Disease-Related Detection with Electrochemical Biosensors: A Review
Abstract
1. Background
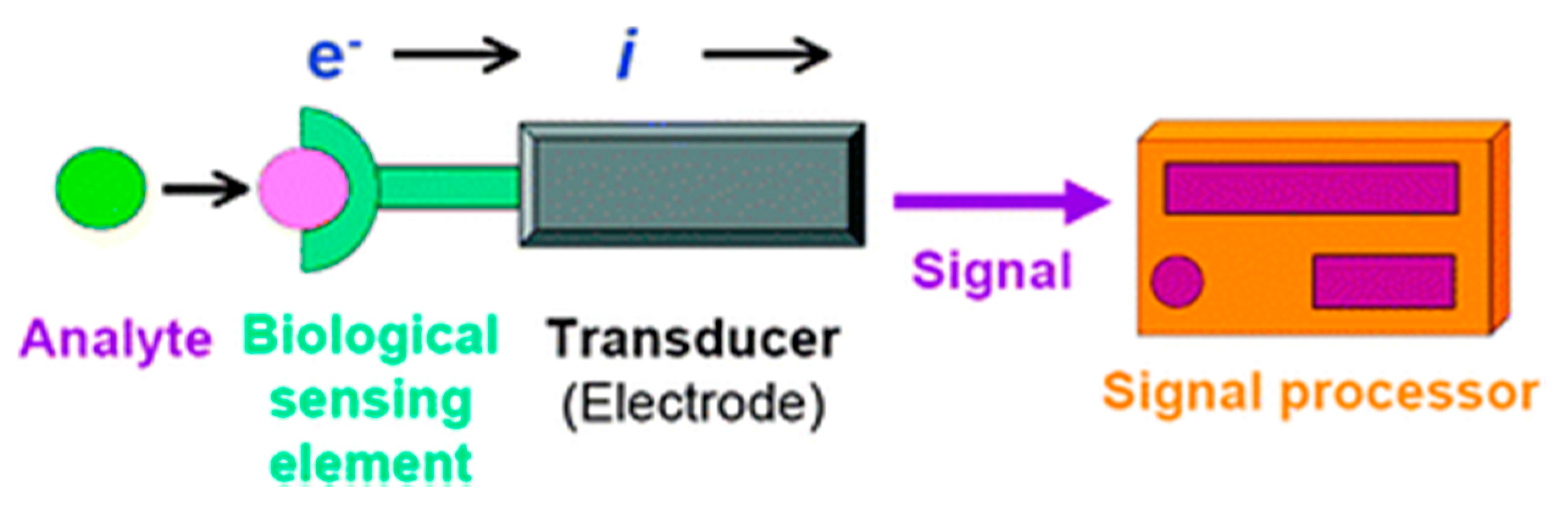
2. Electrochemical Biosensors
2.1. Different Recognition Elements
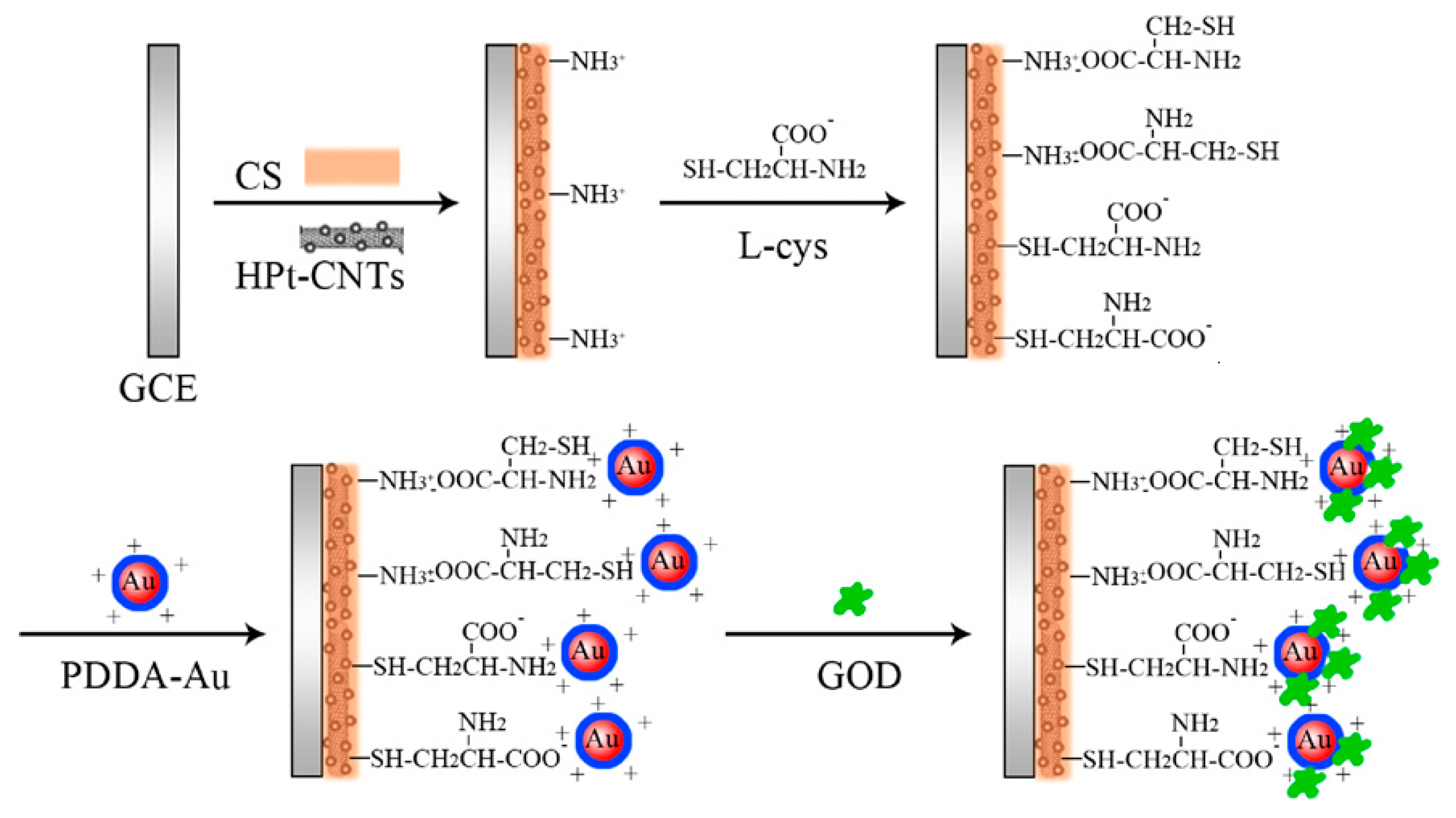
2.2. Immobilization Strategies
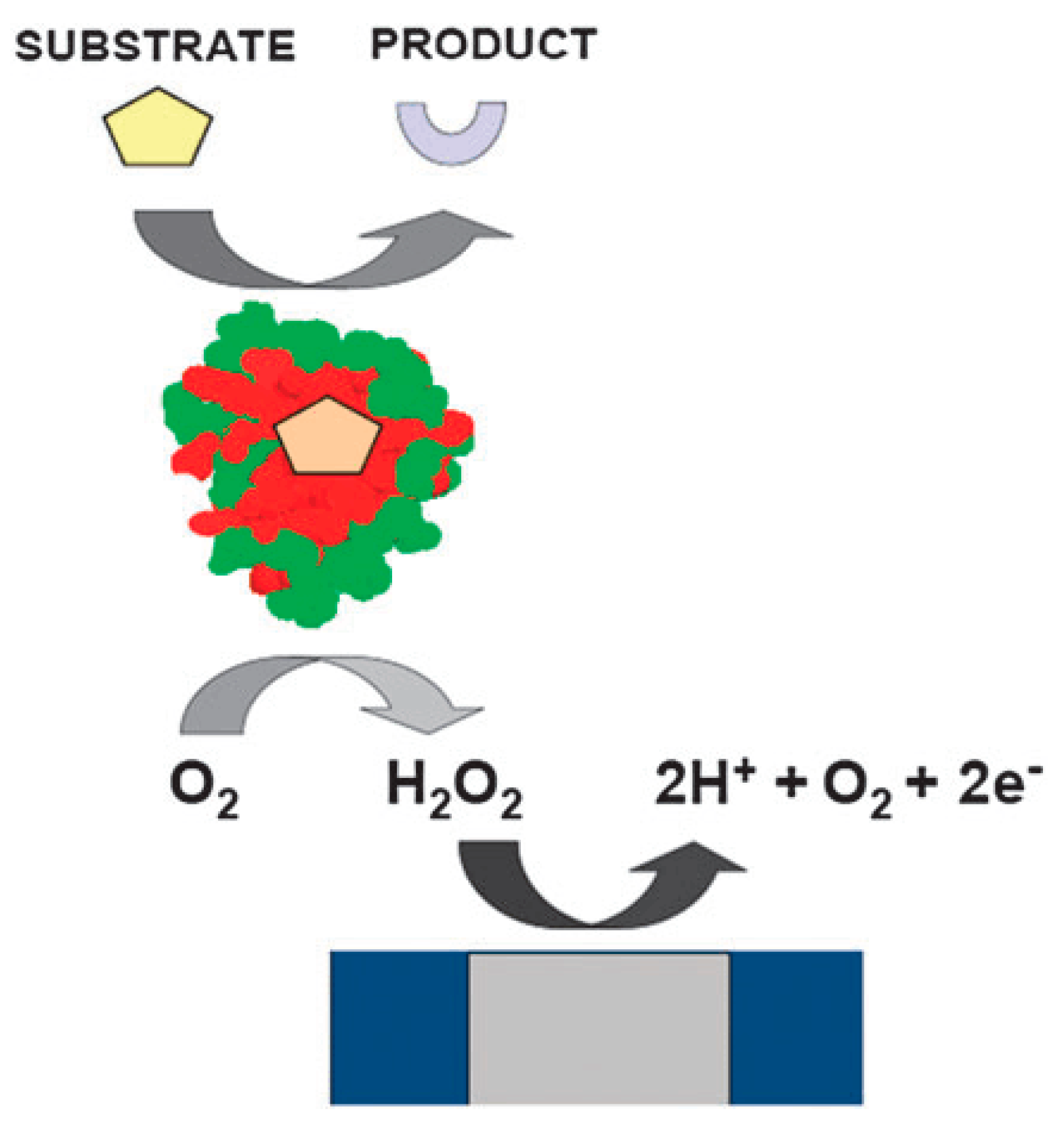
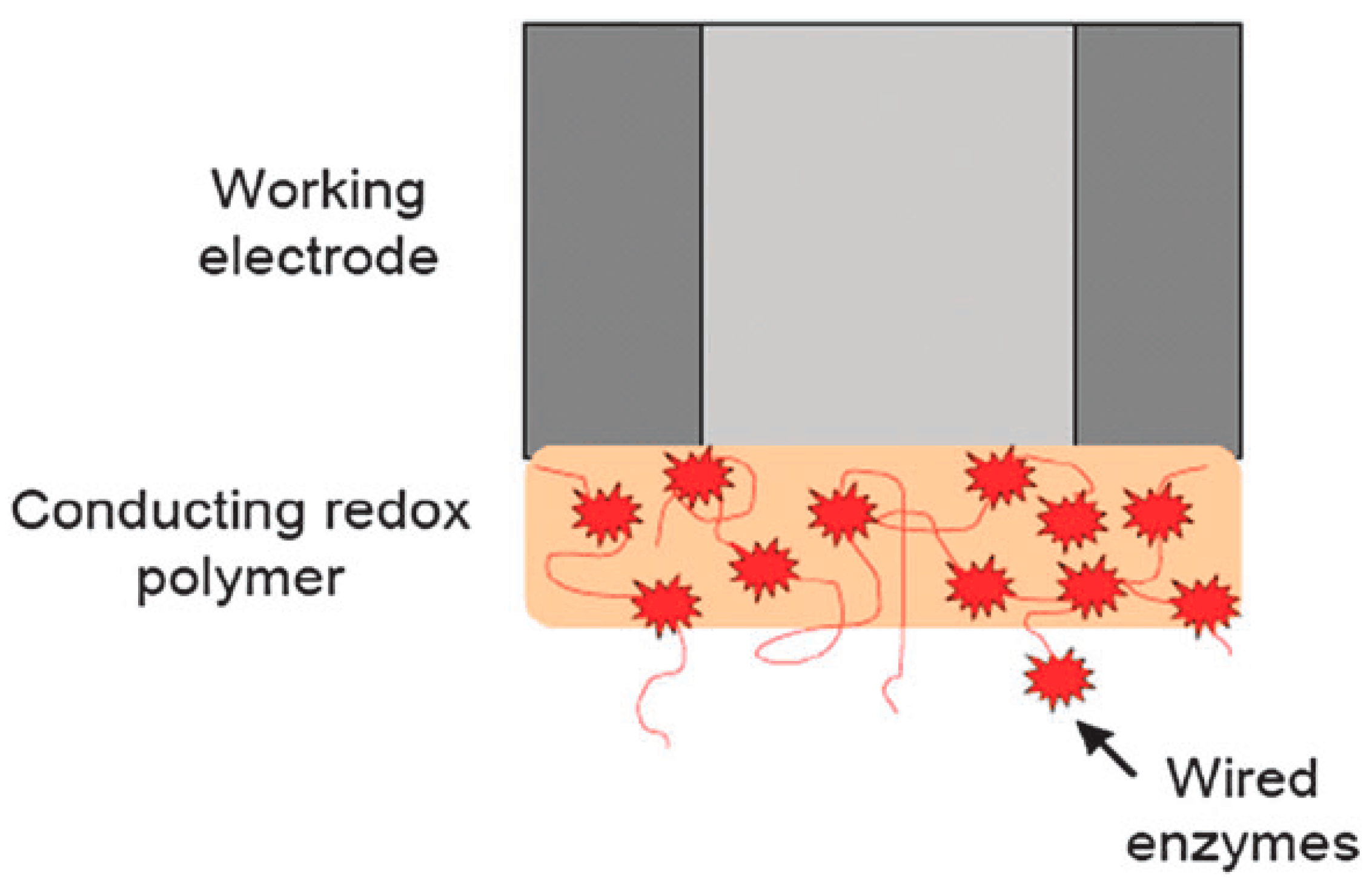
2.3. Electrical Communication between the Biomolecules and the Transducer
2.4. Materials Used in Electrochemical Biosensors
3. Application of Electrochemical Biosensors for Disease
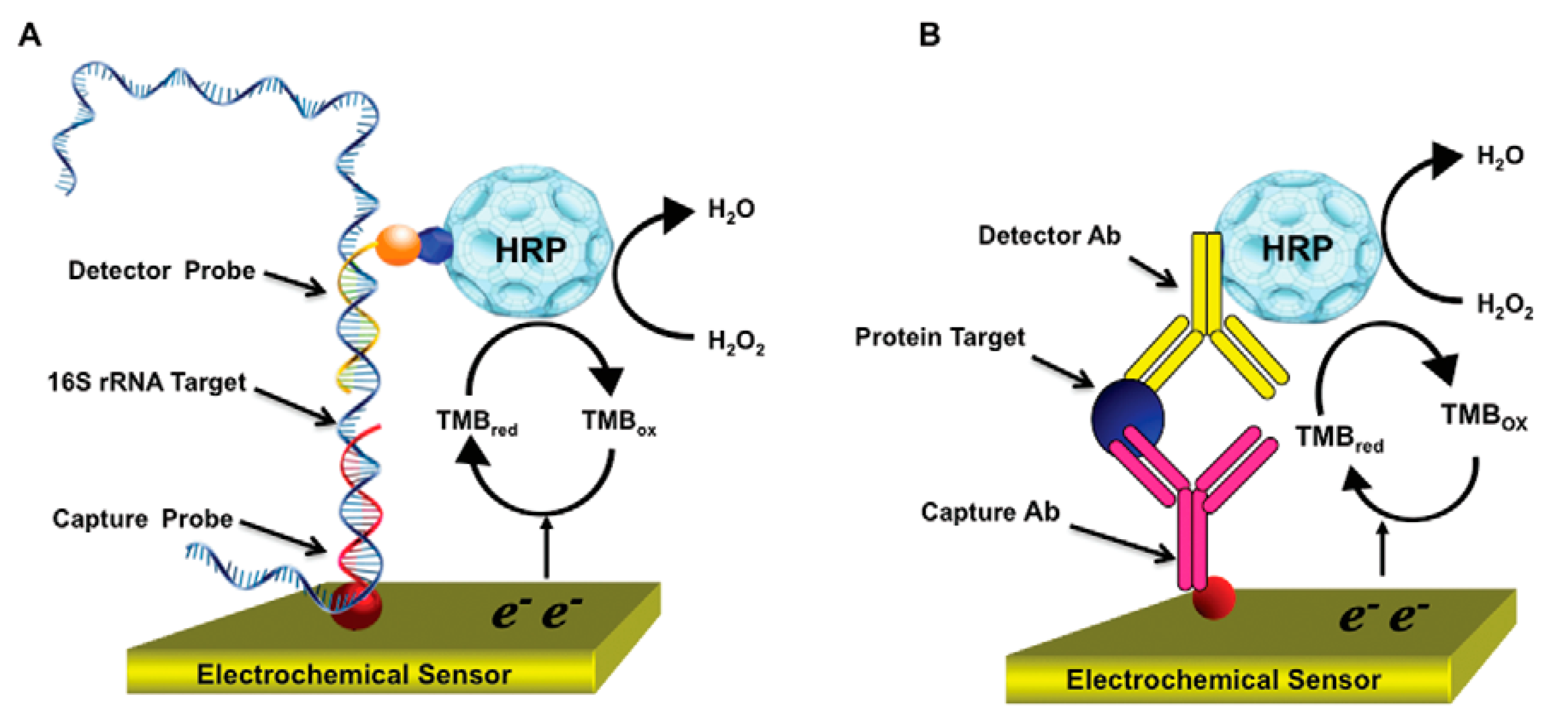
3.1. Antibody as Recognition Biomolecule
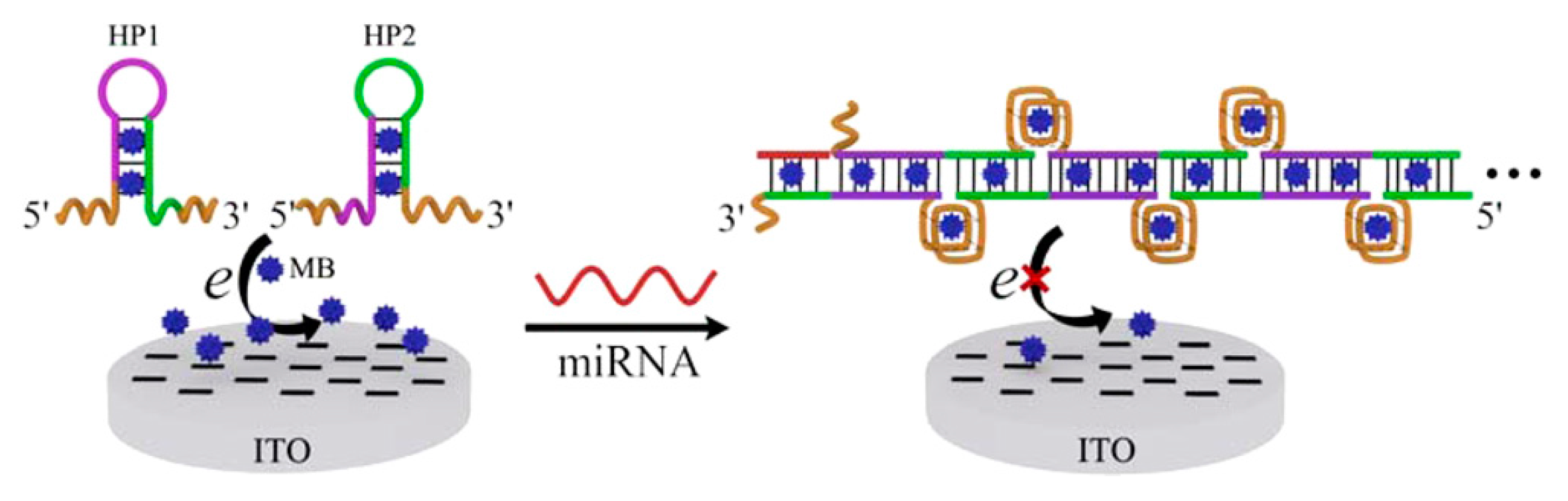
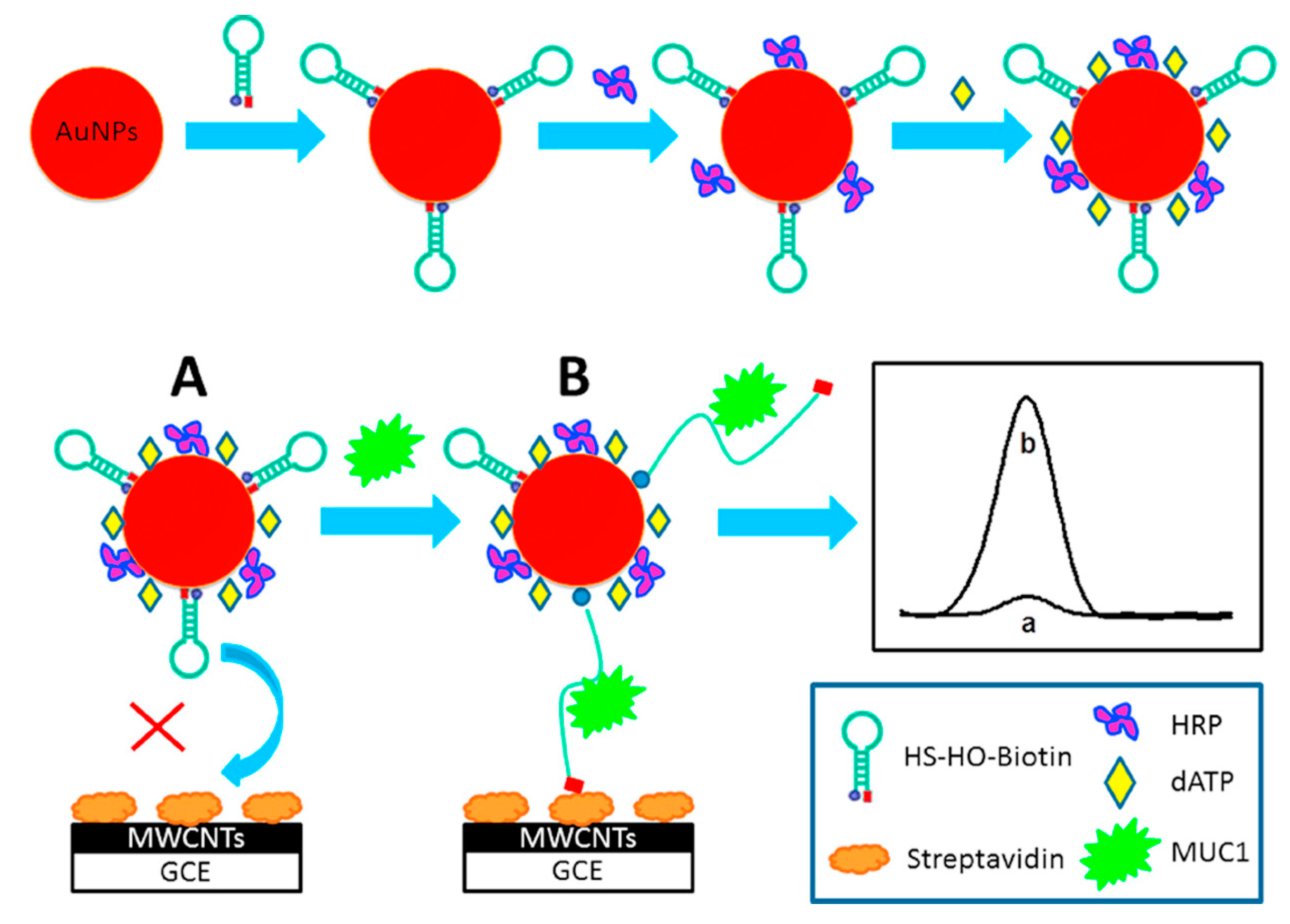
3.2. DNA/microRNA as Biomolecule
3.3. Enzyme as Biomolecules
3.4. Other Biomolecules
4. Conclusions and Outlook
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Trashin, S.; Pchelintsev, N. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Benvidi, A.; Firouzabadi, A.D.; Tezerjani, M.D.; Moshtaghiun, S.M.; Mazloum-Ardakani, M.; Ansarin, A. A highly sensitive and selective electrochemical DNA biosensor to diagnose breast cancer. J. Electroanal. Chem. 2015, 750, 57–64. [Google Scholar] [CrossRef]
- Rafieepour, H.A.; Behpour, M.; Keshavarz, M. A novel label-free electrochemical miRNA biosensor using methylene blue as redox indicator: Application to breast cancer biomarker miRNA-21. Biosens. Bioelectron. 2016, 77, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.Y.; Cheng, K.L.; Mcguffin-Cawley, J.D.; Shieu, F.S.; Samia, A.C.; Gupta, S.; Cooney, M.; Thompson, C.L.; Liu, C.C. Detection of Alpha-Methylacyl-CoA Racemase (AMACR), a Biomarker of Prostate Cancer, in Patient Blood Samples Using a Nanoparticle Electrochemical Biosensor. Biosensors 2012, 2, 377–387. [Google Scholar] [CrossRef] [PubMed]
- De la Escosura-Muñiz, A.; Maltez-da Costa, M.; Sánchez-Espinel, C.; Díaz-Freitas, B.; Fernández-Suarez, J.; González-Fernández, Á.; Merkoçi, A. Gold nanoparticle-based electrochemical magnetoimmunosensor for rapid detection of anti-hepatitis B virus antibodies in human serum. Biosens. Bioelectron. 2010, 26, 1710–1714. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Analytical Electrochemistry, 3rd ed.; Wiley: Hoboken, NJ, USA, 2006. [Google Scholar]
- Chaubey, A.; Malhotra, B.D. Mediated biosensors. Biosens. Bioelectron. 2002, 17, 441–456. [Google Scholar] [CrossRef]
- Eggins, B. Chemical Sensors and Biosensors. Anal. Chem. 2002, 84, 685–707. [Google Scholar]
