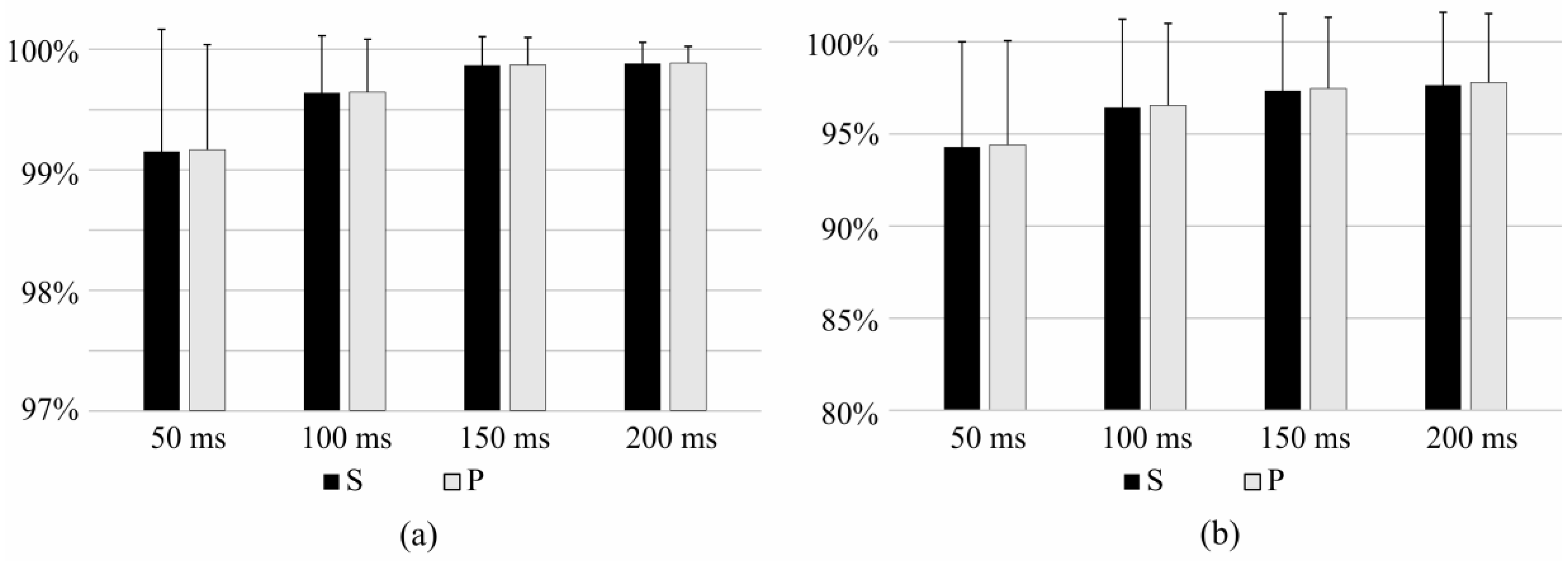
3.1. Bandwidth and Time Window Selection
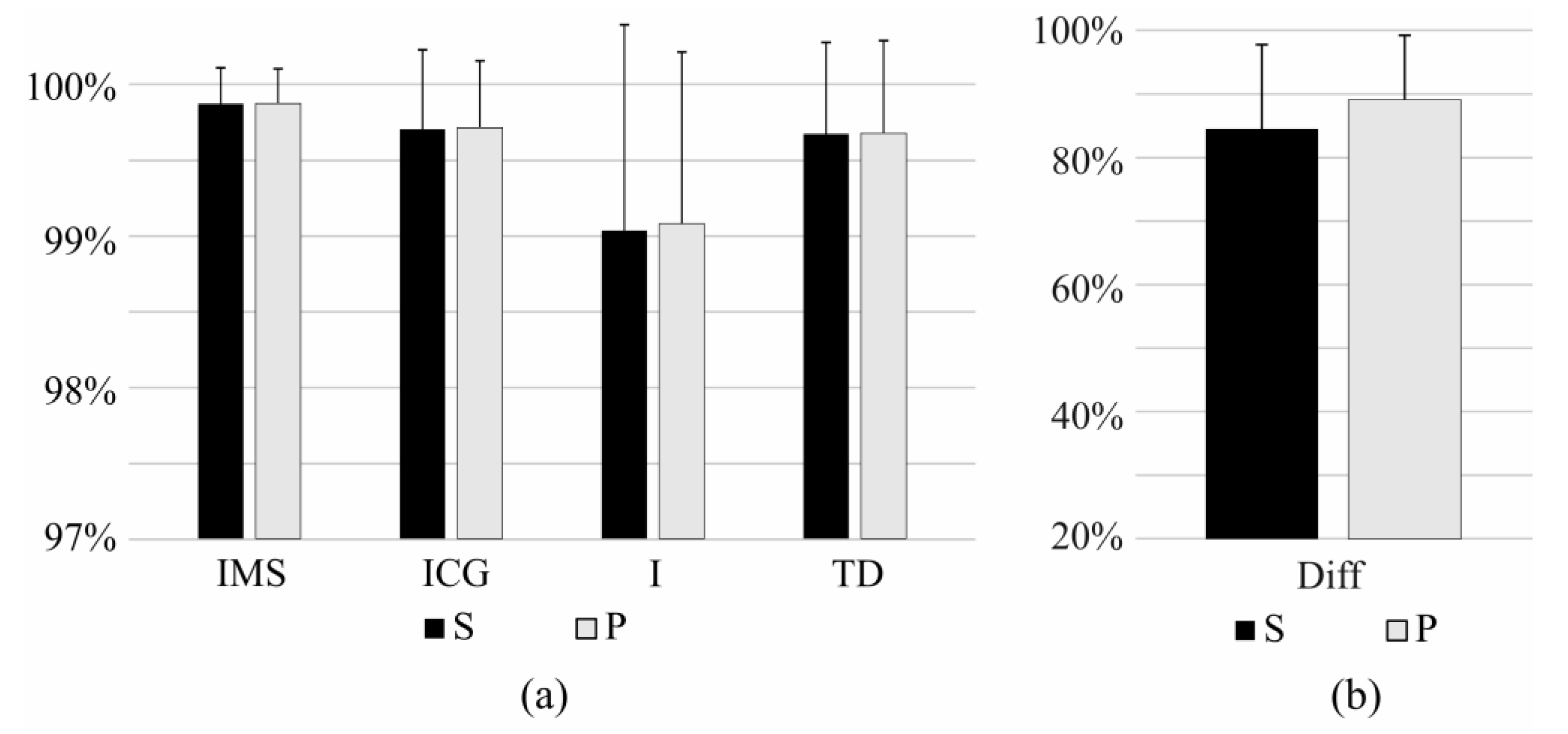
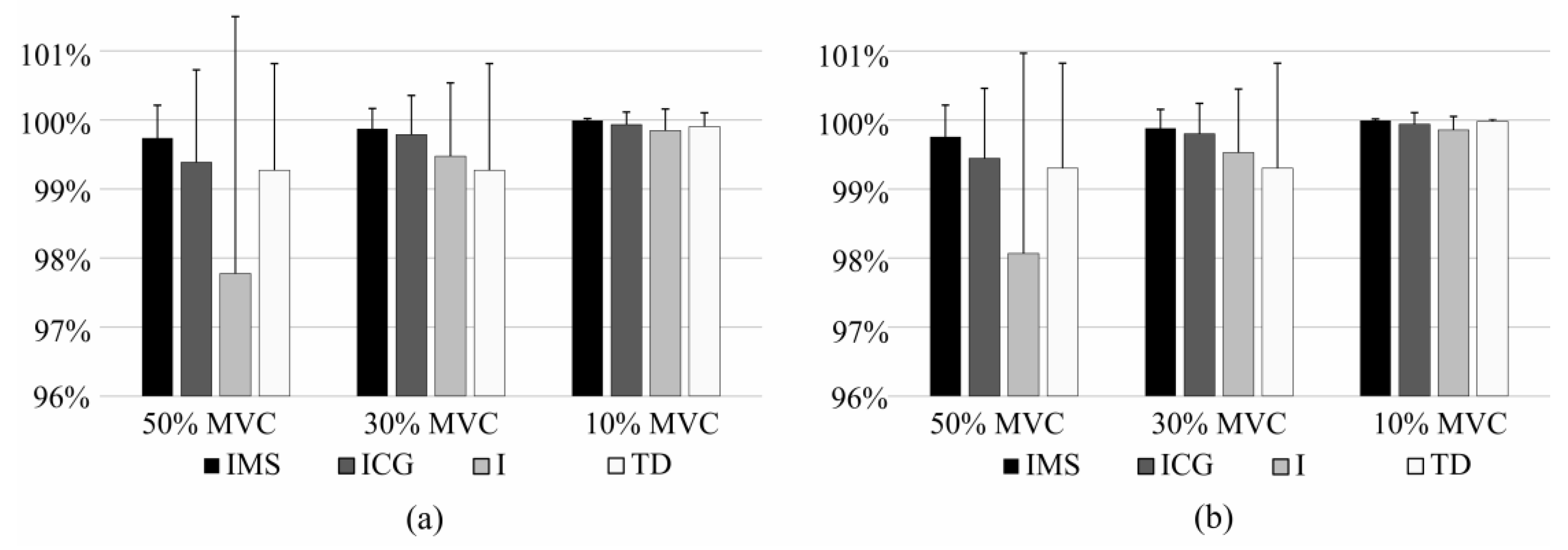
3.2. Short-Term Identification
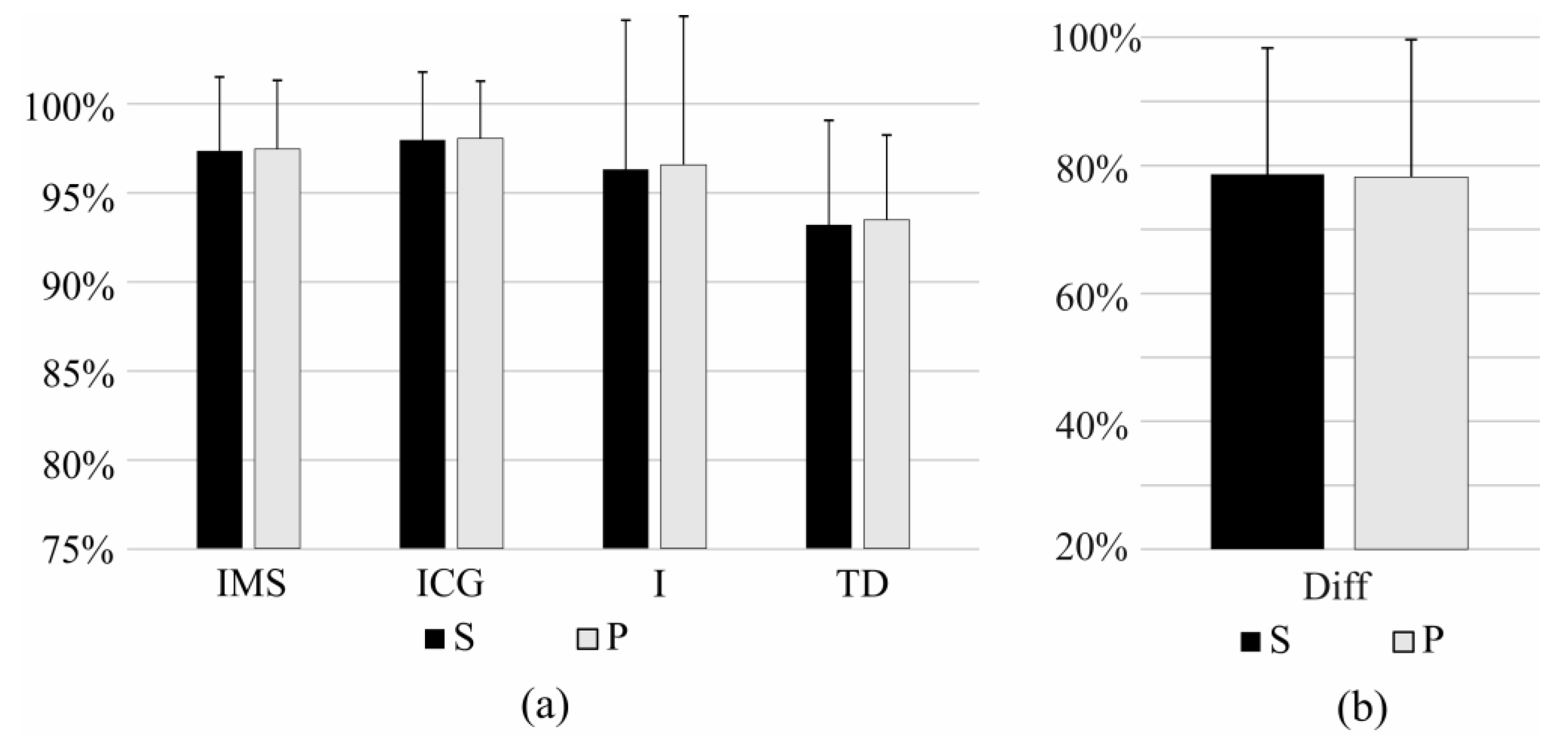
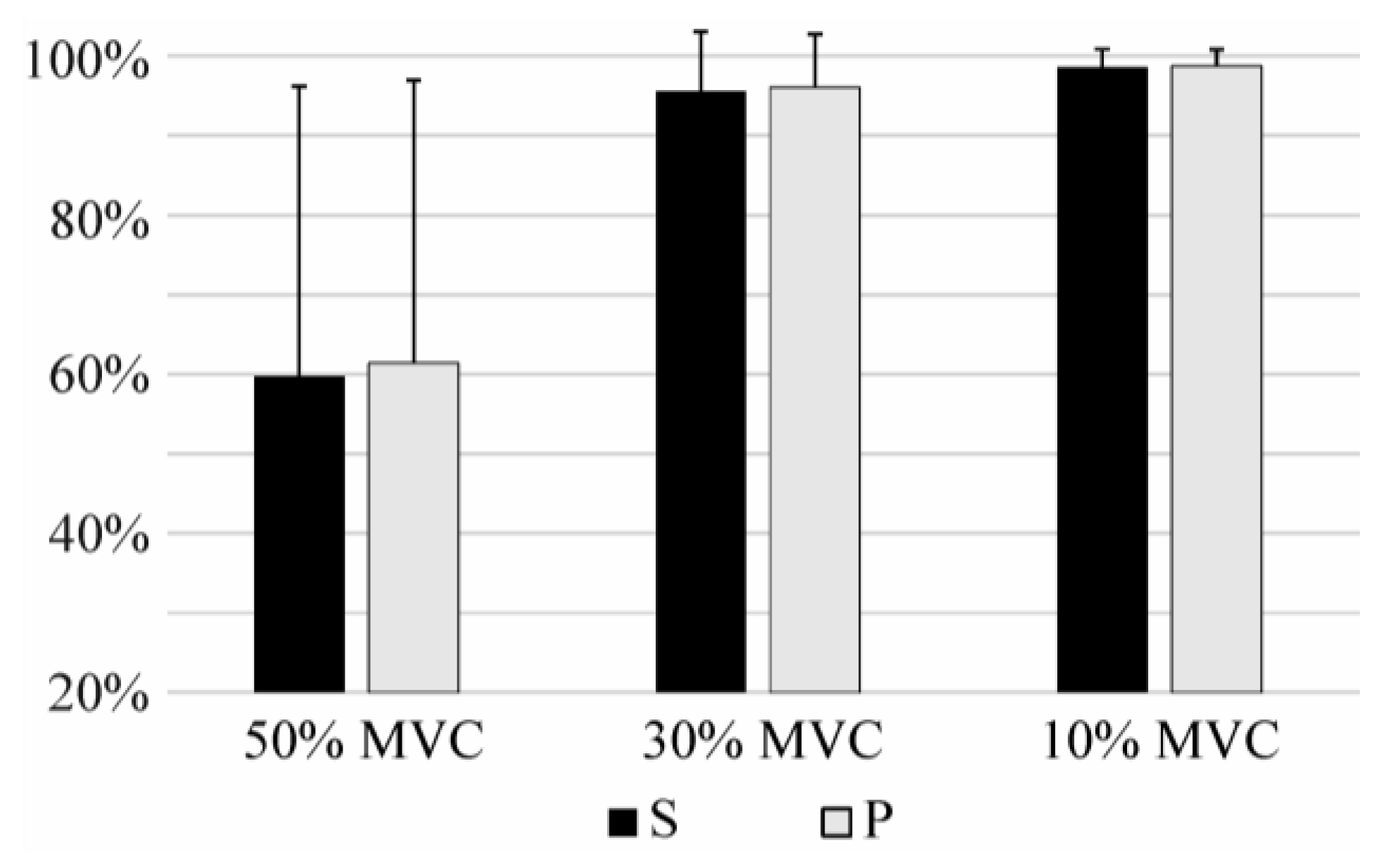
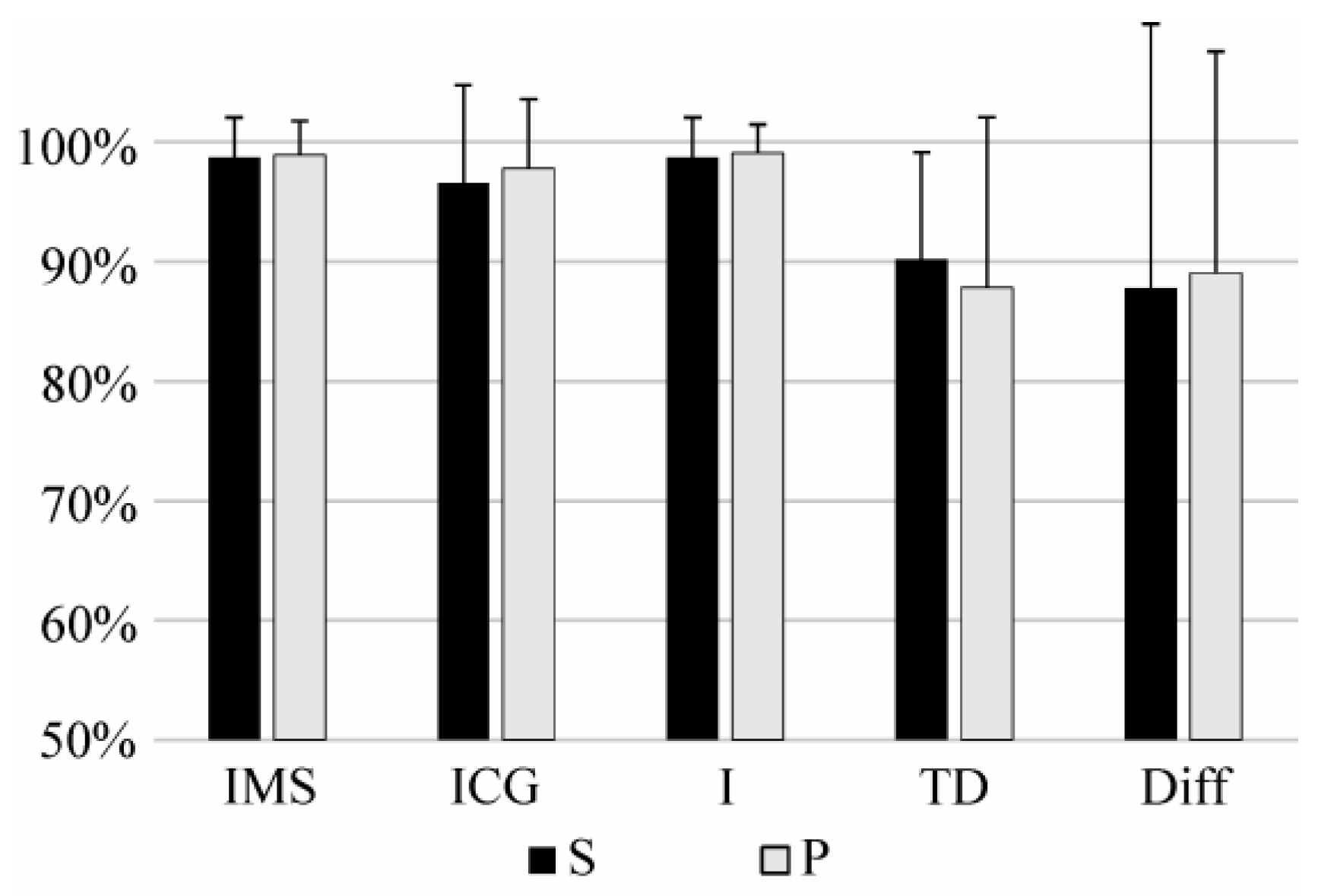
3.3. Long-Term Identification
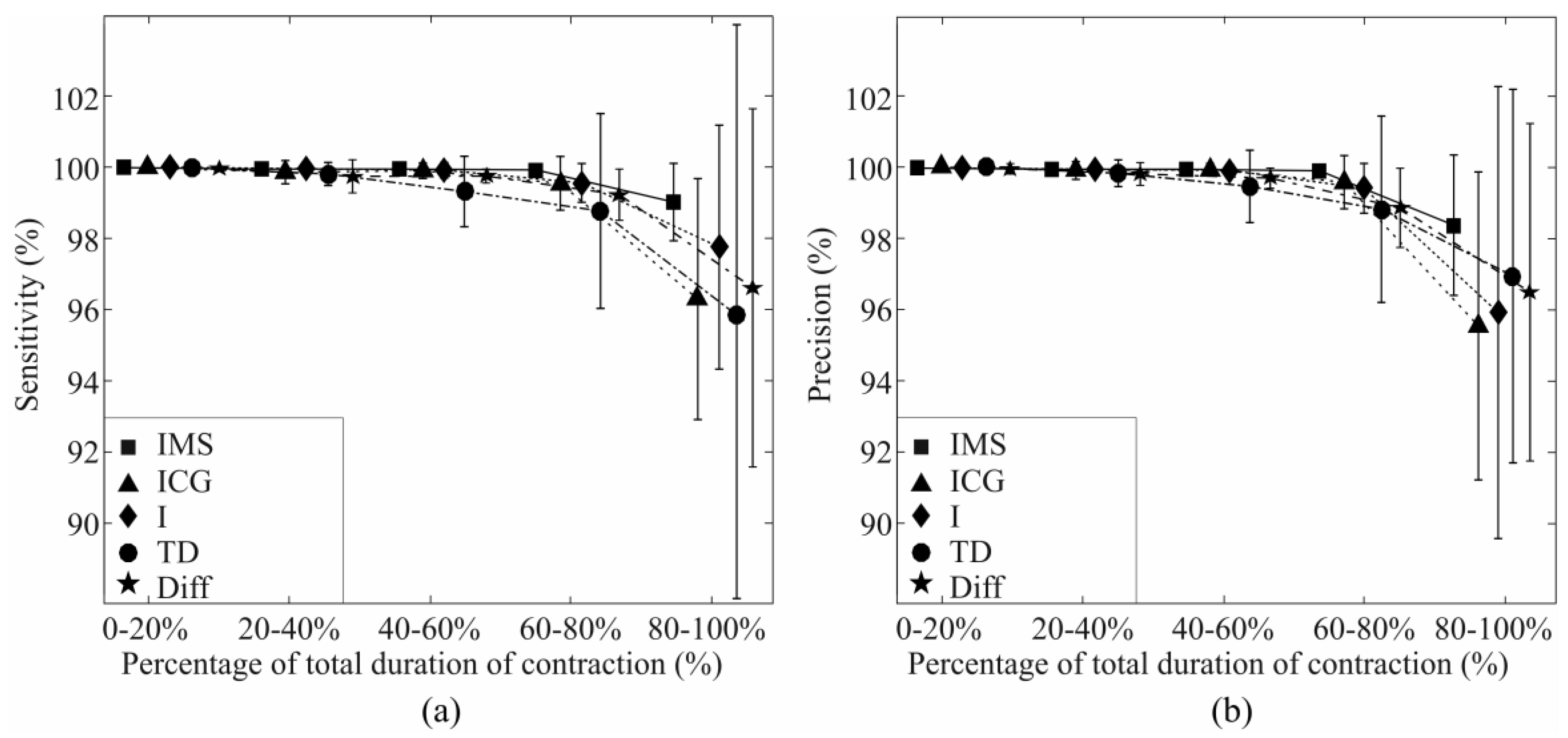
3.4. Identification During Fatigue
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
Appendix B
References
- Farina, D.; Holobar, A.; Merletti, R.; Enoka, R.M. Decoding the neural drive to muscles from the surface electromyogram. Clin. Neurophysiol. 2010, 121, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Nazmi, N.; Abdul Rahman, M.; Yamamoto, S.I.; Ahmad, S.; Zamzuri, H.; Mazlan, S. A Review of Classification Techniques of EMG Signals during Isotonic and Isometric Contractions. Sensors 2016, 16, 1304. [Google Scholar] [CrossRef] [PubMed]
- Hakonen, M.; Piitulainen, H.; Visala, A. Current state of digital signal processing in myoelectric interfaces and related applications. Biomed. Signal Process. Control 2015, 18, 334–359. [Google Scholar] [CrossRef]
- Parker, P.; Englehart, K.; Hudgins, B. Myoelectric signal processing for control of powered limb prostheses. J. Electromyogr. Kinesiol. 2006, 16, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Schultz, A.E.; Kuiken, T.A. Quantifying pattern recognition—based myoelectric control of multifunctional transradial prostheses. IEEE Trans. Neural Syst. Rehabil. Eng. 2010, 18, 185–192. [Google Scholar] [PubMed]
