An Automatic Localization Algorithm for Ultrasound Breast Tumors Based on Human Visual Mechanism
Abstract
:1. Introduction
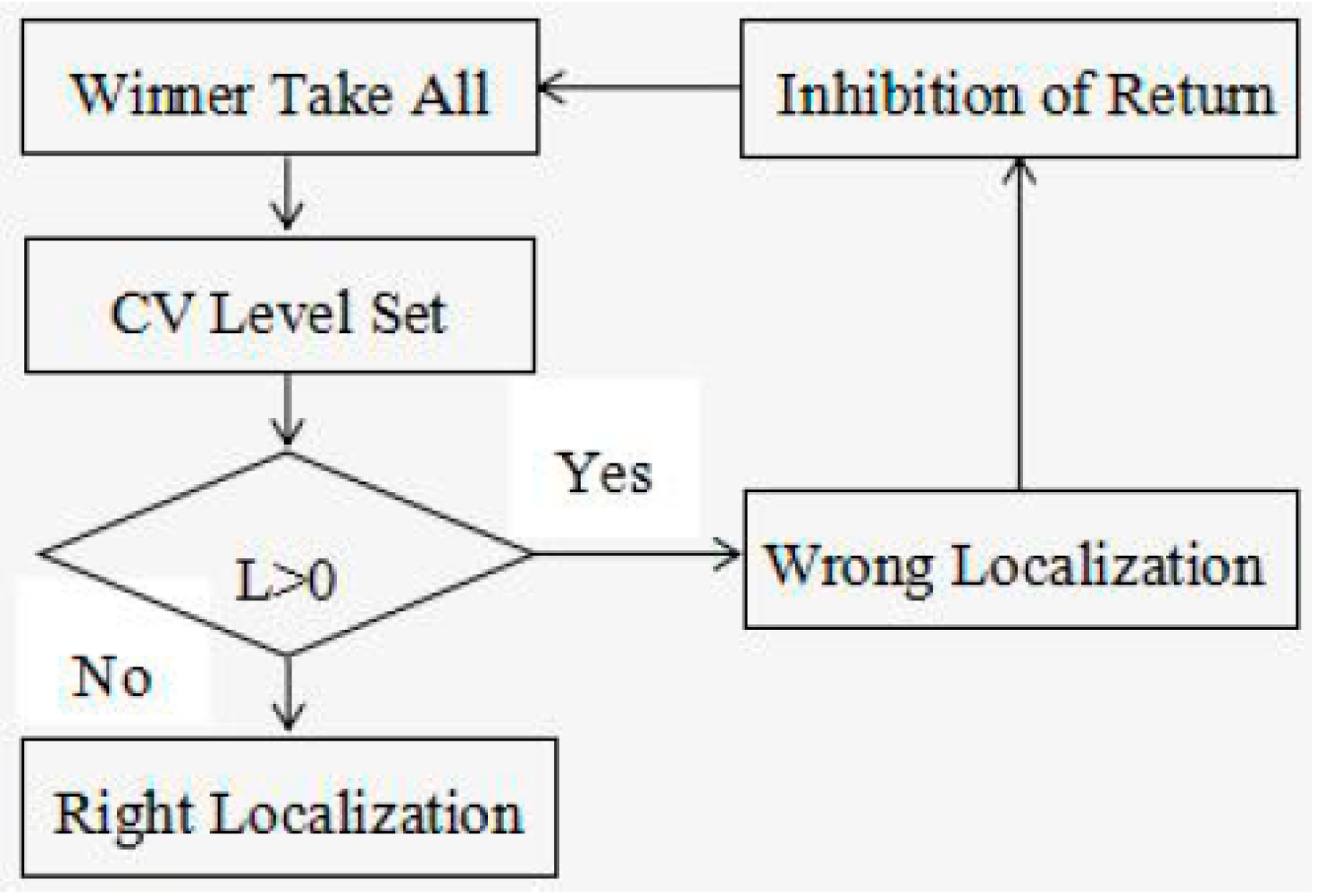
2. Model Architecture
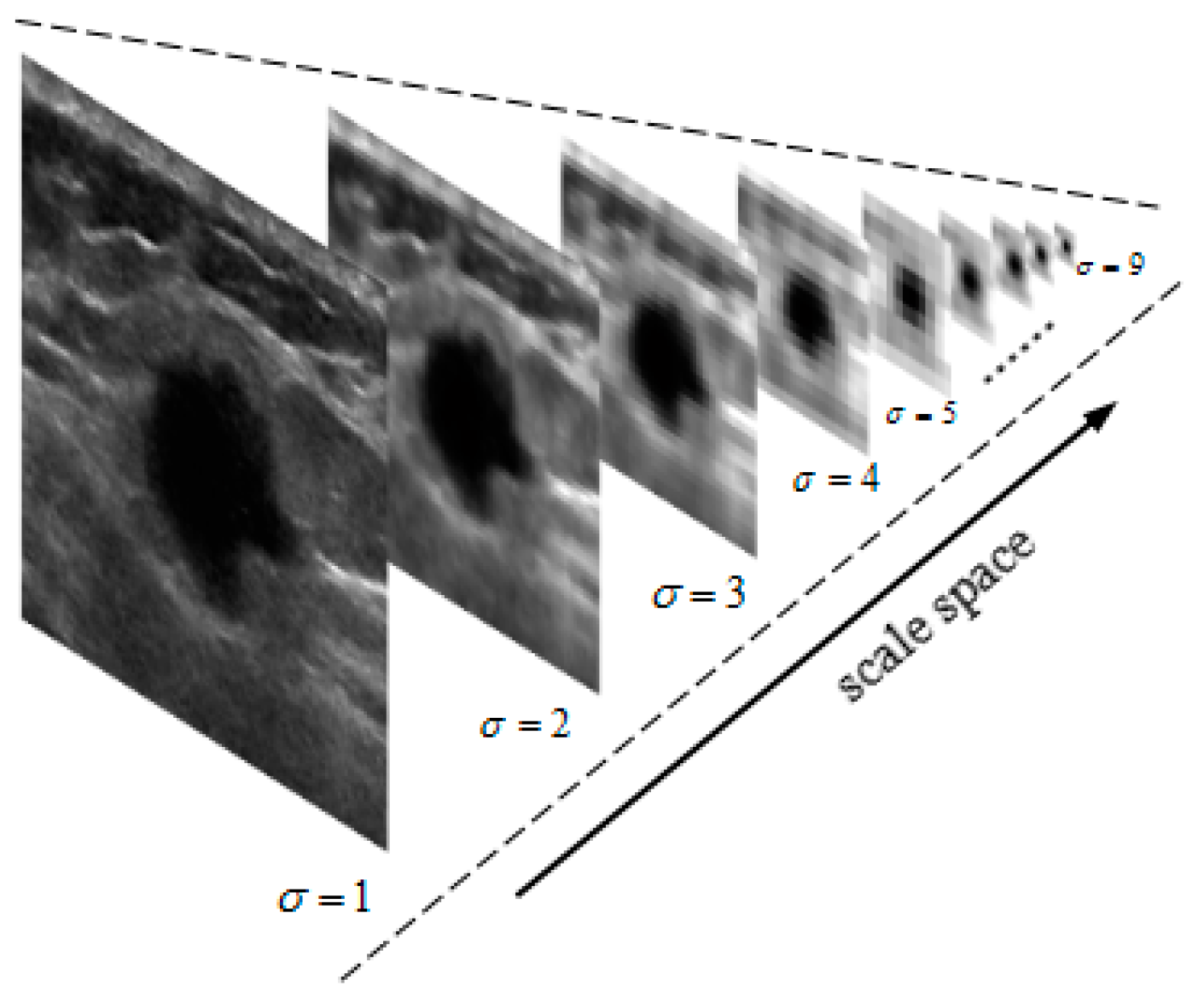
2.1. Gaussian Pyramid Image
2.2. Saliency Map
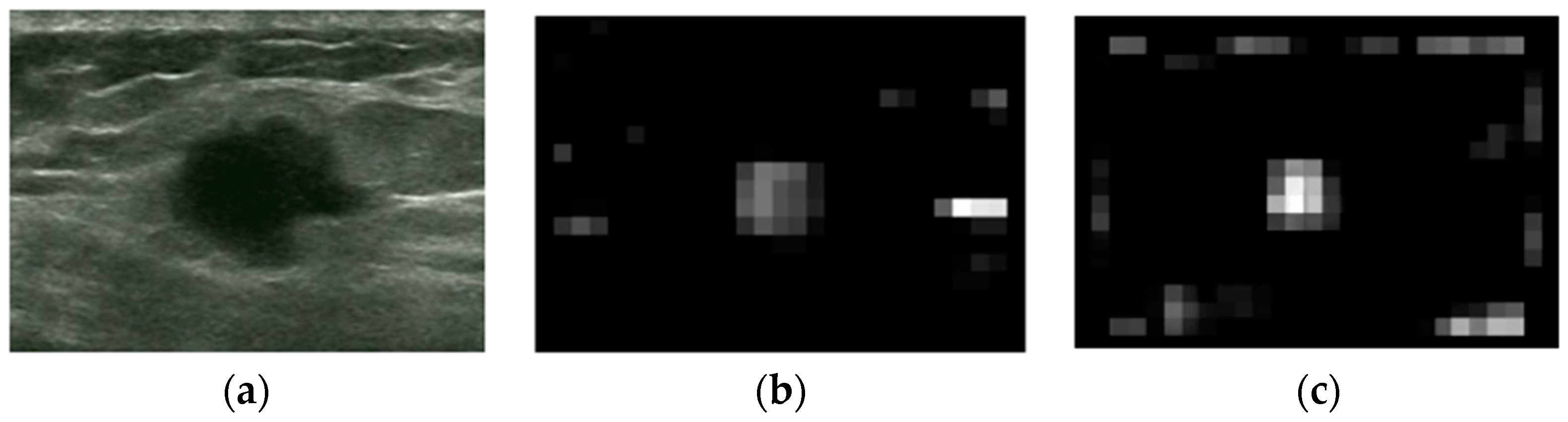
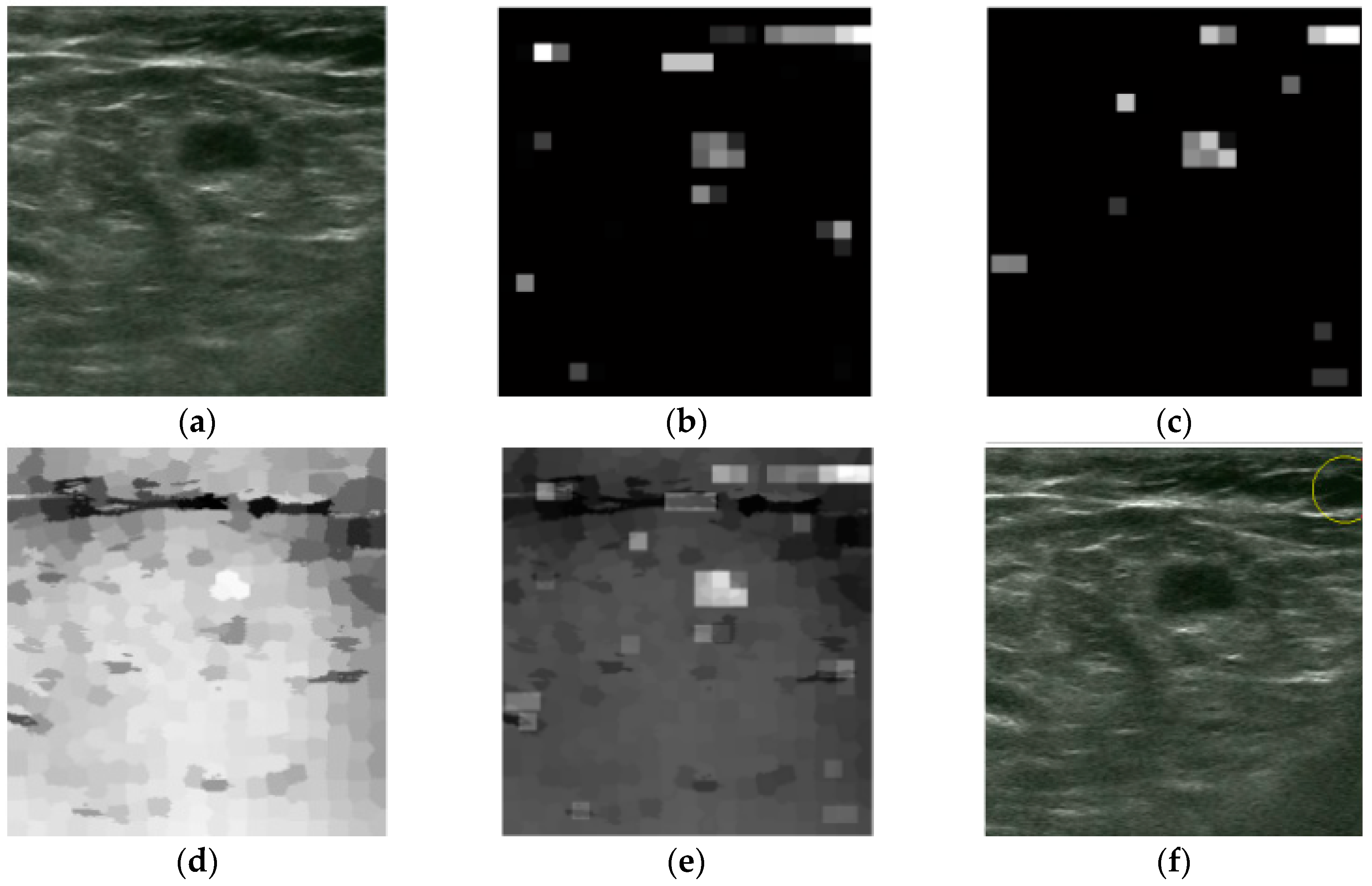
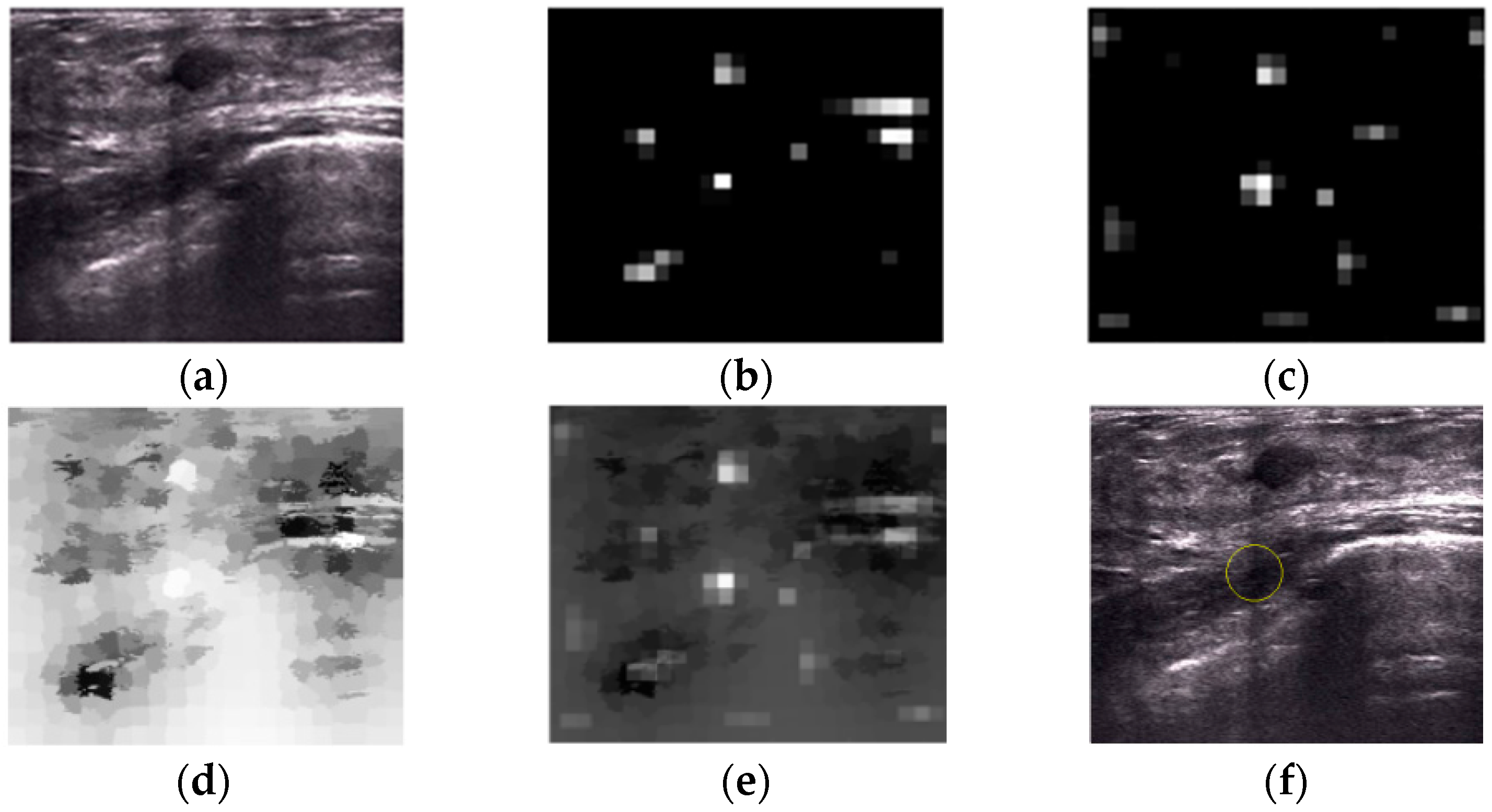
2.2.1. Intensity and Blackness Ratio Features
2.2.2. Pyramid Feature Map
2.2.3. Generating Saliency Map
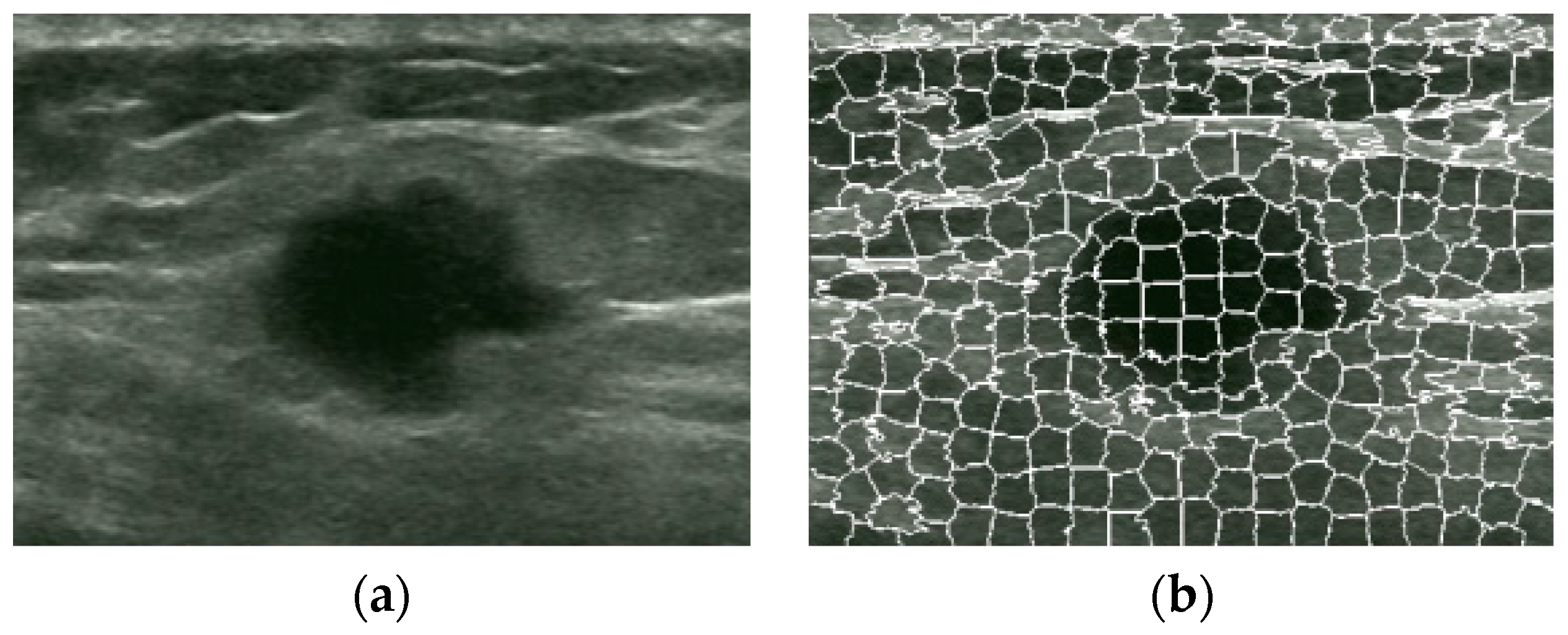
2.3. Superpixel Saliency
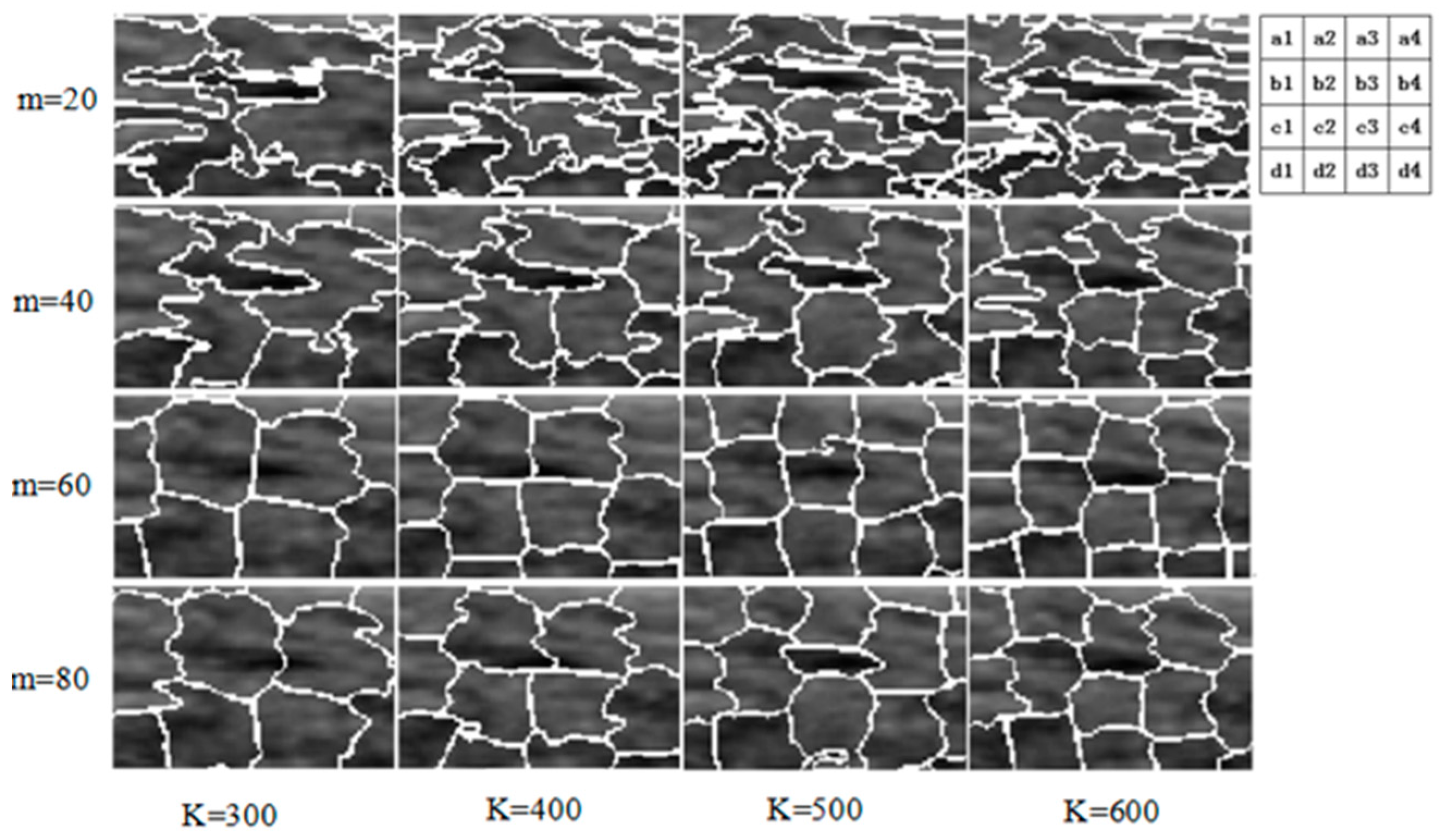
2.3.1. Superpixel
2.3.2. Superpixel Saliency Map
2.4. Combined Saliency Map
2.5. Winner Take All
2.6. Post-Processing Operation
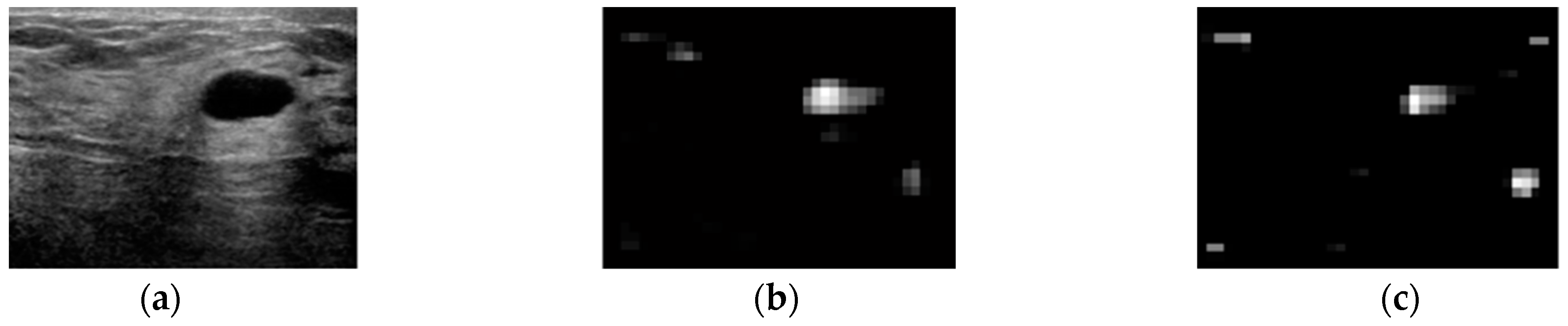
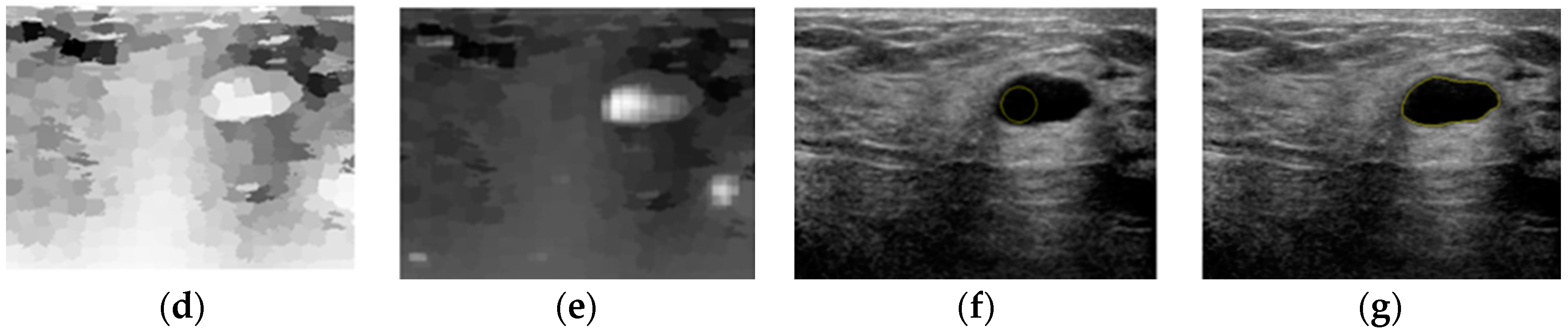
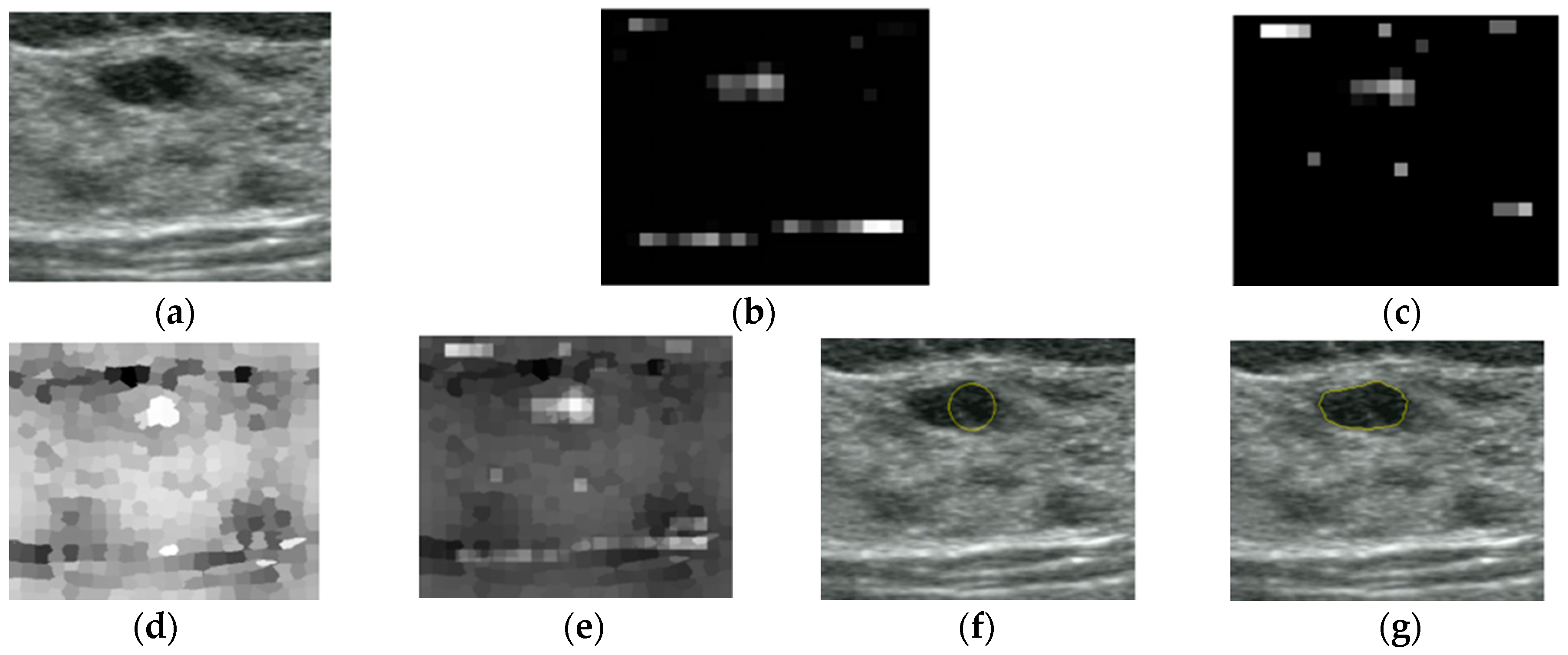
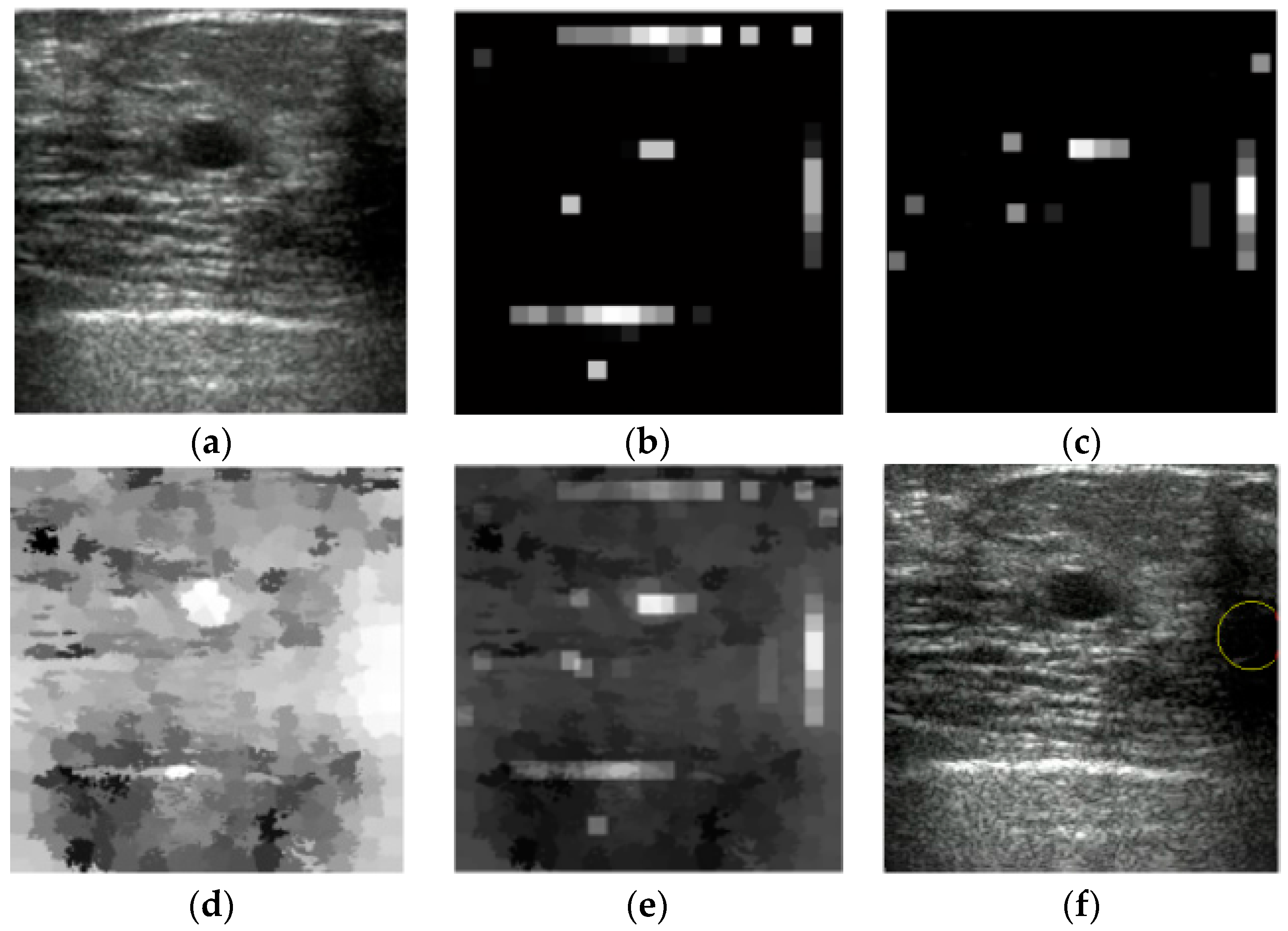