- Banerjee, P.; Bhunia, A.K. Mammalian cell-based biosensors for pathogens and toxins. Trends Biotechnol. 2009, 27, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Kara, P.; Erzurumlu, Y.; Kirmizibayrak, P.B.; Ozsoz, M. Electrochemical aptasensor design for label free cytosensing of human non-small cell lung cancer. J. Electroanal. Chem. 2016, 775, 337–341. [Google Scholar] [CrossRef]
- Teles, F.R.R.; Fonseca, L.P. Applications of polymers for biomolecule immobilization in electrochemical biosensors. Mater. Sci. Eng. C 2008, 28, 1530–1543. [Google Scholar] [CrossRef]
- Herrmann, S.; Leshem, B.; Landes, S.; Rager-Zisman, B.; Marks, R.S. Chemiluminescent optical fiber immunosensor for the detection of anti-West Nile virus IgG *. Talanta 2005, 66, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Naimushin, A.N.; Soelberg, S.D.; Bartholomew, D.U.; Elkind, J.L.; Furlong, C.E. A portable surface plasmon resonance (SPR) sensor system with temperature regulation. Sens. Actuators B Chem. 2003, 96, 253–260. [Google Scholar] [CrossRef]
- Mauriz, E.; Calle, A.; Montoya, A.; Lechuga, L.M. Determination of environmental organic pollutants with a portable optical immunosensor. Talanta 2006, 69, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, W.; Lu, L.; Zheng, D.; Chen, H.; Xu, L. Electrochemical immunosensor for rapid detection of chlorpyrifos in agricultural products. Trans. Chin. Soc. Agric. Eng. 2014, 30, 278–283. [Google Scholar]
- Shankaran, D.R.; Gobi, K.V.; Miura, N. Recent advancements in surface plasmon resonance immunosensors for detection of small molecules of biomedical, food and environmental interest. Sens. Actuators B Chem. 2007, 121, 158–177. [Google Scholar] [CrossRef]
- Akanda, M.R.; Aziz, M.A.; Jo, K.; Tamilavan, V.; Hyun, M.H.; Kim, S.; Yang, H. Optimization of Phosphatase- and Redox Cycling-Based Immunosensors and Its Application to Ultrasensitive Detection of Troponin I. Anal. Chem. 2011, 83, 3926–3933. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Zhao, F.; Zhang, M.; Zhang, S. Hybridization biosensor using 2,9-dimethyl-1,10-phenantroline cobalt as electrochemical indicator for detection of hepatitis B virus DNA. Bioelectrochemistry 2008, 72, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Tolle, F.; Wilke, J.; Wengel, J.; Mayer, G. By-product formation in repetitive PCR amplification of DNA libraries during SELEX. PLoS ONE 2014, 9, e114693. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Yan, M.; Lu, J.; Zhang, M.; Yu, F.; Yu, J.; Song, X.; Yu, S. Electrochemical biosensor based on graphene oxide–Au nanoclusters composites for l -cysteine analysis. Biosens. Bioelectron. 