- Marchal-Crespo, L.; Reinkensmeyer, D.J. Review of control strategies for robotic movement training after neurologic injury. J. Neuroeng. Rehabil. 2009, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Broccard, F.D.; Mullen, T.; Chi, Y.M.; Peterson, D.; Iversen, J.R.; Arnold, M.; Kreutz-Delgado, K.; Jung, T.P.; Makeig, S.; Poizner, H.; et al. Closed-loop brain-machine-body interfaces for noninvasive rehabilitation of movement disorders. Ann. Biomed. Eng. 2014, 42, 1573–1593. [Google Scholar] [CrossRef] [PubMed]
- Marateb, H.R.; Farahi, M.; Rojas, M.; Mañanas, M.A.; Farina, D.; Rix, H. Detection of multiple innervation zones from multi-channel surface EMG recordings with low signal-to-noise ratio using graph-cut segmentation. PLoS ONE 2016, 11, e0167954. [Google Scholar] [CrossRef] [PubMed]
- Verikas, A.; Vaiciukynas, E.; Gelzinis, A.; Parker, J.; Olsson, M. Electromyographic patterns during golf swing: Activation sequence profiling and prediction of shot effectiveness. Sensors 2016, 16, 592. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, L.; van der Sluis, C.K.; van Dijk, H.W.; Bongers, R.M.; Scheidt, R. Learning an EMG controlled game: Task-specific adaptations and transfer. PLoS One 2016, 11, e0160817. [Google Scholar] [CrossRef] [PubMed]
- Marateb, H.R.; McGill, K.C.; Webster, J.G. Electromyographic (EMG) decomposition. In Wiley Encyclopedia of Electrical and Electronics Engineering; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1999. [Google Scholar]
- Hargrove, L.J.; Englehart, K.; Hudgins, B. A comparison of surface and intramuscular myoelectric signal classification. IEEE Trans. Biomed. Eng. 2007, 54, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Oskoei, M.A.; Hu, H. Myoelectric control systems—A survey. Biomed. Signal Process. Control 2007, 2, 275–294. [Google Scholar] [CrossRef]
