Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges
Abstract
1. Introduction
2. Infectious Pathogens: Bacteria and Viruses
3. Conventional Methods for the Determination of Infectious Pathogens
4. Electrochemical Affinity Biosensing of Infectious Pathogens in Liquid Biopsies
- (i)
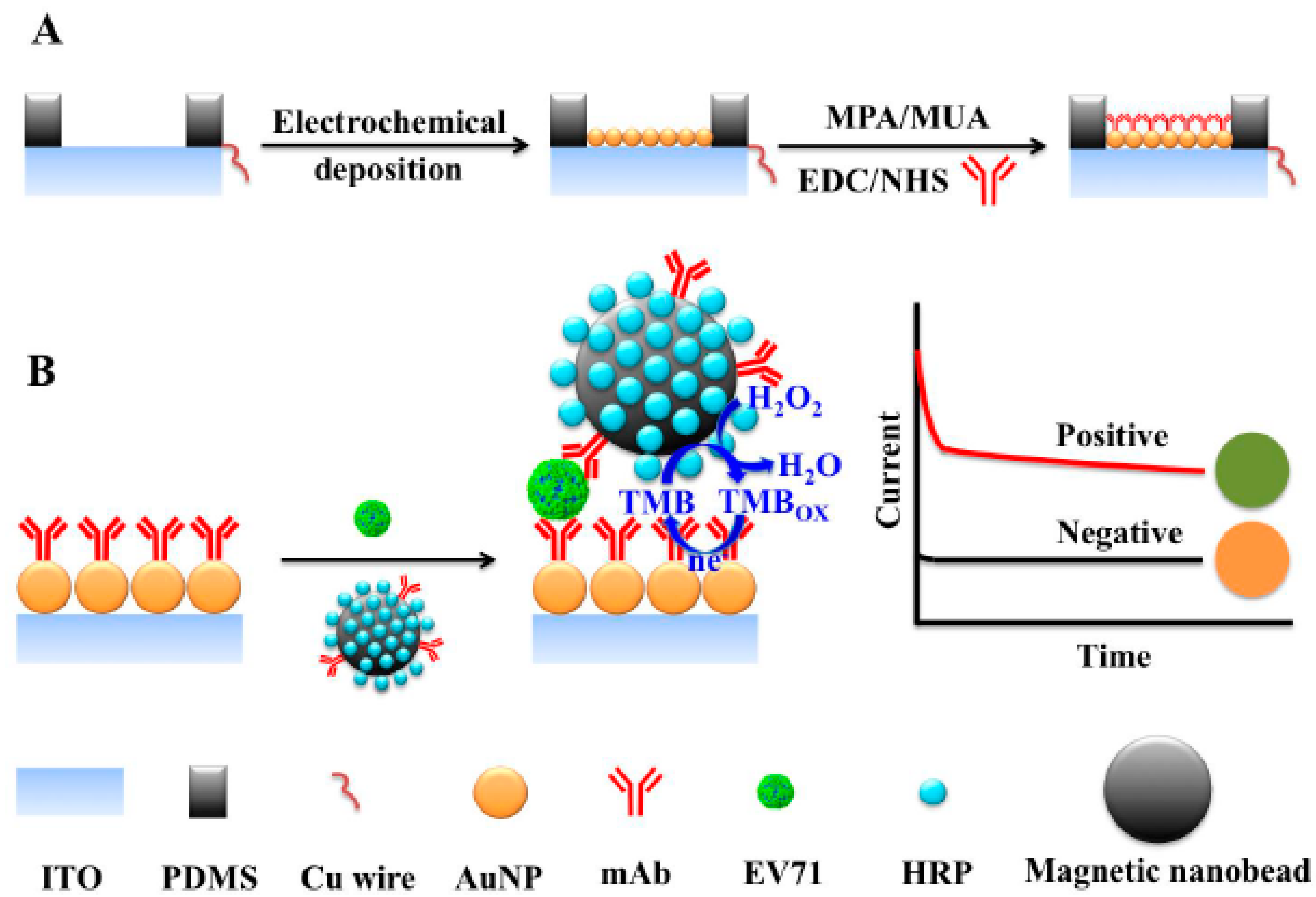
- the whole virus such as human enterovirus 71 [55];
- (ii)
- (iii)
- (iv)
4.1. Minimally Invasive Electrochemical Biosensing of Bacterial Pathogens
4.2. Minimally Invasive Electrochemical Biosensing of Viral Pathogens
5. General Considerations, Challenges and Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for Whole-Cell Bacterial Detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Sepunaru, L.; Plowman, B.J.; Sokolov, S.V.; Young, N.P.; Compton, R.G. Rapid electrochemical detection of single influenza viruses tagged with silver nanoparticles. Chem. Sci. 2016, 7, 3892–3899. [Google Scholar] [CrossRef]
- Sin, M.L.Y.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Kelley, S.O. What Are Clinically Relevant Levels of Cellular and Biomolecular Analytes? ACS Sens. 2017, 2, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Krejcova, L.; Michalek, P.; Merlos Rodrigo, M.; Heger, Z.; Krizkova, S.; Vaculovicova, M.; Hynek, D.; Adam, V.; Kizek, R. Nanoscale virus biosensors: State of the art. Nanobiosens. Dis. Diagn. 2015, 4, 47–66. [Google Scholar]
- Nagel, B.; Dellweg, H.; Gierasch, L.M. Glossary for chemists of terms used in biotechnology. Pure Appl. Chem. 1992, 64, 143–168. [Google Scholar] [CrossRef]
- Grabowska, I.; Malecka, K.; Jarocka, U.; Radecki, J.; Radecka, H. Electrochemical biosensors for detection of avian influenza virus—Current status and future trends. Acta Biochim. Pol. 2014, 61, 471–478. [Google Scholar] [PubMed]
- Bagshaw, S.M.; Lapinsky, S.; Dial, S.; Arabi, Y.; Dodek, P.; Wood, G.; Ellis, P.; Guzman, J.; Marshall, J.; Parrillo, J.E.; et al. Acute Kidney Injury in Septic Shock: Clinical Outcomes and Impact of Duration of Hypotension Prior to Initiation of Antimicrobial Therapy. Intensive Care Med. 2009, 35, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Naze, F.; Le