2012, 31, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Munge, B.S.; Fisher, J.; Millord, L.N.; Krause, C.E.; Dowd, R.S.; Rusling, J.F. Sensitive electrochemical immunosensor for matrix metalloproteinase-3 based on single-wall carbon nanotubes. Analyst 2010, 135, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Nanduri, V.; Sorokulova, I.B.; Samoylov, A.M.; Simonian, A.L.; Petrenko, V.A.; Vodyanoy, V. Phage as a molecular recognition element in biosensors immobilized by physical adsorption. Biosens. Bioelectron. 2007, 22, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, W.; Zhang, S. Development of DNA electrochemical biosensor based on covalent immobilization of probe DNA by direct coupling of sol-gel and self-assembly technologies. Biosens. Bioelectron. 2008, 24, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Immobilization strategies to develop enzymatic biosensors. Biotechnol. Adv. 2012, 30, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Xu, J.; Liu, M.; Li, D.; Lu, L.; Yue, R.; He, H. Vitamin C electrochemical biosensor based on one-step immobilization of ascorbate oxidase in the biocompatible conducting poly(3,4-ethylenedioxythiophene)-lauroylsarcosinate film for agricultural application in crops. J. Electroanal. Chem. 2012, 674, 71–82. [Google Scholar] [CrossRef]
- Yasuoka, S.; Ishida, J.; Kishida, K.; Inui, H. Effects of cerium on the hydrogen absorption-desorption properties of rare earth-Mg-Ni hydrogen-absorbing alloys. J. Power Sources 2017, 346, 56–62. [Google Scholar] [CrossRef]
- Yang, D.F.; Wilde, C.P.; Morin, M. Electrochemical Desorption and Adsorption of Nonyl Mercaptan at Gold Single Crystal Electrode Surfaces. Langmuir 1996, 12, 6570–6577. [Google Scholar] [CrossRef]
- Dixit, C.K.; Vashist, S.K.; Maccraith, B.D.; O’Kennedy, R. Evaluation of apparent non-specific protein loss due to adsorption on sample tube surfaces and/or altered immunogenicity. Analyst 2011, 136, 1406–1411. [Google Scholar] [CrossRef] [PubMed]
- Dixit, C.K.; Ó’Fágáin, C.; O’Kennedy, R. 3 Antibody Immobilization Chemistries for Nanosurfaces: From Lab to Field; Ajeet KumarKaushik, A., Dixit, C.K., Eds.; Apple Academic Press Inc.: Oakville, ON, Canada, 2016; pp. 65–100. [Google Scholar]
- Shen, M.; Rusling, J.; Dixit, C.K. Site-Selective Orientated Immobilization of Antibodies and Conjugates for Immunodiagnostics Development. Methods 2016, 116, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Makaraviciute, A.; Ramanaviciene, A. Site-directed antibody immobilization techniques for immunosensors. Biosens. Bioelectron. 2013, 50, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.K.; Zhao, J.H.; Yan, F.; Zhu, Y.L.; Ju, H.X. Electrochemical behavior and detection of hepatitis B virus DNA PCR production at gold electrode. Biosens. Bioelectron. 2003, 18, 1501–1508. [Google Scholar] [CrossRef]