- Huang, H.; Zhou, P.; Li, G.; Kuiken, T. Spatial filtering improves EMG classification accuracy following targeted muscle reinnervation. Ann. Biomed. Eng. 2009, 37, 1849–1857. [Google Scholar] [CrossRef] [PubMed]
- Celadon, N.; Došen, S.; Binder, I.; Ariano, P.; Farina, D. Proportional estimation of finger movements from high-density surface electromyography. J. Neuroeng. Rehabil. 2016, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Ameri, A.; Englehart, K.B.; Parker, P.A. A comparison between force and position control strategies in myoelectric prostheses. In Proceedings of the 2012 Annual International Conference of the IEEE on Engineering in Medicine and Biology Society (EMBC), San Diego, CA, USA, 28 August–1 September 2012; pp. 1342–1345. [Google Scholar]
- Li, Z.; Wang, B.; Yang, C.; Xie, Q.; Su, C.Y. Boosting-based EMG patterns classification scheme for robustness enhancement. IEEE J. Biomed. Heal. Inform. 2013, 17, 545–552. [Google Scholar] [CrossRef]
- Jordanic, M.; Rojas-Martínez, M.; Mañanas, M.A.; Alonso, J.F. Spatial distribution of HD-EMG improves identification of task and force in patients with incomplete spinal cord injury. J. Neuroeng. Rehabil. 2016, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Jordanić, M.; Rojas-Martínez, M.; Mañanas, M.A.; Alonso, J.F. Prediction of isometric motor tasks and effort levels based on high-density EMG in patients with incomplete spinal cord injury. J. Neural Eng. 2016, 13, 046002. [Google Scholar] [CrossRef] [PubMed]