Roux, K.; Schuffenecker, I.; Zeller, H.; Staikowsky, F.; Grivard, P.; Michault, A.; Laurent, P. Simultaneous Detection and Quantitation of Chikungunya, Dengue and West Nile Viruses by Multiplex rt-PCT Assays and Dengue Virus Typing Using High Resolution Melting. J. Virol. Methods 2009, 162, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.K.; Chao, D.Y.; Kao, C.L.; Wu, H.C.; Liu, Y.C.; Li, C.M.; Lin, S.C.; Ho, S.T.; Huang, J.H.; King, C.C. High Levels of Plasma Dengue Viral Load During Defervescence in Patients with Dengue Hemorrhagic Fever: Implications for Pathogenesis. Virology 2003, 305, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Poloni, T.R.; Oliveira, A.S.; Alfonso, H.L.; Galvao, L.R.; Amarilla, A.A.; Poloni, D.F.; Figueiredo, L.T.; Aquino, V.H. Detection of Dengue Virus in Saliva and Urine by Real Time rt-PCR. Virol. J. 2010, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Hue, K.D.; Tuan, T.V.; Thi, H.T.; Bich, C.T.; Anh, H.H.; Wills, B.A.; Simmons, C.P. Validation of an Internally Controlled One-Step Real-Time Multiplex rt-PCR Assay for the Detection and Quantitation of Dengue Virus Rna in Plasma. J. Virol. Methods 2011, 177, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Rasche, A.; Baronti, C.; Aldabbagh, S.; Cadar, D.; Reusken, C.B.E.M.; Pas, S.D.; Goorhuis, A.; Schinkel, J.; Molenkamp, R.; et al. Clinical Comparison, Standardization, and Optimization of Zika Virus Molecular Detection. Bull. WHO 2016, 94, 880. [Google Scholar] [PubMed]
- Mansuy, J.M.; Dutertre, M.; Mengelle, C.; Fourcade, C.; Marchou, B.; Delobel, P.; Izopet, J.; Martin-Blondel, G. Zika Virus: High Infectious Viral Load in Semen, a New Sexually Transmitted Pathogen? Lancet Infect. Dis. 2016, 16, 405. [Google Scholar] [CrossRef]
- Lee, N.; Chan, P.K.; Hui, D.S.; Rainer, T.H.; Wong, E.; Choi, K.W.; Lui, G.C.; Wong, B.C.; Wong, R.Y.; Lam, W.Y.; et al. Viral Loads and Duration of Viral Shedding in Adult Patients Hospitalized with Influenza. J. Infect. Dis. 2009, 200, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Ngaosuwankul, N.; Noisumdaeng, P.; Komolsiri, P.; Pooruk, P.; Chokephaibulkit, K.; Chotpitayasunondh, T.; Sangsajja, C.; Chuchottaworn, C.; Farrar, J.; Puthavathana, P. Influenza