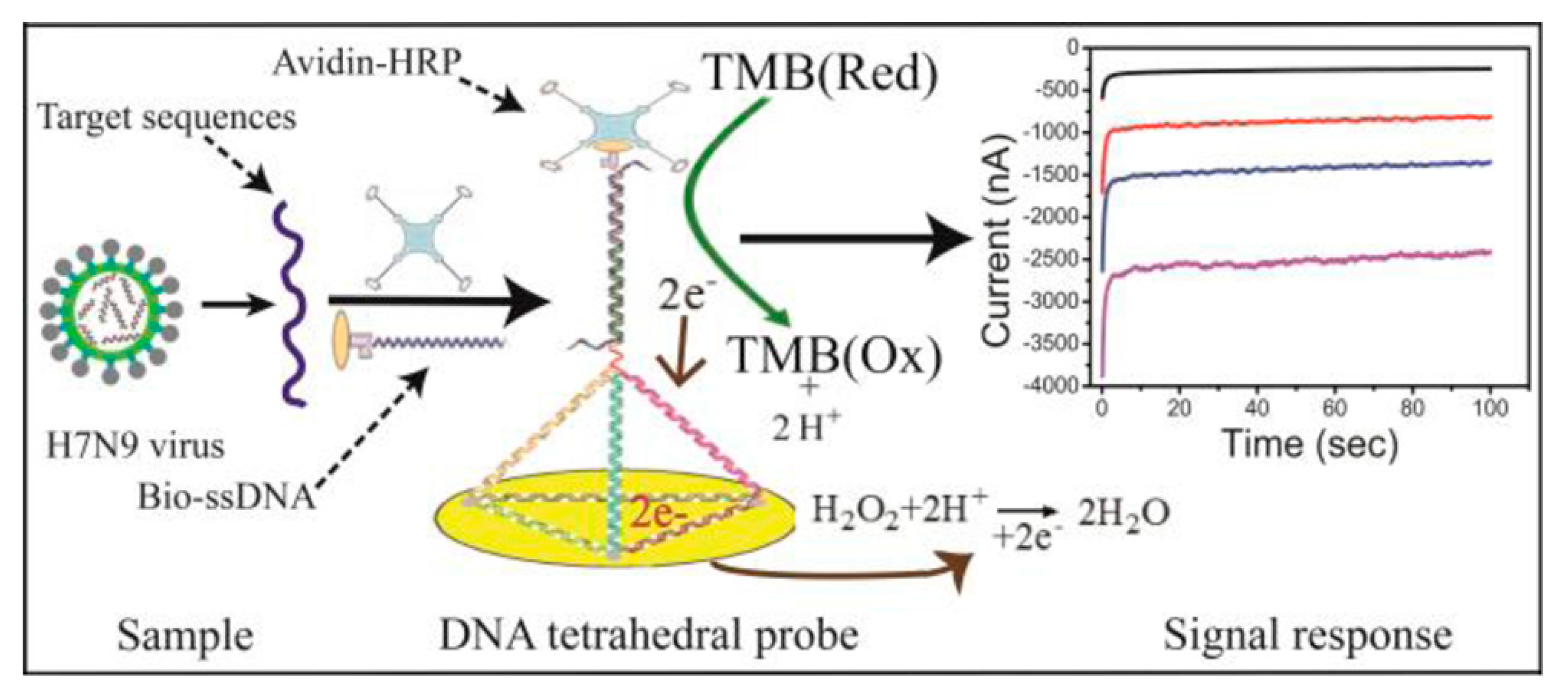
- Dong, S.; Zhao, R.; Zhu, J.; Lu, X.; Li, Y.; Qiu, S.; Jia, L.; Jiao, X.; Song, S.; Fan, C. Electrochemical DNA Biosensor Based on a Tetrahedral Nanostructure Probe for the Detection of Avian Influenza A (H7N9) Virus. ACS Appl. Mater. Interfaces 2015, 7, 8834–8842. [Google Scholar] [CrossRef] [PubMed]
- Kara, P.; Meric, B.; Zeytinoglu, A.; Ozsoz, M. Electrochemical DNA biosensor for the detection and discrimination of herpes simplex Type I and Type II viruses from PCR amplified real samples. Anal. Chim. Acta 2004, 518, 69–76. [Google Scholar] [CrossRef]
- Lin, X.H.; Wu, P.; Chen, W.; Zhang, Y.F.; Xia, X.H. Electrochemical DNA biosensor for the detection of short DNA species of Chronic Myelogenous Leukemia by using methylene blue. Talanta 2007, 72, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, R.; Chaia, Y.; Li, W.; Zhuo, Y.; Yuan, Y.; Li, J. Direct electron transfer: Electrochemical glucose biosensor based on hollow Pt nanosphere functionalized multiwall carbon nanotubes. J. Mol. Catal. B 2011, 71, 146–151. [Google Scholar] [CrossRef]
- Freire, R.S.; Pessoa, C.A.; Mello, L.D.; Kubota, L.T. Direct electron transfer: An approach for electrochemical biosensors with higher selectivity and sensitivity. J. Braz. Chem. Soc. 2003, 14, 72–76. [Google Scholar] [CrossRef]
- Zhang, W.; Li, G. Third-generation biosensors based on the direct electron transfer of proteins. Anal. Sci. Int. J. Jan. Soc. Anal. Chem. 2004, 20, 603–609. [Google Scholar] [CrossRef]
- Hrapovic, S.; Liu, Y.; Male, K.B.; Luong, J.H. Electrochemical biosensing platforms using platinum nanoparticles and carbon nanotubes. Anal. Chem. 2004, 76, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Chen, J.; Yao, S.; Nie, L.; Deng, G.; Kuang, Y. Amperometric glucose biosensor based on adsorption of glucose oxidase at platinum nanoparticle-modified carbon nanotube electrode. Anal. Biochem. 2004, 331, 89–97. [Google Scholar] [CrossRef]
- Lim, S.H.; Wei, J.; Lin, J.; Li, Q.; Kuayou, J. A glucose biosensor based on electrodeposition of palladium nanoparticles and glucose oxidase onto Nafion-solubilized carbon nanotube electrode. Biosens. Bioelectron. 2005, 20, 2341–2346. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.N.; Rusling, J.F.; Papadimitrakopoulos, F. Carbon Nanotubes for Electronic and Electrochemical Detection of Biomolecules. Adv. Mater. 2007, 19, 3214–3228. [Google Scholar] [CrossRef] [PubMed]