- Vidovic, M.M.C.; Hwang, H.J.; Amsuss, S.; Hahne, J.M.; Farina, D.; Muller, K.R. Improving the robustness of myoelectric pattern recognition for upper limb prostheses by covariate shift adaptation. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 24, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Hahne, J.M.; Dähne, S.; Hwang, H.J.; Müller, K.R.; Parra, L.C. Concurrent adaptation of human and machine improves simultaneous and proportional myoelectric control. IEEE Trans. Neural Syst. Rehabil. Eng. 2015, 23, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sheng, X.; Zhang, D.; Jiang, N.; Zhu, X. Towards zero retraining for myoelectric control based on common model component analysis. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 24, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Tkach, D.; Huang, H.; Kuiken, T.A. Study of stability of time-domain features for electromyographic pattern recognition. J. Neuroeng. Rehabil. 2010, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Scheme, E.; Englehart, K. Training strategies for mitigating the effect of proportional control on classification in pattern recognition-based myoelectric control. J. Prosthet. Orthot. 2013, 25, 76–83. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, D.; Sheng, X.; Li, S.; Zhu, X. Invariant surface EMG feature against varying contraction level for myoelectric control based on muscle coordination. IEEE J. Biomed. Heal. Inf. 2015, 19, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Merletti, R.; Botter, A.; Troiano, A.; Merlo, E.; Minetto, M.A. Technology and instrumentation for detection and conditioning of the surface electromyographic signal: State of the art. Clin. Biomech. 