a Viral Loads in Respiratory Samples Collected from Patients Infected with Pandemic H1N1, Seasonal H1N1 and H3N2 Viruses. Virol. J. 2010, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Chan, K.H.; Li, I.W.; Tsang, T.Y.; Tse, H.; Chan, J.F.; Hung, I.F.; Lai, S.T.; Leung, C.W.; Kwan, Y.W.; et al. Viral Load in Patients Infected with Pandemic H1N1 2009 Influenza a Virus. J. Med. Virol. 2010, 82, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yagupsky, P.; Nolte, F.S. Quantitative Aspects of Septicemia. Clin. Microbiol. Rev. 1990, 3, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Blakemore, R.; Nabeta, P.; Davidow, A.L.; Vadwai, V.; Tahirli, R.; Munsamy, V.; Nicol, M.; Jones, M.; Persing, D.H.; Hillemann, D.; et al. A Multisite Assessment of the Quantitative Capabilities of the Xpert Mtb/Rif Assay. Am. J. Respir. Crit. Care Med. 2011, 184, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Honeyborne, I.; Mtafya, B.; Phillips, P.P.; Hoelscher, M.; Ntinginya, E.N.; Kohlenberg, A.; Rachow, A.; Rojas-Ponce, G.; McHugh, T.D.; Heinrich, N. The Molecular Bacterial Load Assay Replaces Solid Culture for Measuring Early Bactericidal Response to Antituberculosis Treatment. J. Clin. Microbiol. 2014, 52, 3064–3067. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Sonn, G.A.; Sin, M.L.Y.; Mach, K.E.; Shih, M.-C.; Gau, V.; Wong, P.K.; Liao, J.C. Electrochemical immunosensor detection of urinary lactoferrin in clinical samples for urinary tract infection diagnosis. Biosens. Bioelectron. 2010, 26, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.L.; Gaido, L. Laboratory Diagnosis of Urinary Tract Infections in Adult Patients. Clin. Infect. Dis. 2004, 38, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Nidzworski, D.; Pranszke, P.; Grudniewska, M.; Król, E.; Gromadzka, B. Universal biosensor for detection of influenza virus. Biosens. Bioelectron. 2014, 59, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Radecka, H.; Radecki, J. Label-free Electrochemical Immunosensors for Viruses and Antibodies Detection-Review. J. Mex. Chem. Soc. 2015, 59, 269–275. [Google Scholar]