- Claussen, J.C.; Franklin, A.D.; Haque, A.U.; Porterfield, D.M.; Fisher, T.S. Electrochemical Biosensor of Nanocube-Augmented Carbon Nanotube Networks. Acs Nano 2009, 3, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Rakhi, R.B.; Sethupathi, K.; Ramaprabhu, S. A glucose biosensor based on deposition of glucose oxidase onto crystalline gold nanoparticle modified carbon nanotube electrode. J. Phys. Chem. B 2009, 113, 3190–3194. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Nanomaterial-based electrochemical biosensors. Analyst 2005, 130, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Bo, G.P.; Kim, S.B.; Yong, J.P. Effects of surface area on electrochemical performance of Li[Ni0.2Li0.2Mn0.6]O2 cathode material. J. Appl. Electrochem. 2009, 39, 1059–1066. [Google Scholar] [CrossRef]
- Tamiya, E. Nano-materials for LSPR and electrochemical biosensors. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 449–575. [Google Scholar] [CrossRef]
- Sanvicens, N.; Pastells, C.; Pascual, N.; Marco, M.P. Nanoparticle-based biosensors for detection of pathogenic bacteria. TrAC Trends Anal. Chem. 2009, 28, 1243–1252. [Google Scholar] [CrossRef]
- Merkoçi, A.; Aldavert, M.; Martín, S.; Alegret, S. New materials for electrochemical sensing V: Nanoparticles for DNA labeling. TrAC Trends Anal. Chem. 2005, 24, 341–349. [Google Scholar] [CrossRef]
- Rusling, J.F.; Bishop, G.W.; Doan, N.; Papadimitrakopoulos, F. Nanomaterials and biomaterials in electrochemical arrays for protein detection. J. Mater. Chem. B 2014, 2, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.T.; Wu, M.F.; Wang, G.J.; Hsieh, S.L. Nanostructured electrochemical biosensor for th0065 detection of the weak binding between the dengue virus and the CLEC5A receptor. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Hu, S. Electrochemical sensors based on metal and semiconductor nanoparticles. Microchim. Acta 2009, 165, 1–22. [Google Scholar] [CrossRef]
- Katz, E.; Willner, I.; Wang, J. Electroanalytical and Bioelectroanalytical Systems Based on Metal and Semiconductor Nanoparticles. Electroanalysis 2010, 16, 19–44. [Google Scholar] [CrossRef]
- Authier, L.; Céline Grossiord, A.; Brossier, P.; Limoges, B. Gold Nanoparticle-Based Quantitative Electrochemical Detection of Amplified Human Cytomegalovirus DNA Using Disposable Microband Electrodes. Anal. Chem. 2001, 73, 4450–4456. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, Y.; Liu, Y.; Shen, G.; Yu, R. Platinum nanoparticles-doped sol–gel/carbon nanotubes composite electrochemical sensors and biosensors. Biosens. Bioelectron. 2006, 21, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Carbon-Nanotube Based Electrochemical Biosensors: A Review. Electroanalysis 2005, 17, 7–14. [Google Scholar] [CrossRef]