2009, 24, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Du, Y.; Jin, W.; Wei, W.; Hu, Y.; Li, J. Gesture recognition by instantaneous surface EMG images. Sci. Rep. 2016, 6, 36571. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Jin, W.; Wei, W.; Hu, Y.; Geng, W. Surface EMG-based inter-session gesture recognition enhanced by deep domain adaptation. Sensors 2017, 17, 458. [Google Scholar] [CrossRef] [PubMed]
- Hahne, J.M.; Graimann, B.; Muller, K.R. Spatial filtering for robust myoelectric control. IEEE Trans. Biomed. Eng. 2012, 59, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Holtermann, A.; Roeleveld, K.; Karlsson, J.S. Inhomogeneities in muscle activation reveal motor unit recruitment. J. Electromyogr. Kinesiol. 2005, 15, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Tucker, K.; Falla, D.; Graven-Nielsen, T.; Farina, D. Electromyographic mapping of the erector spinae muscle with varying load and during sustained contraction. J. Electromyogr. Kinesiol. 2009, 19, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Vieira, T.M.M.; Merletti, R.; Mesin, L. Automatic segmentation of surface EMG images: Improving the estimation of neuromuscular activity. J. Biomech. 2010, 43, 2149–2158. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Martínez, M.; Mañanas, M.A.; Alonso, J.F.; Merletti, R. Identification of isometric contractions based on High Density EMG maps. J. Electromyogr. Kinesiol. 2013, 23, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Hermens, H.; Freriks, B. SENIAM 9: European Recommendations for Surface ElectroMyoGraphy, Results of the SENIAM Project (CD); Roessingh Research and Development: Enschede, The Netherlands, 1999. [Google Scholar]