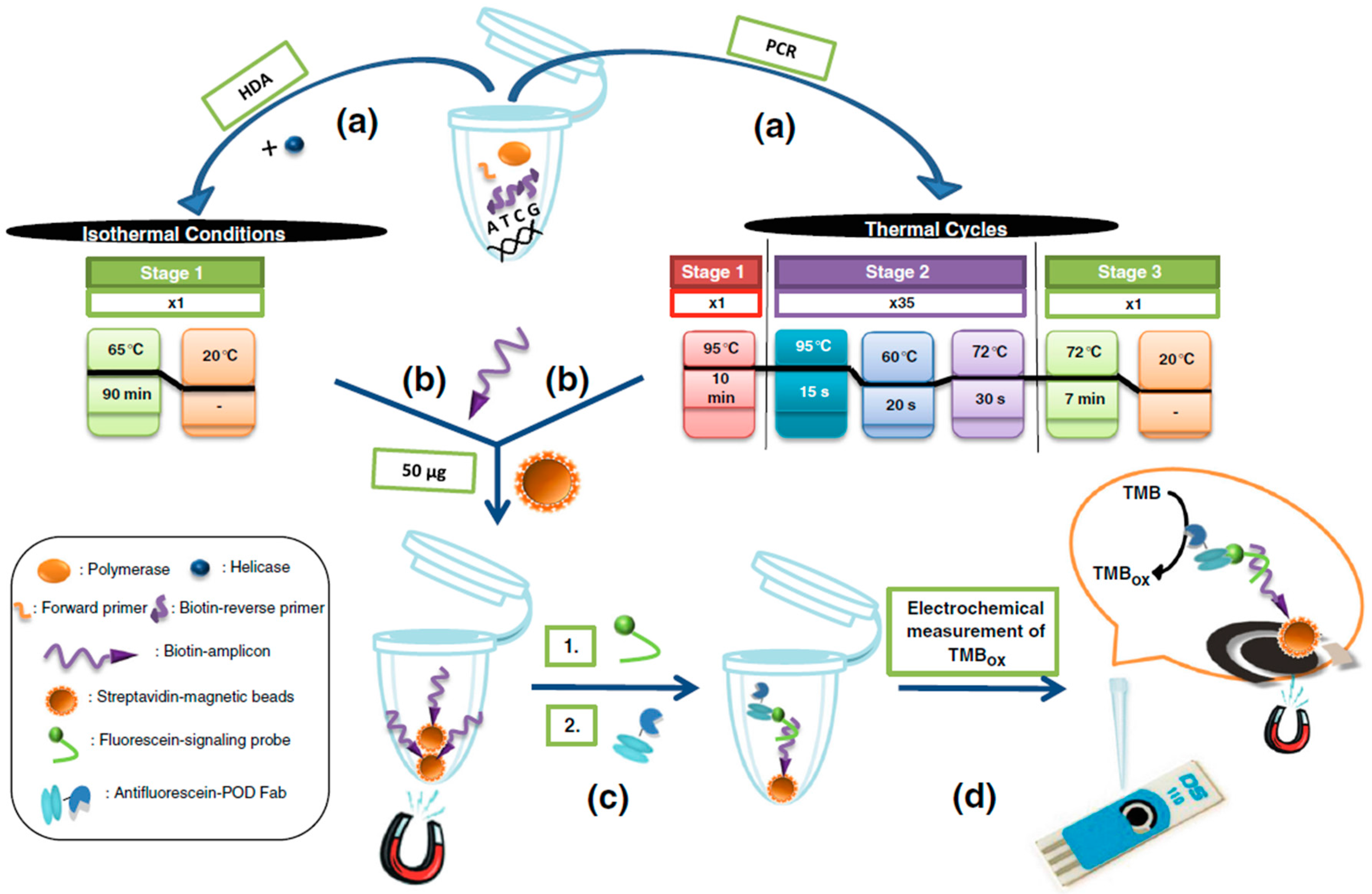
- Barreda-García, S.; González-Álvarez, M.J.; de los Santos-Álvarez, N.; Palacios-Gutiérrez, J.J.; Miranda-Ordieres, A.J. Attomolar quantitation of Mycobacterium tuberculosis by asymmetric helicase-dependent isothermal DNA-amplification and electrochemical detection. Biosens. Bioelectron. 2015, 68, 122–128. [Google Scholar]
- Pinto, A.M.; Gonçalves, I.C.; Magalhães, F.D. Graphene-based materials biocompatibility: A review. Colloids Surf. B 2013, 111, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, N.J.; Okon, S.L. Nanomaterial-based electrochemical immunosensors for clinically significant biomarkers. Materials 2014, 7, 4669–4709. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Leong, M.C.; Leong, E.Z.W.; Kuan, W.S.; Leong, D.T. Clinically Relevant Detection of Streptococcus pneumoniae with DNA Antibody Nanostructures. Anal. Chem. 2017, 89, 6900–6906. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.; Bonner, A.; Cohen, D.M.; Birkhahn, R.; Yogev, R.; Triner, W.; Cohen, J.; Palavecino, E.; Selvarangan, R. Multicenter clinical evaluation of the novel Alere i Influenza A&B isothermal nucleic acid amplification test. J. Clin. Virol. 2014, 61, 81–86. [Google Scholar] [PubMed]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Barreda-García, S.; Miranda-Castro, R.; de los Santos Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Solid-phase helicase dependent amplification and electrochemical detection of Salmonella on highly stable oligonucleotide-modified ITO electrodes. Chem. Commun. 2017, 53, 9721–9724. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Pedrero, M.; García, J.L.; García, E.; García, P.; Pingarrón, J.M. Development of amperometric magnetogenosensors coupled to asymmetric PCR for the specific detection of Streptococcus pneumoniae. Anal. Bioanal. Chem. 2011, 399, 2413–2420. [Google Scholar] [CrossRef] [PubMed]