- Kara, P.; de la Escosura-Muñiz, A.; Maltez-da Costa, C.M.; Guix, M.; Ozsoz, M.; Merkoçi, A. Aptamers based electrochemical biosensor for protein detection using carbon nanotubes platforms. Biosens. Bioelectron. 2010, 26, 1715–1718. [Google Scholar] [CrossRef] [PubMed]
- Patolsky, F.; Weizmann, Y.; Willner, I. Long-range electrical contacting of redox enzymes by SWCNT connectors. Angew. Chem. 2004, 43, 2113–2117. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Cella, L.N.; Chen, W.; Myung, N.V.; Mulchandani, A. Carbon nanotubes-based chemiresistive immunosensor for small molecules: Detection of nitroaromatic explosives. Biosens. Bioelectron. 2010, 26, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Fei, L.; Choi, K.S.; Park, T.J.; Sang, Y.L.; Seo, T.S. Graphene-based electrochemical biosensor for pathogenic virus detection. BioChip J. 2011, 5, 123–128. [Google Scholar]
- Altintas, Z.; Tothill, I. Biomarkers and biosensors for the early diagnosis of lung cancer. Sens. Actuators B Chem. 2013, 188, 988–998. [Google Scholar] [CrossRef]
- Jin, B.; Wang, P.; Mao, H.; Hu, B.; Zhang, H.; Cheng, Z.; Wu, Z.; Bian, X.; Jia, C.; Jing, F. Multi-nanomaterial electrochemical biosensor based on label-free graphene for detecting cancer biomarkers. Biosens. Bioelectron. 2014, 55, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of Biomarker Development for Early Detection of Cancer. J. Natl. Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Ju, H. Electrochemical and chemiluminescent immunosensors for tumor markers. Biosens. Bioelectron. 2005, 20, 1461–1470. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, N.; Hallaj, R.; Salimi, A. A highly sensitive electrochemical immunosensor for hepatitis B virus surface antigen detection based on Hemin/G-quadruplex horseradish peroxidase-mimicking DNAzyme-signal amplification. Biosens. Bioelectron. 2017, 94, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Healy, D.A.; Hayes, C.J.; Leonard, P.; Mckenna, L.; O’Kennedy, R. Biosensor developments: Application to prostate-specific antigen detection. Trends Biotechnol. 2007, 25, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Dixit, C.K.; Kadimisetty, K.; Otieno, B.A.; Tang, C.; Malla, S.; Krause, C.E.; Rusling, J.F. Electrochemistry-based Approaches to Low Cost, High Sensitivity, Automated, Multiplexed Protein Immunoassays for Cancer Diagnostics. Analyst 2016, 141, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Kadimisetty, K.; Malla, S.; Sardesai, N.; Joshi, A.A.; Faria, R.C.; Lee, N.; Rusling, J.F. Automated Multiplexed ECL Immunoarrays for Cancer Biomarker Proteins. Anal. Chem. 2015, 87, 4472–4478. [Google Scholar] [CrossRef] [PubMed]