- Rojas-Martínez, M.; Mañanas, M.A.; Alonso, J.F. High-density surface EMG maps from upper-arm and forearm muscles. J. Neuroeng. Rehabil. 2012, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Valle, S.; Li, W.; Qin, S.J. Selection of the number of principal components: The variance of the reconstruction error criterion with a comparison to other methods. Ind. Eng. Chem. Res. 1999, 38, 4389–4401. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Mosteller, F. A k-sample slippage test for an extreme population. Ann. Math. Stat. 1948, 19, 58–65. [Google Scholar] [CrossRef]
- Mohebian, M.R.; Marateb, H.R.; Mansourian, M.; Mañanas, M.A.; Mokarian, F. A hybrid computer-aided-diagnosis system for prediction of breast cancer recurrence (HPBCR) using optimized ensemble learning. Comput. Struct. Biotechnol. J. 2017, 15, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Farina, D.; Jiang, N.; Rehbaum, H.; Holobar, A.; Graimann, B.; Dietl, H.; Aszmann, O.C. The extraction of neural information from the surface EMG for the control of upper-limb prostheses: Emerging avenues and challenges. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Sensinger, J.W.; Lock, B.A.; Kuiken, T.A. Adaptive pattern recognition of myoelectric signals: exploration of conceptual framework and practical algorithms. IEEE Trans. Neural Syst. Rehabil. Eng. 2009, 17, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, L.J.; Li, G.; Englehart, K.B.; Hudgins, B.S. Principal