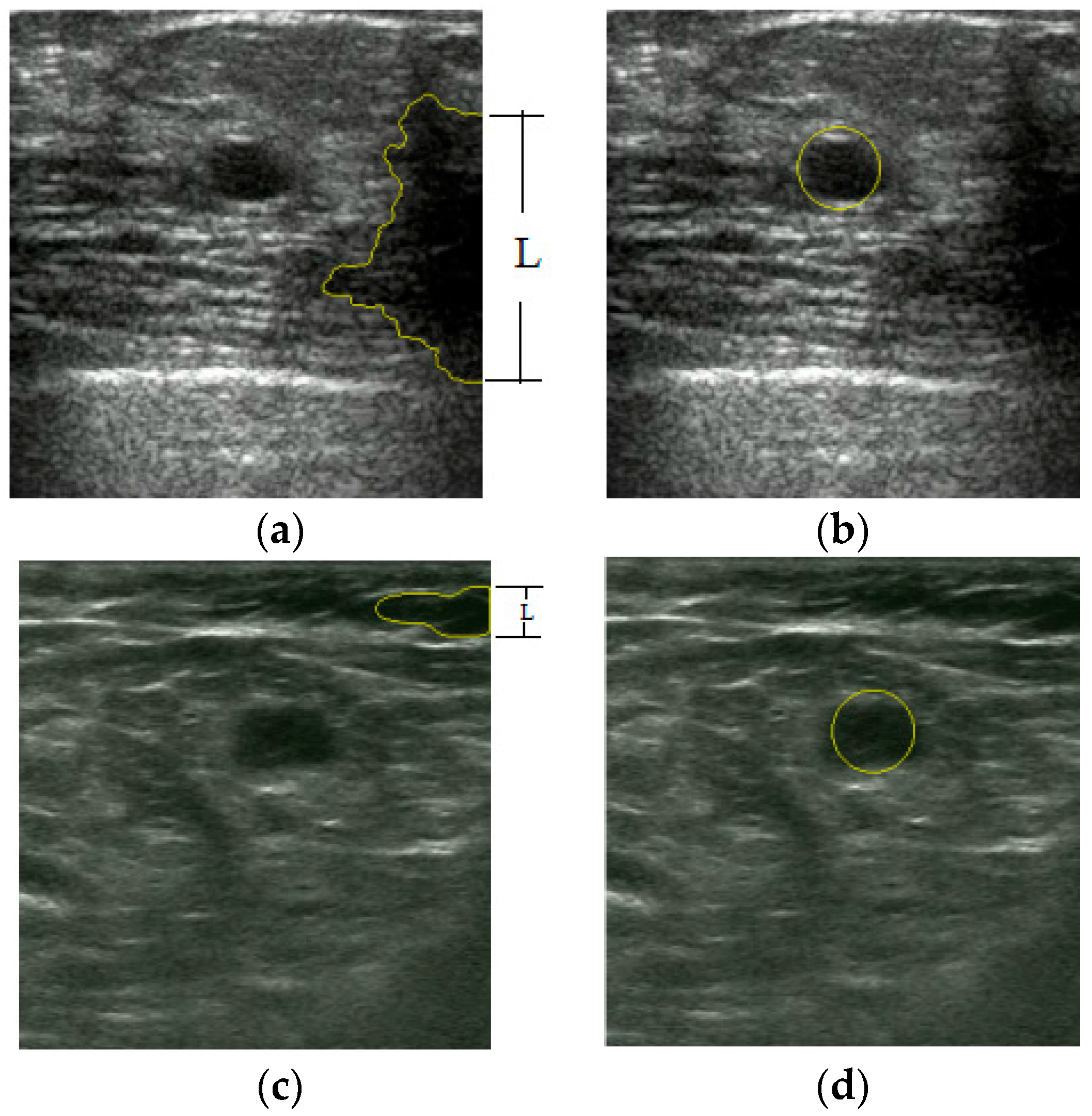
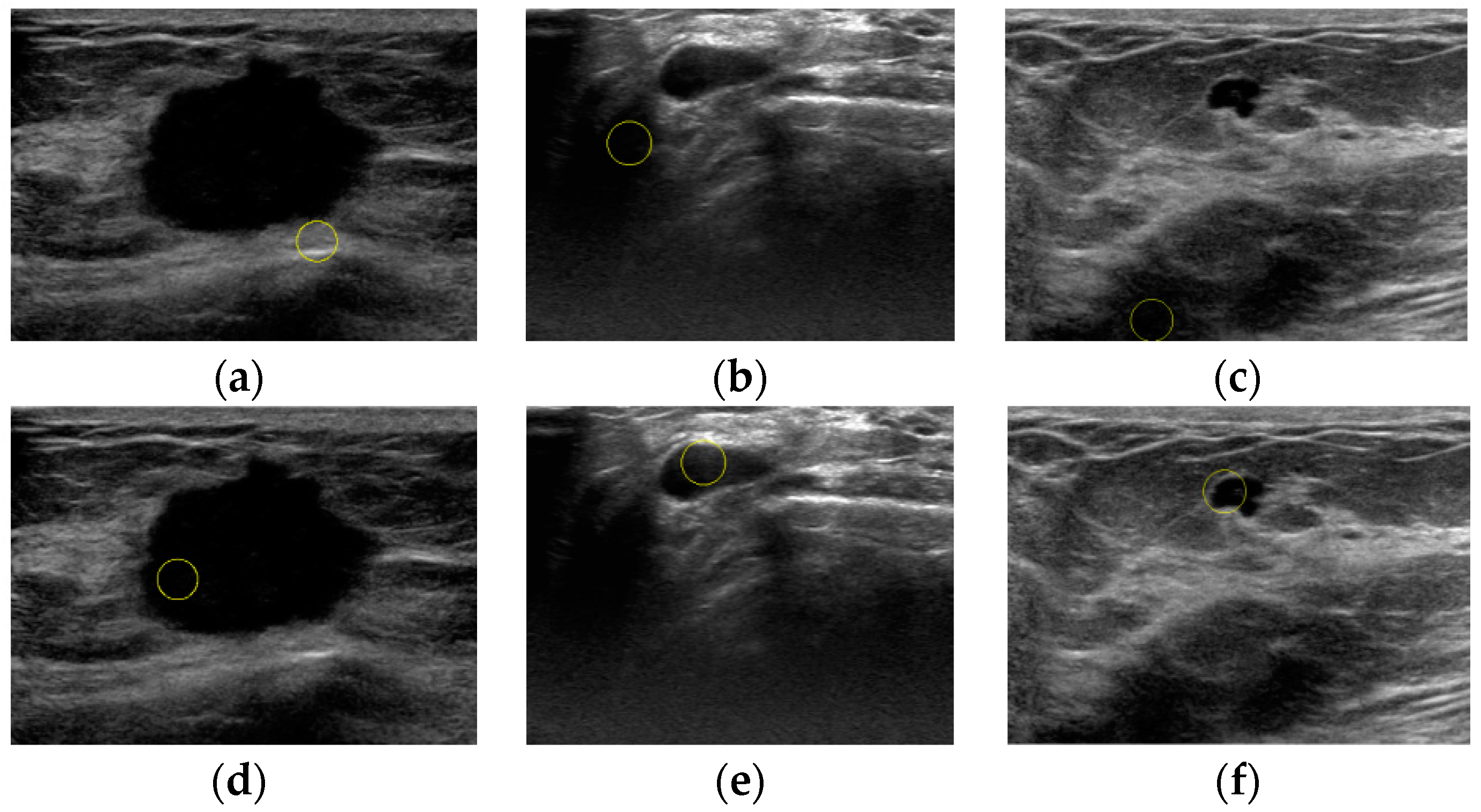
3. Results
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, X.; Song, J.L.; Wang, S.H.; Zhao, J.W.; Chen, Y.Q. Learning to Diagnose Cirrhosis with Liver Capsule Guided Ultrasound Image Classification. Sensors 2017, 17, 149. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Sengur, A.; Tian, J.W. A novel breast ultrasound image segmentation algorithm based on neutrosophic similarity score and level set. Comput. Methods Progr. Biomed. 2015, 123, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Huynh, B.; DrUKker, K.; Giger, M. MO-DE-207B-06: Computer-Aided Diagnosis of Breast Ultrasound Images Using Transfer Learning From Deep Convolutional Neural Networks. Med. Phys. 2016, 43, 3705. [Google Scholar] [CrossRef]