- Dulay, S.; Lozano-Sánchez, P.; Iwuoha, E.; Katakis, I.; O’Sullivan, C.K. Electrochemical detection of celiac disease-related anti-tissue transglutaminase antibodies using thiol based surface chemistry. Biosens. Bioelectron. 2011, 26, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Dahlbom, I.; Agardh, D.; Hansson, T. Protein A and protein G ELISA for the detection of IgG autoantibodies against tissue transglutaminase in childhood celiac disease. Clin. Chim. Acta 2008, 395, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Pividori, M.I.; Lermo, A.; Bonanni, A.; Alegret, S.; Del, V.M. Electrochemical immunosensor for the diagnosis of celiac disease. Anal. Biochem. 2009, 388, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Barnaba, V.; Franco, A.; Alberti, A.; Balsano, C.; Benvenuto, R.; Balsano, F. Recognition of hepatitis B virus envelope proteins by liver-infiltrating T lymphocytes in chronic HBV infection. J. Immunol. 1989, 143, 2650–2655. [Google Scholar] [PubMed]
- Vento, S.; Hegarty, J.E.; Alberti, A.; O'Brien, C.J.; Alexander, G.J.; Eddleston, A.L.; Williams, R. T lymphocyte sensitization to HBcAg and T cell-mediated unresponsiveness to HBsAg in hepatitis B virus-related chronic liver disease. Hepatology 2010, 5, 192–197. [Google Scholar] [CrossRef]
- Luo, Y.; Lo, C.M.; Cheung, C.K.; Lau, G.K.; Fan, S.T.; Wong, J. Identification of hepatitis B virus-specific lymphocytes in human liver grafts from HBV-immune donors. Liver Transplant. 2007, 13, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Dusheiko, G.M.; Hoofnagle, J.H.; Cooksley, W.G.; James, S.P.; Jones, E.A. Synthesis of antibodies to hepatitis B virus by cultured lymphocytes from chronic hepatitis B surface antigen carriers. J. Clin. Investig. 1983, 71, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Reverté, L.; Prieto-Simón, B.; Campàs, M. New advances in electrochemical biosensors for the detection of toxins: Nanomaterials, magnetic beads and microfluidics systems. A review. Anal. Chim. Acta 2016, 908, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Y. Nanomaterial labels in electrochemical immunosensors and immunoassays. Sci. Total Environ. 2007, 74, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Dong, S. Biomolecule-nanoparticle hybrids for electrochemical biosensors. Cheminform 2009, 40, 96–109. [Google Scholar] [CrossRef]