- Sotillo, A.; Pedrero, M.; de Pablos, M.; García, J.L.; García, E.; García, P.; Pingarrón, J.M.; Mingorance, J.; Campuzano, S. Clinical evaluation of a disposable amperometric magneto-genosensor for the detection and identification of Streptococcus pneumoniae. J. Microbiol. Meth. 2014, 103, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Barreda-García, S.; Miranda-Castro, R.; de los Santos Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Comparison of isothermal helicase-dependent amplification and PCR for the detection of Mycobacterium tuberculosis by an electrochemical genomagnetic assay. Anal. Bioanal. Chem. 2016, 408, 8603–8610. [Google Scholar]
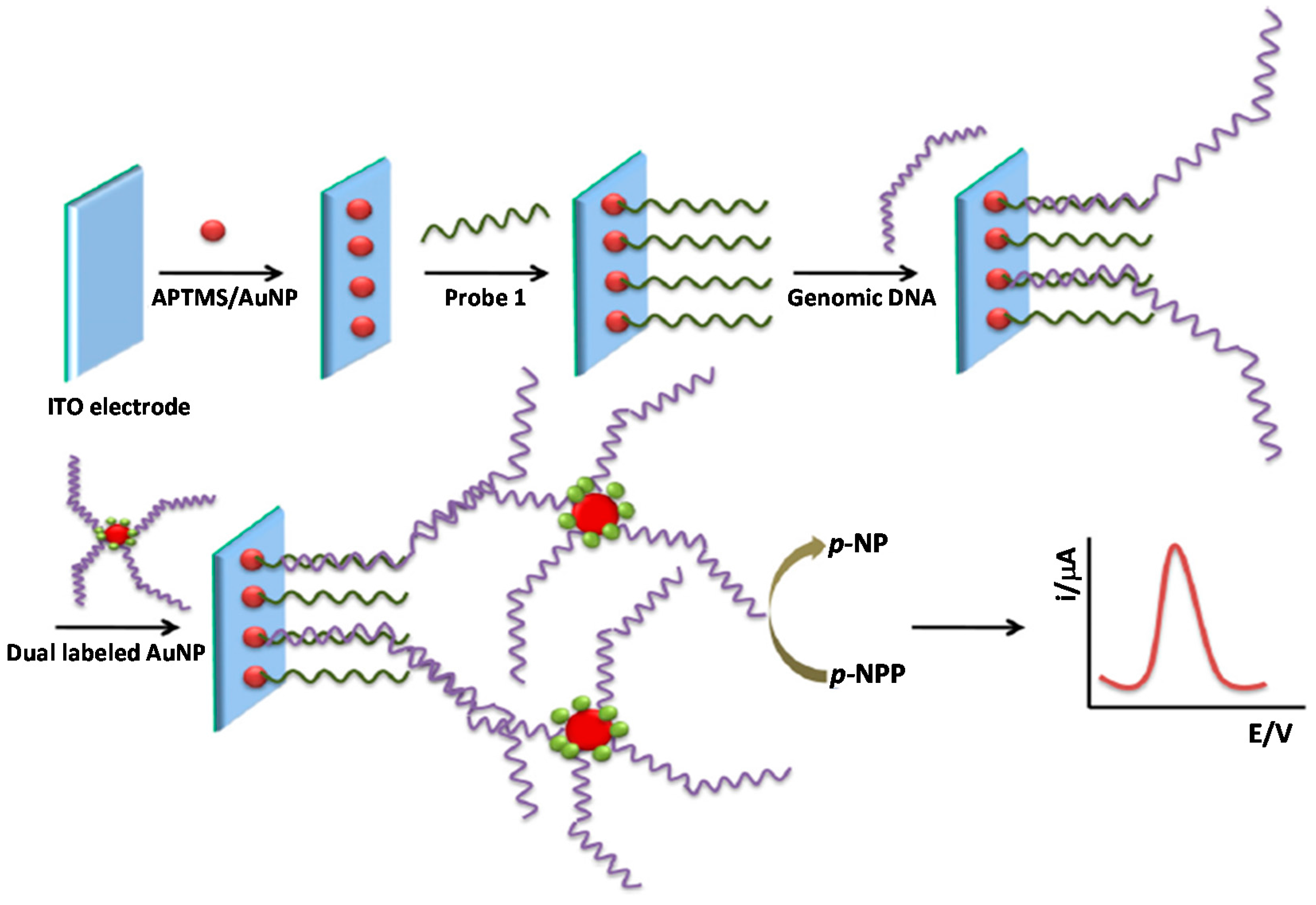
- De la Escosura-Muñiz, A.; Baptista-Pires, L.; Serrano, L.; Altet, L.; Francino, O.; Sánchez, A.; Merkoçi, A. Magnetic Bead/Gold Nanoparticle Double-Labeled Primers for Electrochemical Detection of Isothermal Amplified Leishmania DNA. Small 2016, 12, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiong, Y.; Fang, L.; Jiang, L.; Huang, H.; Deng, J.; Liang, W.; Zheng, J. An Electrochemical Strategy using Multifunctional Nanoconjugates for Efficient Simultaneous Detection of Escherichia Coli O157: H7 and Vibrio cholerae O1. Theranostics 2017, 7, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Esteban-Fernández de Ávila, B.; Yuste, J.; Pedrero, M.; García, J.L.; García, P.; García, E.; Pingarrón, J.M. Disposable amperometric magnetoimmunosensors for the specific detection of Streptococcus pneumoniae. Biosens. Bioelectron. 2010, 26, 1225–1230. [Google Scholar] [CrossRef] [PubMed]
- Gayathri, C.H.; Mayuri, P.; Sankaran, K.; Kumar, A.S. An electrochemical immunosensor for efficient detection of uropathogenic E. coli based on thionine dye immobilized chitosan/functionalized-MWCNT modified electrode. Biosens. Bioelectron. 2016, 82, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Campuzano, S.; Halford, C.; Haake, D.A.; Wang, J. Ternary surface monolayers for ultrasensitive (zeptomole) amperometric detection of nucleic acid hybridization without signal amplification. Anal. Chem. 2010, 82, 8830–8837. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lu, Y.; Gau, V.; Liao, J.C.; Wong, P.K. Rapid Antimicrobial Susceptibility Testing with Electrokinetics Enhanced Biosensors for Diagnosis of Acute Bacterial Infections. Ann. Biomed. Eng. 2014, 42, 2314–2321. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Jeffries, L.; Mach, K.E.; Craft, D.W.; Thomas, N.J.; Gau, V.; Liao, J.C.; Wong, P.K. A Multiplex Electrochemical Biosensor for Bloodstream Infection Diagnosis. J. Lab. Autom. 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mach, K.E.; Mohan, R.; Patel, S.; Wong, P.K.; Hsieh, M.; Liao, J.C. Development of a Biosensor-Based Rapid Urine Test for Detection of Urogenital Schistosomiasis. PLoS Negl. Trop. Dis. 2015, 9, e0003845. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Sekine, S.; Uenoyama, T.; Wada, M.; Ikeuchi, T.; Saito, M.; Yamaguchi, Y.; Tamiya, E. Quantitative detection for Porphyromonas gingivalis in tooth pocket and saliva by portable electrochemical DNA sensor linked with PCR. Electroanalysis 2014, 26, 2686–2692. [Google Scholar] [CrossRef]
- Thiruppathiraja, C.; Kamatchiammal, S.; Adaikkappan, P.; Santhosh, D.J.; Alagar, M. Specific detection of Mycobacterium sp. genomic DNA using dual labeled gold nanoparticle based electrochemical biosensor. Anal. Biochem. 2011, 417, 73–79. [Google Scholar] [CrossRef] [PubMed]
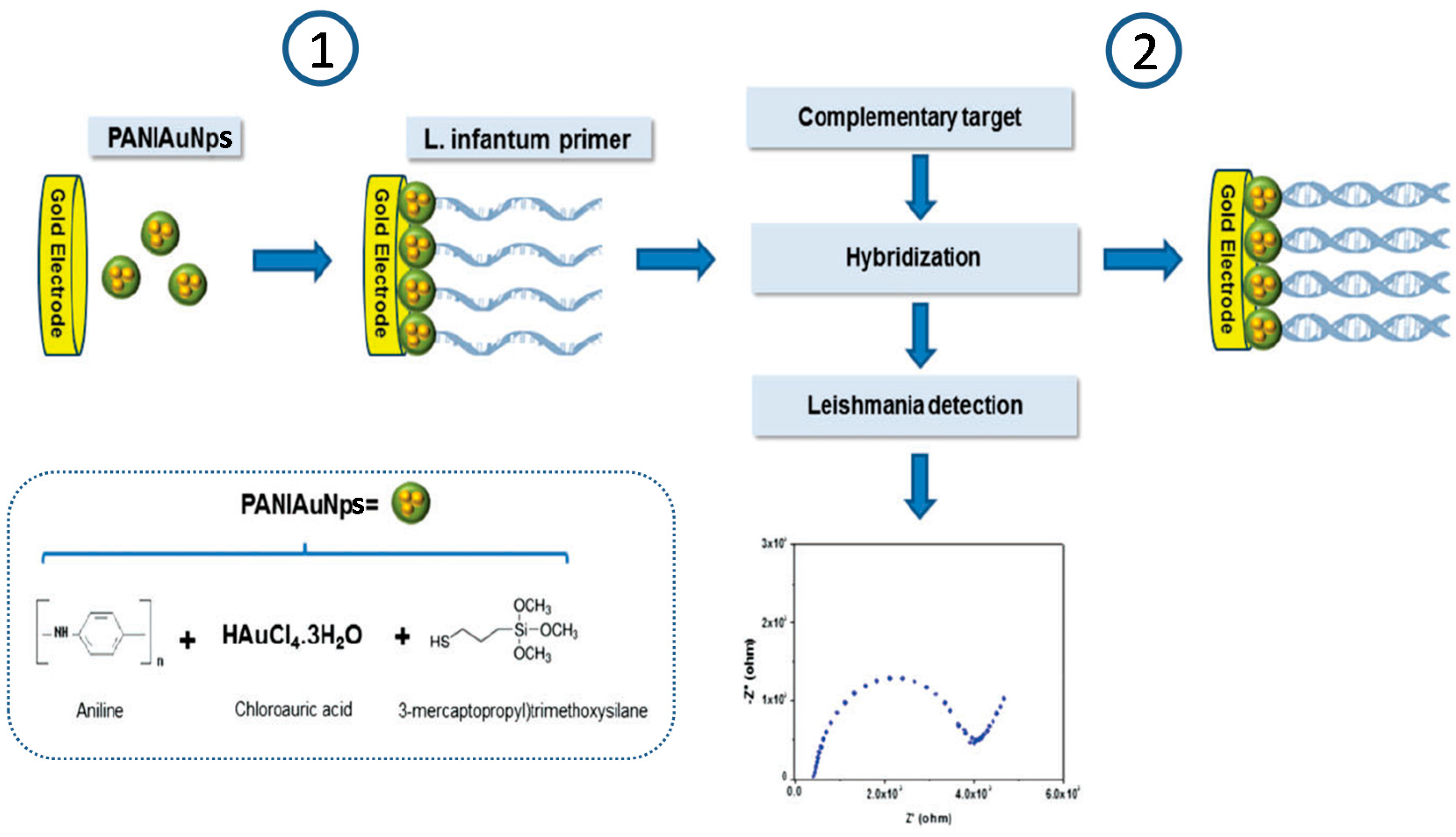
- Garcia, M.F.K.; Andrade, C.A.S.; de Melo, C.P.; Gomes, D.S.; Silva, L.G.; Dias, R.V.; Balbino, V.Q.; Oliveira, M.D.L. Impedimetric sensor for Leishmania infantum genome based on gold nanoparticles dispersed in polyaniline matrix. J. Chem. Technol. Biotechnol. 2016, 91, 2810–2816. [Google Scholar] [CrossRef]
- Ning, G.; Nai-Xing, L.; Tian-Hua, L.; Lei, Z.; Min-Jun, N. A Non-enzyme Amperometric Immunosensor for Rapid Determination of Human Immunodeficiency Virus p24 Based on Magnetism Controlled Carbon Nanotubes Modified Printed Electrode. Chin. J. Anal. Chem. 2010, 38, 1556–1562. [Google Scholar]
- Gan, N.; Hou, J.; Hu, F.; Zheng, L.; Ni, M.; Cao, Y. An Amperometric Immunosensor Based on a Polyelectrolyte/ Gold Magnetic Nanoparticle Supramolecular Assembly—Modified Electrode for the Determination of HIV p24 in Serum. Molecules 2010, 15, 5053–5065. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Jia, L.; Li, B.; Situ, B.; Liu, Q.; Wang, Q.; Gan, N. A Sandwich HIV p24 Amperometric Immunosensor Based on a Direct Gold Electroplating-Modified Electrode. Molecules 2012, 17, 5988–6000. [Google Scholar] [CrossRef] [PubMed]