- Prabusankarlal, K.M.; Thirumoorthy, P.; Manavalan, R. Computer Aided Breast Cancer Diagnosis Techniques in Ultrasound: A Survey. J. Med. Imaging Health Inform. 2014, 4, 331–349. [Google Scholar] [CrossRef]
- Drukker, K.; Giger, M.L.; Vyborny, C.J.; Mendelson, E.B. Computerized detection and classification of cancer on breast ultrasound. Acad. Radiol. 2004, 11, 526–535. [Google Scholar] [CrossRef]
- Huang, Q.; Yang, F.; Liu, L.; Li, X. Automatic segmentation of breast lesions for interaction in ultrasonic computer-aided diagnosis. Inf. Sci. 2015, 314, 293–310. [Google Scholar] [CrossRef]
- Huang, Q.; Luo, Y.; Zhang, Q. Breast ultrasound image segmentation: A survey. Int. J. Comput. Assist. Radiol. Surg. 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Wang, Y. Automatic Boundary Detection in Breast Ultrasound Images Based on Improved Pulse Coupled Neural Network and Active Contour Model. In Proceedings of the 5th International Conference on Bioinformatics and Biomedical Engineering, Wuhan, China, 10–12 May 2011. [Google Scholar]
- Huang, Y.L.; Chen, D.R. Automatic contouring for breast tumors in 2-D sonography. In Proceedings of the 27th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Shanghai, China, 17–18 January 2006. [Google Scholar]
- Hao, X.; Bruce, C.J.; Pislaru, C.; Greenleaf, J.F. Segmenting high-frequency intracardiac ultrasound images of myocardium into infarcted, ischemic, and normal regions. IEEE Trans. Med. Imaging 2001, 20, 1373–1383. [Google Scholar] [PubMed]
- Madabhushi, A.; Metaxas, D.N. Combining low, high level and empirical domain knowledge for automated segmentation of ultrasonic breast lesions. IEEE Trans. Med. Imaging 2003, 22, 55–169. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Cheng, H.D.; Wang, Y. A novel automatic seed point selection algorithm for breast ultrasound images. In Proceedings of the 19th International Conferences on Pattern Recognition, Tampa, FL, USA, 8–11 December 2008. [Google Scholar]
- Xu, J.; Gao, X. Fully automatic detection and segmentation algorithm for ultrasound breast images using SVM and level set. J. Comput.-Aided Des. Comput. Gr. 2012, 24, 662–668. [Google Scholar] [CrossRef]
- Yap, M.H.; Edirisinghe, E.A.; Bez, H.E. Automatic lesion boundary detection in ultrasound breast images. In Proceedings of the 2007 Medical Imaging International Society for Optics and Photonics, San Diego, CA, USA, 17 February 2007. [Google Scholar]
- Liu, B.; Cheng, H.D.; Huang, J.; Tian, J.; Liu, J.; Tang, J. Automated segmentation of ultrasonic breast lesions using statistical texture classification and active contour based on probability distance. Ultrasound Med. Biol. 2008, 35, 1309–1324. [Google Scholar] [CrossRef] [PubMed]
- Itti, L.; Koch, C.; Niebur, E. A Model of Saliency-Based Visual Attention for Rapid Scene Analysis. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 1254–1259. [Google Scholar] [CrossRef]
- Peterson, H.A.; Rajala, S.A.; Delp, E.J. Human visual system properties applied to image segmentation for image compression. In Proceedings of the IEEE Global Telecommunications Conference, Phoenix, AZ, USA, 2–5 December 1991. [Google Scholar]
- Chantelau, K. Segmentation of moving images by the human visual system. Biol. Cybern. 1997, 77, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Rajala, S.A. Segmentation based image coding using fractals and the human visual system. In Proceedings of the 1990 International Conference on Acoustics, Speech, and Signal Processing, Albuquerque, NM, USA, 3–6 April 1990. [Google Scholar]