components analysis preprocessing for improved classification accuracies in pattern-recognition-based myoelectric control. IEEE Trans. Biomed. Eng. 2009, 56, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Samuel, O.W.; Zhang, X.; Wang, H.; Fang, P.; Li, G. A motion-classification strategy based on sEMG-EEG signal combination for upper-limb amputees. J. Neuroeng. Rehabil. 2017, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Zhang, X.; Zhang, Y.T.; Li, G. A novel channel selection method for multiple motion classification using high-density electromyography. Biomed. Eng. Online 2014, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Hennig, C.; Meila, M.; Murtagh, F.; Rocci, R. Handbook of Cluster Analysis; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Fukunaga, K.; Hostetler, L. The estimation of the gradient of a density function, with applications in pattern recognition. IEEE Trans. Inf. Theory 1975, 21, 32–40. [Google Scholar] [CrossRef]
- Parzen, E. On Estimation of a Probability Density Function and Mode. Ann. Math. Stat. 1962, 33, 1065–1076. [Google Scholar] [CrossRef]
- Cacoullos, T. Estimation of a multivariate density. Ann. Inst. Stat. Math. 1966, 18, 179–189. [Google Scholar] [CrossRef]
| Task | Sensitivity | Precision |
|---|---|---|
| Flexion | 99.7 ± 0.5% | 99.9 ± 0.2% |
| Extension | 99.9 ± 0.1% | 99.9 ± 0.1% |
| Supination | 99.9 ± 0.2% | 99.7 ± 0.5% |
| Pronation | 99.9 ± 0.1% | 99.9 ± 0.1% |
| Average | 99.9 ± 0.2% | 99.9 ± 0.2% |
| Task | Sensitivity (%) | Precision (%) |
|---|---|---|
| Flexion 10% MVC | 98.2 ± 2.8% | 99.9 ± 0.3% |
| Flexion 30% MVC | 98.7 ± 1.1% | 97.0 ± 3.1% |