- Gan, N.; Du, X.; Cao, Y.; Hu, F.; Li, T.; Jiang, Q. An Ultrasensitive Electrochemical Immunosensor for HIV p24 Based on Fe3O4@SiO2 Nanomagnetic Probes and Nanogold Colloid-Labeled Enzyme–Antibody Copolymer as Signal Tag. Materials 2013, 6, 1255–1269. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhou, R.; Zhao, K.; Chen, H.; Hu, Y. Magnetic Beads-Based Electrochemical Immunosensor for Detection of Pseudorabies Virus Antibody in Swine Serum. Talanta 2011, 87, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Bhimji, A.; Zaragoza, A.A.; Live, L.S.; Kelley, S.O. Electrochemical Enzyme-Linked Immunosorbent Assay Featuring Proximal Reagent Generation: Detection of Human Immunodeficiency Virus Antibodies in Clinical Samples. Anal. Chem. 2013, 85, 6813–6819. [Google Scholar] [CrossRef] [PubMed]
- Jarocka, U.; Sawicka, R.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecki, J.; Radecka, H. Electrochemical immunosensor for detection of antibodies against influenza A virus H5N1 in hen serum. Biosens. Bioelectron. 2014, 55, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Jarocka, U.; Sawicka, R.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Sączyńska, V.; Porębska, A.; Dehaen, W.; Radecki, J.; Radecka, H. A biosensor based on electroactive dipyrromethene-Cu(II) layer deposited onto gold electrodes for the detection of antibodies against avian influenza virus type H5N1 in hen sera. Anal. Bioanal. Chem. 2015, 407, 7807–7814. [Google Scholar] [CrossRef] [PubMed]
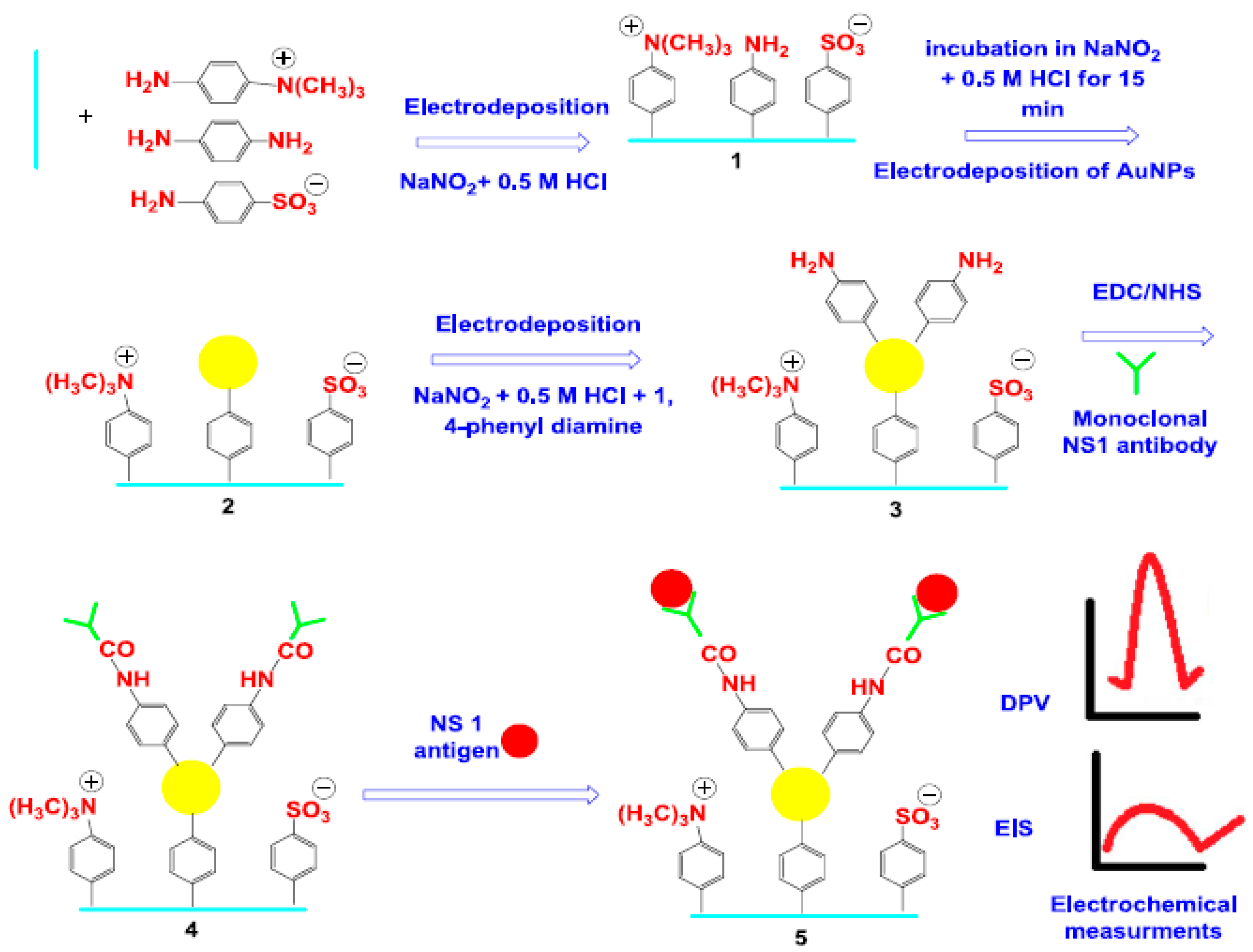
- Darwish, N.T.; Alrawi, A.H.; Sekaran, S.D.; Alias, Y.; Khor, S.M. Electrochemical Immunosensor Based on Antibody-Nanoparticle Hybrid for Specific Detection of the Dengue Virus NS1 Biomarker. J. Electrochem. Soc. 2016, 163, B19–B25. [Google Scholar] [CrossRef]
- Hou, Y.-H.; Wang, J.-J.; Jiang, Y.-Z.; Lv, C.; Xia, L.; Hong, S.-L.; Lin, M.; Lin, Y.; Zhang, Z.-L.; Pang, D.-W. A Colorimetric and Electrochemical Immunosensor for Point-of-Care Detection of Enterovirus 71. Biosens. Bioelectron. 2017. [Google Scholar] [CrossRef] [PubMed]