- Tsai, C.S.; Chang, C.C. An improvement to image segment based on human visual system for object-based coding. Fundam. Inform. 2003, 58, 167–178. [Google Scholar]
- Walther, D.; Koch, C. Modeling attention to salient proto-objects. Neural Netw. Off. J. Int. Neural Netw. Soc. 2006, 19, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Ullman, S. Matters of Intelligence, Shifts in Selective Visual Attention: Towards the Underlying Neural Circuitry; Springer: Heidelberg, Germany, 1987; Volume 188, pp. 115–141. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Strunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [PubMed]


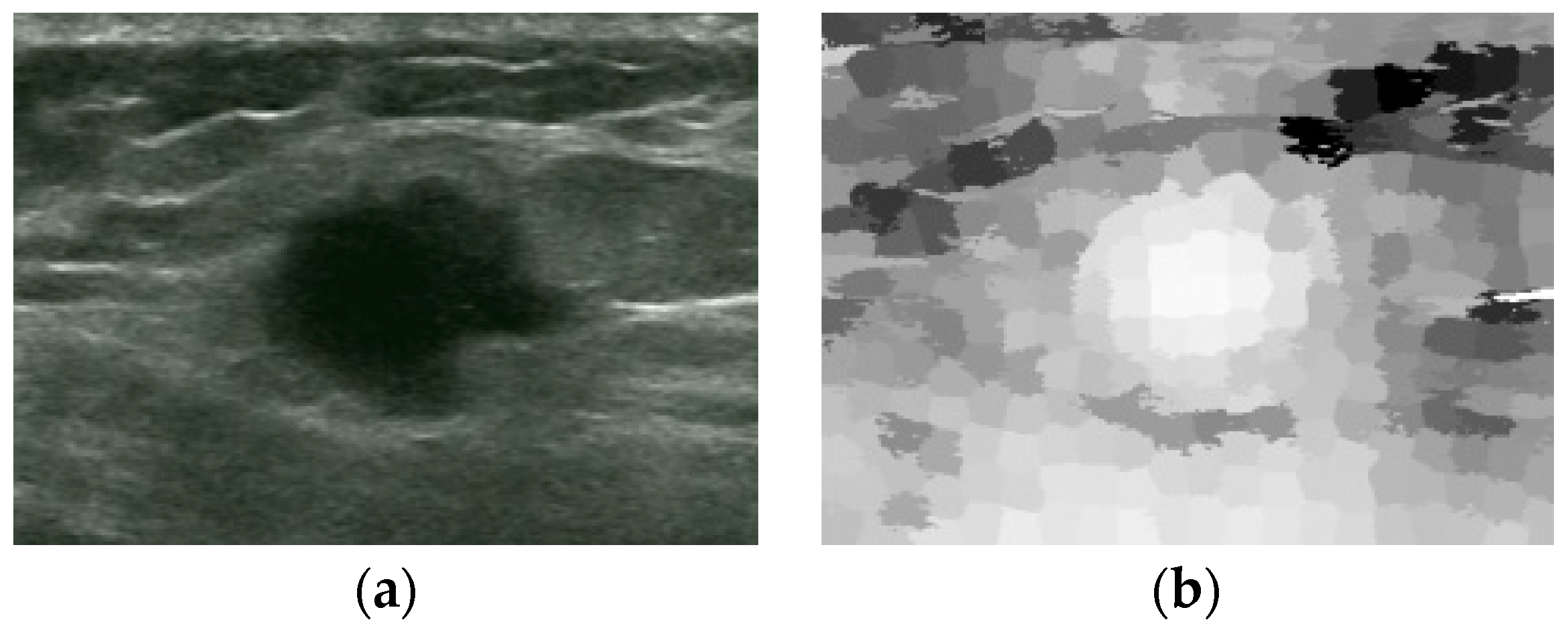
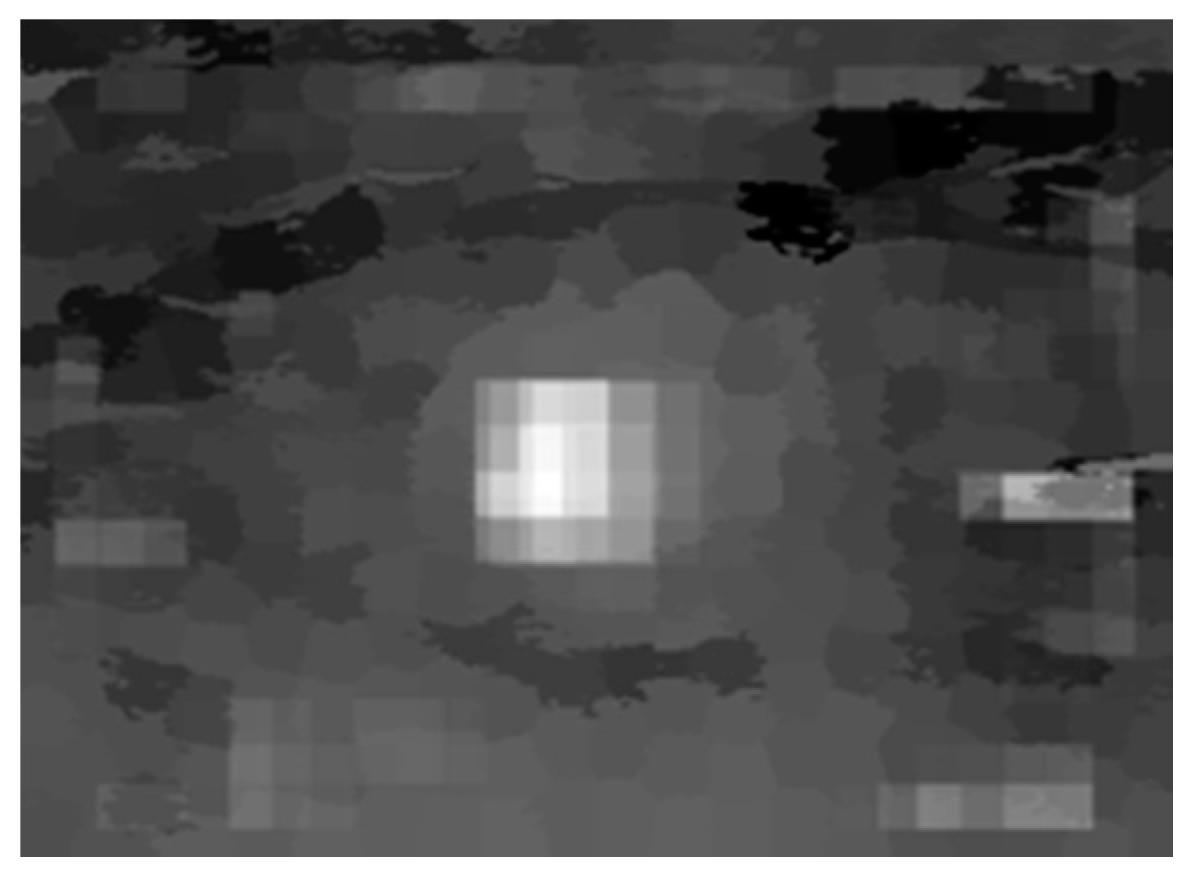
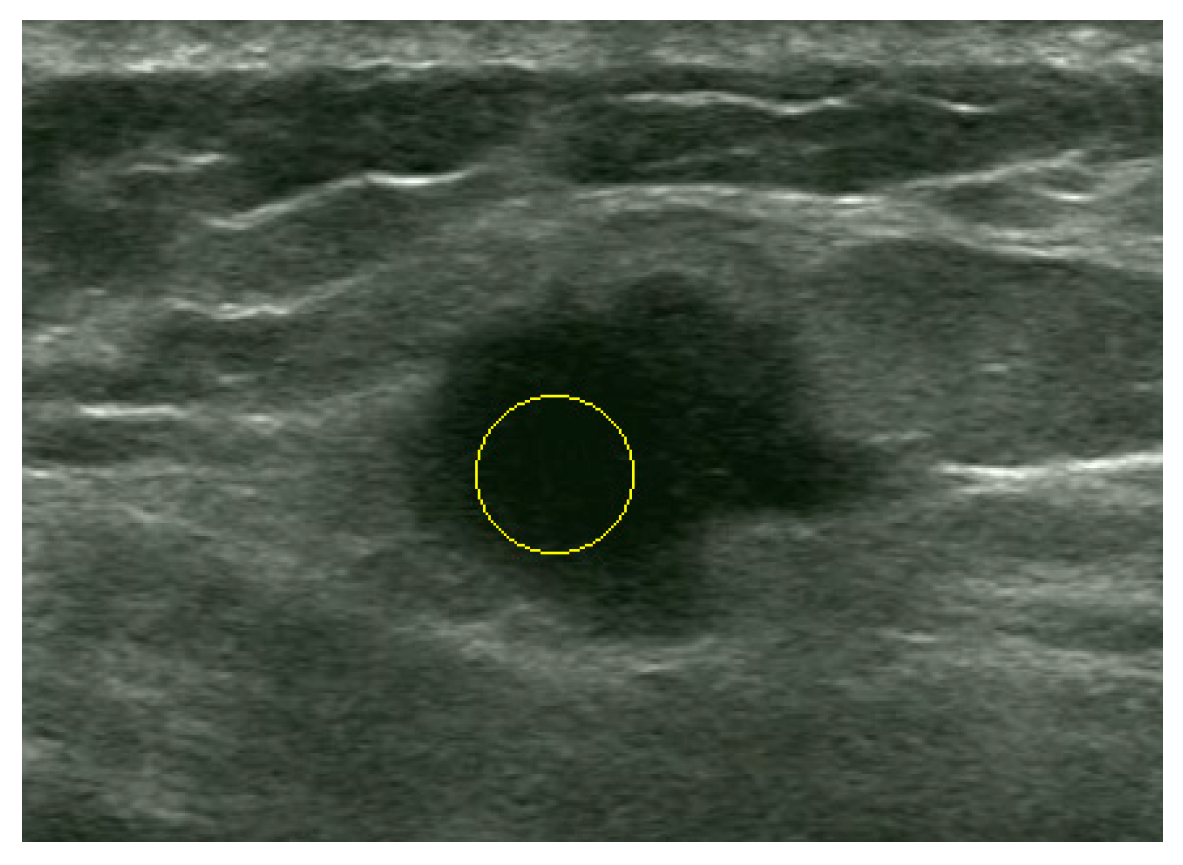


| Proposed Method | Liu’s Method | ||
|---|---|---|---|
| Right Localization | Wrong Localization | Sum | |
| Right Localization | 325 | 51 | 376 |
| Wrong Localization | 0 | 24 | 24 |
| Sum | 325 | 75 | 400 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Chen, K.; Lin, J. An Automatic Localization Algorithm for Ultrasound Breast Tumors Based on Human Visual Mechanism. Sensors 2017, 17, 1101. https://doi.org/10.3390/s17051101
Xie Y, Chen K, Lin J. An Automatic Localization Algorithm for Ultrasound Breast Tumors Based on Human Visual Mechanism. Sensors. 2017; 17(5):1101. https://doi.org/10.3390/s17051101
Chicago/Turabian StyleXie, Yuting, Ke Chen, and Jiangli Lin. 2017. "An Automatic Localization Algorithm for Ultrasound Breast Tumors Based on Human Visual Mechanism" Sensors 17, no. 5: 1101. https://doi.org/10.3390/s17051101
APA StyleXie, Y., Chen, K., & Lin, J. (2017). An Automatic Localization Algorithm for Ultrasound Breast Tumors Based on Human Visual Mechanism. Sensors, 17(5), 1101. https://doi.org/10.3390/s17051101