| Flexion 50% MVC | 97.7 ± 2.9% | 98.6 ± 1.1% |
| Extension 10% MVC | 99.7 ± 0.6% | 99.6 ± 1.1% |
| Extension 30% MVC | 97.4 ± 3.4% | 97.5 ± 2.1% |
| Extension 50% MVC | 97.7 ± 2.3% | 98.2 ± 2.9% |
| Supination 10% MVC | 99.7 ± 0.5% | 99.9 ± 0.2% |
| Supination 30% MVC | 95.2 ± 7.1% | 96.0 ± 5.1% |
| Supination 50% MVC | 96.6 ± 4.9% | 95.4 ± 6.3% |
| Pronation 10% MVC | 99.8 ± 0.2% | 99.4 ± 1.1% |
| Pronation 30% MVC | 93.8 ± 12.3% | 93.9 ± 11.3% |
| Pronation 50% MVC | 93.7 ± 11.9% | 94.2 ± 11.9% |
| Average | 97.4 ± 4.2% | 97.5 ± 3.9% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jordanić, M.; Rojas-Martínez, M.; Mañanas, M.A.; Alonso, J.F.; Marateb, H.R. A Novel Spatial Feature for the Identification of Motor Tasks Using High-Density Electromyography. Sensors 2017, 17, 1597. https://doi.org/10.3390/s17071597
Jordanić M, Rojas-Martínez M, Mañanas MA, Alonso JF, Marateb HR. A Novel Spatial Feature for the Identification of Motor Tasks Using High-Density Electromyography. Sensors. 2017; 17(7):1597. https://doi.org/10.3390/s17071597
Chicago/Turabian StyleJordanić, Mislav, Mónica Rojas-Martínez, Miguel Angel Mañanas, Joan Francesc Alonso, and Hamid Reza Marateb. 2017. "A Novel Spatial Feature for the Identification of Motor Tasks Using High-Density Electromyography" Sensors 17, no. 7: 1597. https://doi.org/10.3390/s17071597
APA StyleJordanić, M., Rojas-Martínez, M., Mañanas, M. A., Alonso, J. F., & Marateb, H. R. (2017). A Novel Spatial Feature for the Identification of Motor Tasks Using High-Density Electromyography. Sensors, 17(7), 1597. https://doi.org/10.3390/s17071597