- McQuistan, A.; Zaitouna, A.J.; Echeverria, E.; Lai, R.Y. Use of thiolated oligonucleotides as anti-fouling diluents in electrochemical peptide-based sensors. Chem. Commun. 2014, 50, 4690–4692. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.; Souza, E.; Ferreira, D.; Zanforlin, D.; Bezerra, W.; Borba, M.A.; Arruda, M.; Lopes, K.; Nascimento, G.; Martins, D.; et al. A Sensitive and Selective Label-Free Electrochemical DNA Biosensor for the Detection of Specific Dengue Virus Serotype 3 Sequences. Sensors 2015, 15, 15562–15577. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, M.; Durikova, H.; Vojtesek, B.; Anton, M.; Jandakova, E.; Hrstka, R. Electrochemical chip-based genomagnetic assay for detection of high-risk human papillomavirus DNA. Biosens. Bioelectron. 2016, 83, 300–305. [Google Scholar] [CrossRef] [PubMed]
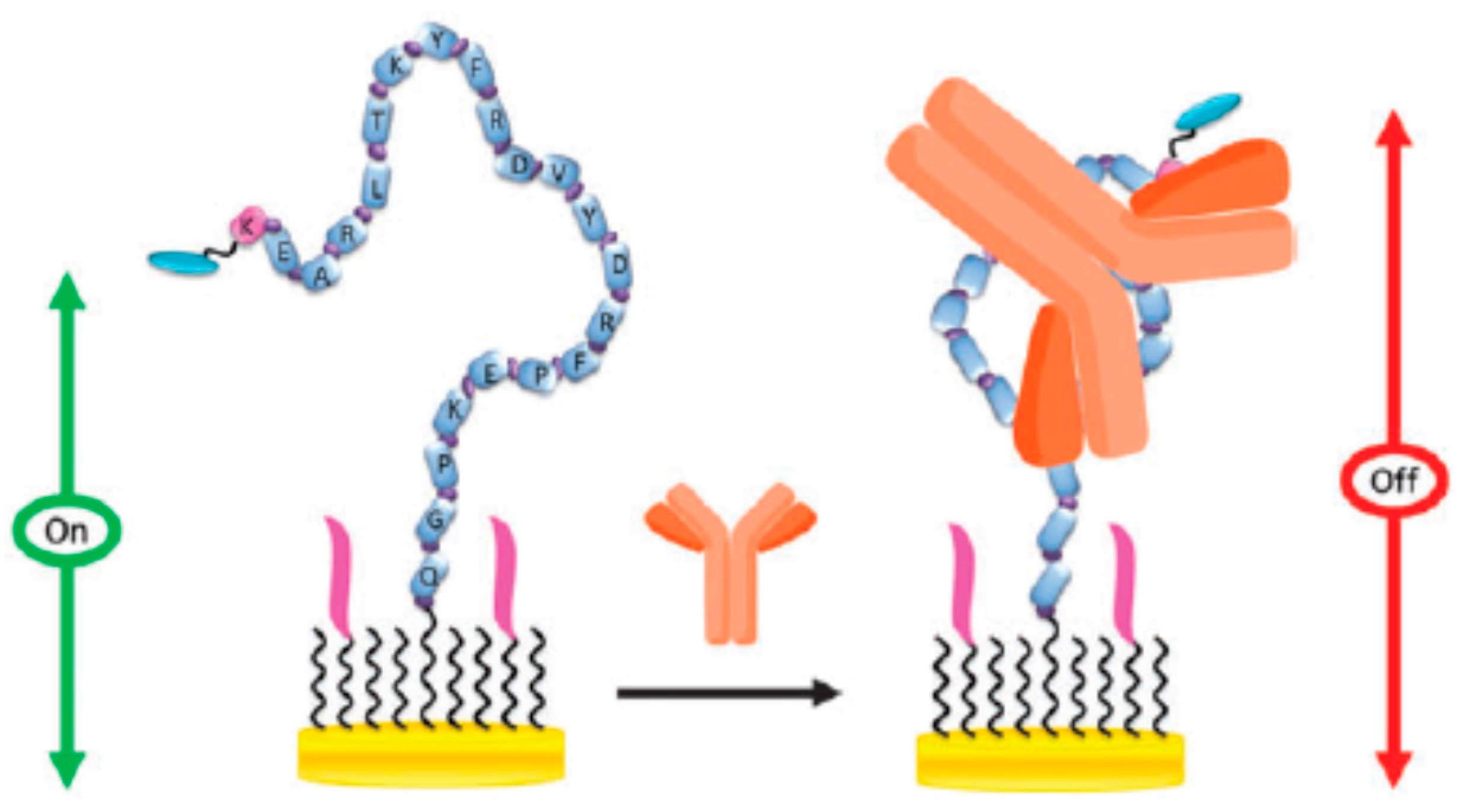
- Hwang, H.; Ryu, M.; Park, C.Y.; Ahn, J.; Park, H.G.; Choi, C.; Ha, S.-D.; Park, T.J.; Park, J.P. High sensitive and selective electrochemical biosensor: Label-free detection of human norovirus using affinity peptide as molecular binder. Biosens. Bioelectron. 2017, 87, 164–170. [Google Scholar] [CrossRef] [PubMed]

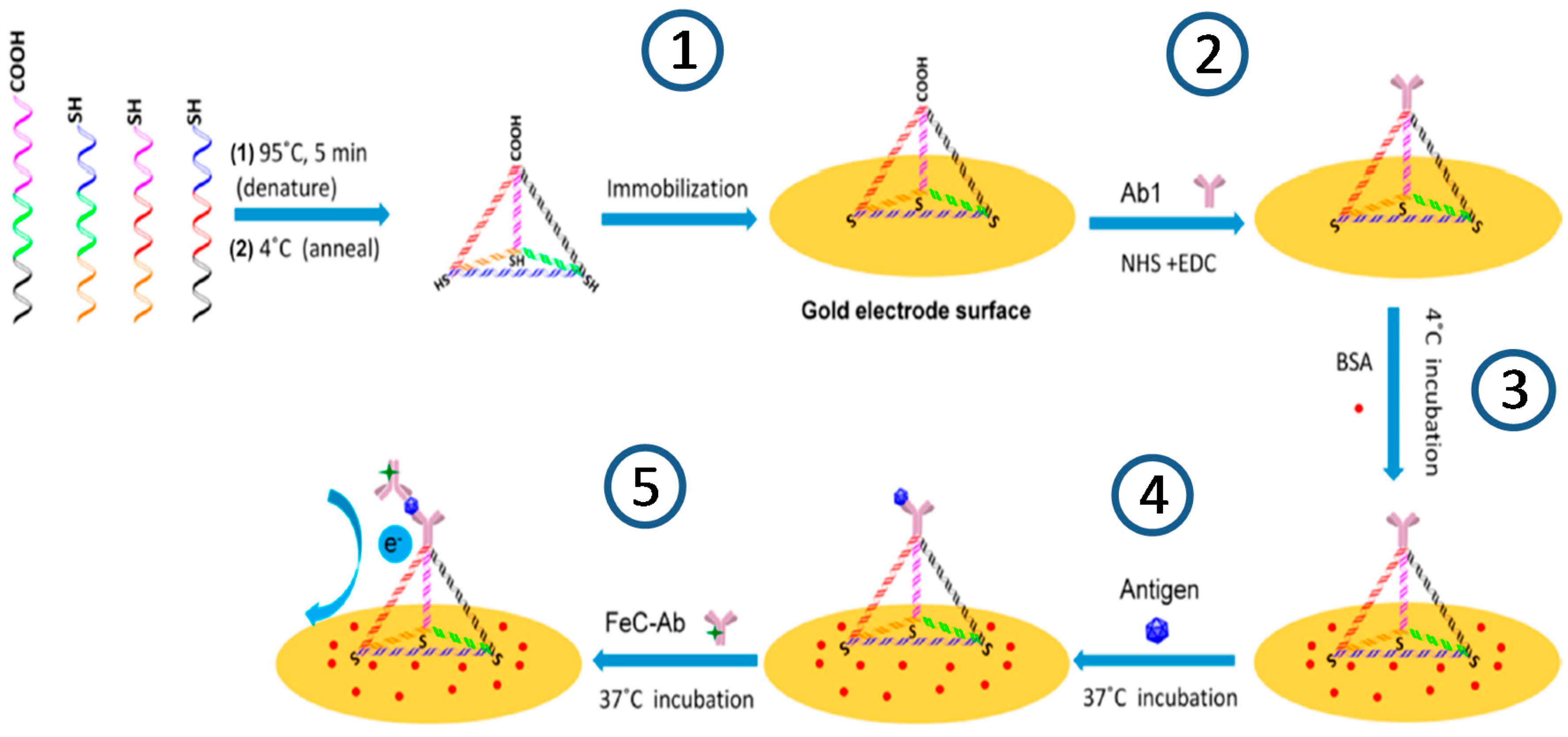
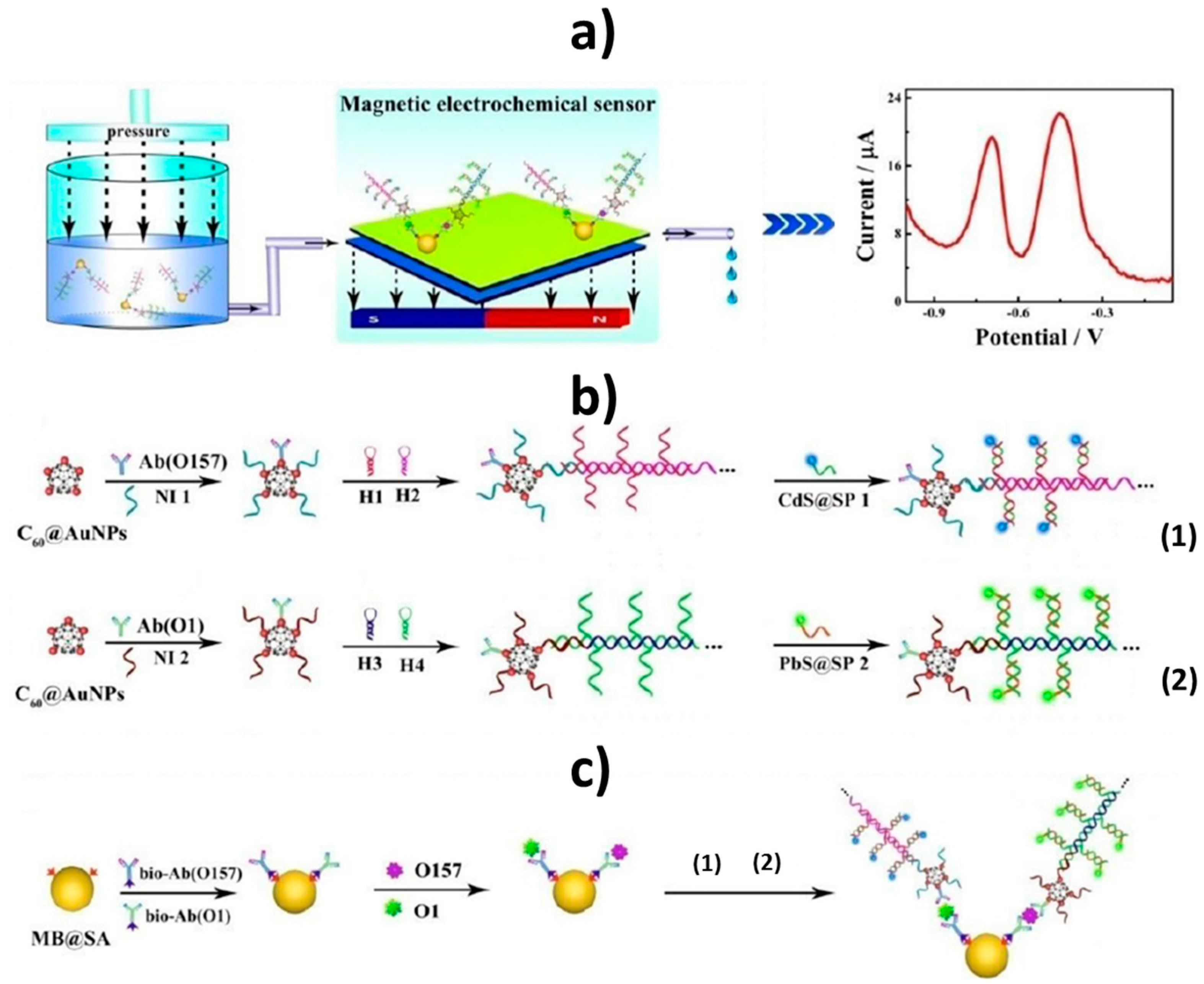
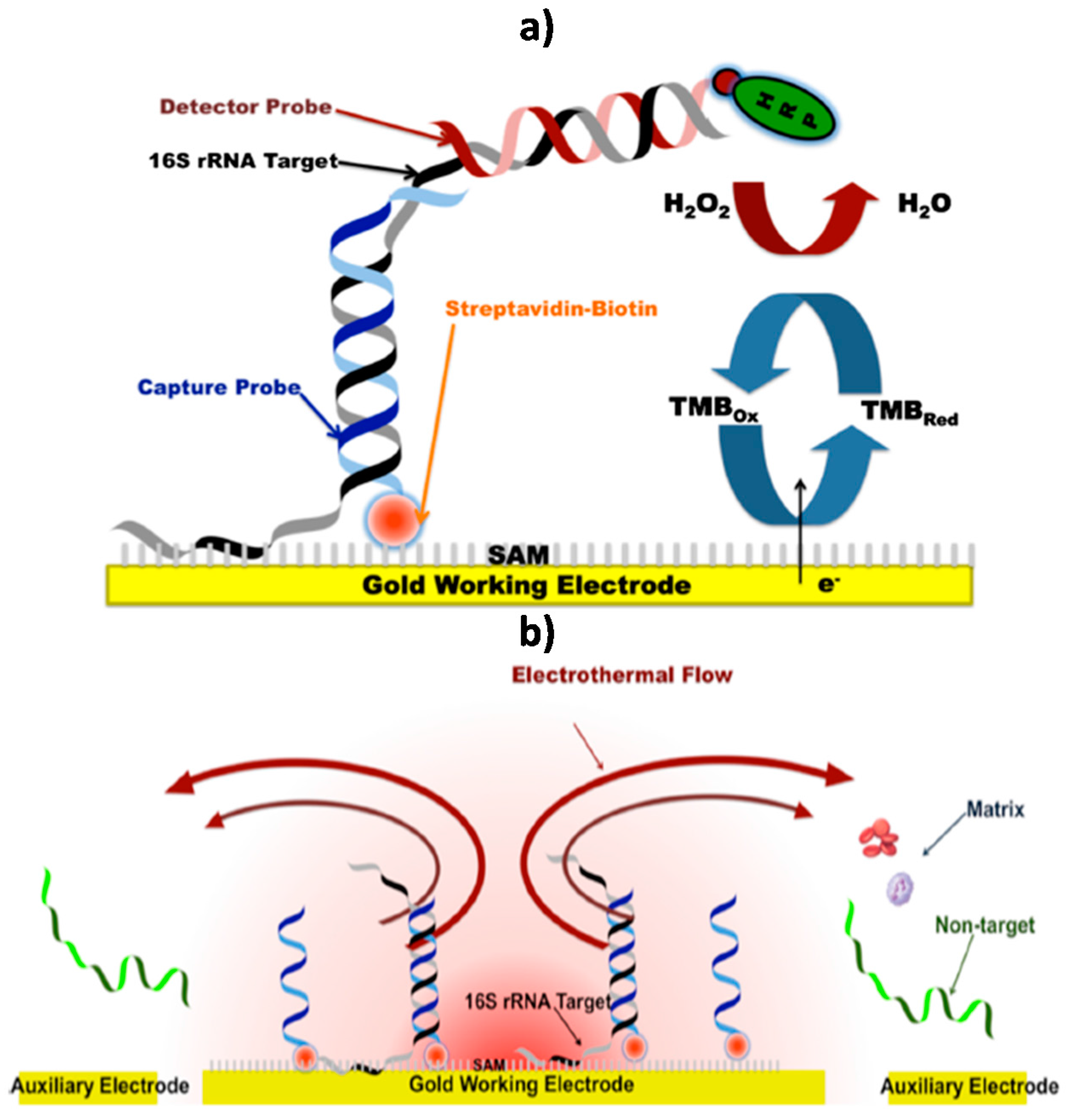
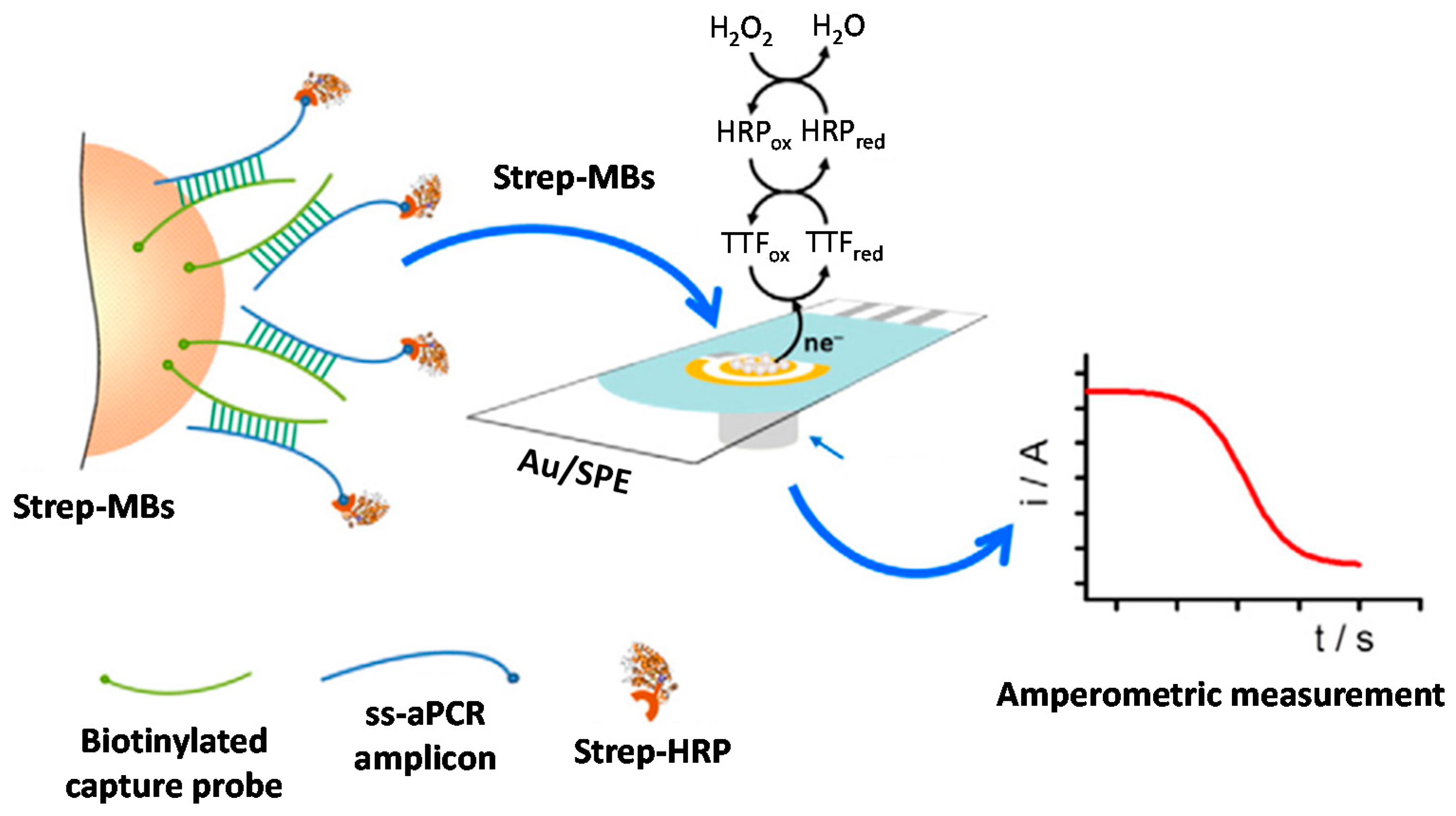

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges. Sensors 2017, 17, 2533. https://doi.org/10.3390/s17112533
Campuzano S, Yáñez-Sedeño P, Pingarrón JM. Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges. Sensors. 2017; 17(11):2533. https://doi.org/10.3390/s17112533
Chicago/Turabian StyleCampuzano, Susana, Paloma Yáñez-Sedeño, and José Manuel Pingarrón. 2017. "Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges" Sensors 17, no. 11: 2533. https://doi.org/10.3390/s17112533
APA StyleCampuzano, S., Yáñez-Sedeño, P., & Pingarrón, J. M. (2017). Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges. Sensors, 17(11), 2533. https://doi.org/10.3390/s17112533