- Li, J.; Gao, H.; Chen, Z.; Wei, X.; Yang, C.F. An electrochemical immunosensor for carcinoembryonic antigen enhanced by self-assembled nanogold coatings on magnetic particles. Anal. Chim. Acta 2010, 665, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tian, J.; Zhao, Y.; Zhao, S. Ag/Au nanoparticles coated graphene electrochemical sensor for ultrasensitive analysis of carcinoembryonic antigen in clinical immunoassay. Sens. Actuators B Chem. 2015, 206, 570–576. [Google Scholar] [CrossRef]
- Tang, D.; Ren, J. In situ amplified electrochemical immunoassay for carcinoembryonic antigen using horseradish peroxidase-encapsulated nanogold hollow microspheres as labels. Anal. Chem. 2008, 80, 8064. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Tang, J.; Dai, Z.; Yan, F.; Ju, H.; Murr, N.E. A disposable electrochemical immunosensor for flow injection immunoassay of carcinoembryonic antigen. Biosens. Bioelectron. 2006, 22, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Jia, X.; Chen, X.; Ma, Z. Simultaneous electrochemical detection of multiple tumor markers using metal ions tagged immunocolloidal gold. Biosens. Bioelectron. 2014, 56, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Chen, J.; Nie, L.; Kuang, Y.; Yao, S. A label-free electrochemical immunoassay for carcinoembryonic antigen (CEA) based on gold nanoparticles (AuNPs) and nonconductive polymer film. Biosens. Bioelectron. 2007, 22, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, J.; Wang, T.; Li, D.; Xi, F.; Wang, J.; Wang, E. Three-dimensional electrochemical immunosensor for sensitive detection of carcinoembryonic antigen based on monolithic and macroporous graphene foam. Biosens. Bioelectron. 2015, 65, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, C.; Qi, H.; Gao, Q.; Zhang, C. Electrochemical lectin-based biosensor array for detection and discrimination of carcinoembryonic antigen using dual amplification of gold nanoparticles and horseradish peroxidase. Sens. Actuators B Chem. 2016, 235, 575–582. [Google Scholar] [CrossRef]
- Aydinlik, S.; Ozkan-Ariksoysal, D.; Kara, P.; Sayiner, A.A.; Ozsoz, M. Electrochemical DNA biosensor using a disposable electrochemical printed (DEP) chip for the detection of SNPs from unpurified PCR am for the detection of influenza B virus from PCR samples using gold nanoparticle-adsorbed disposable graphite electrode and Meldola's blue as an intercalator. Anal. Methods 2011, 3, 1607–1614. [Google Scholar]
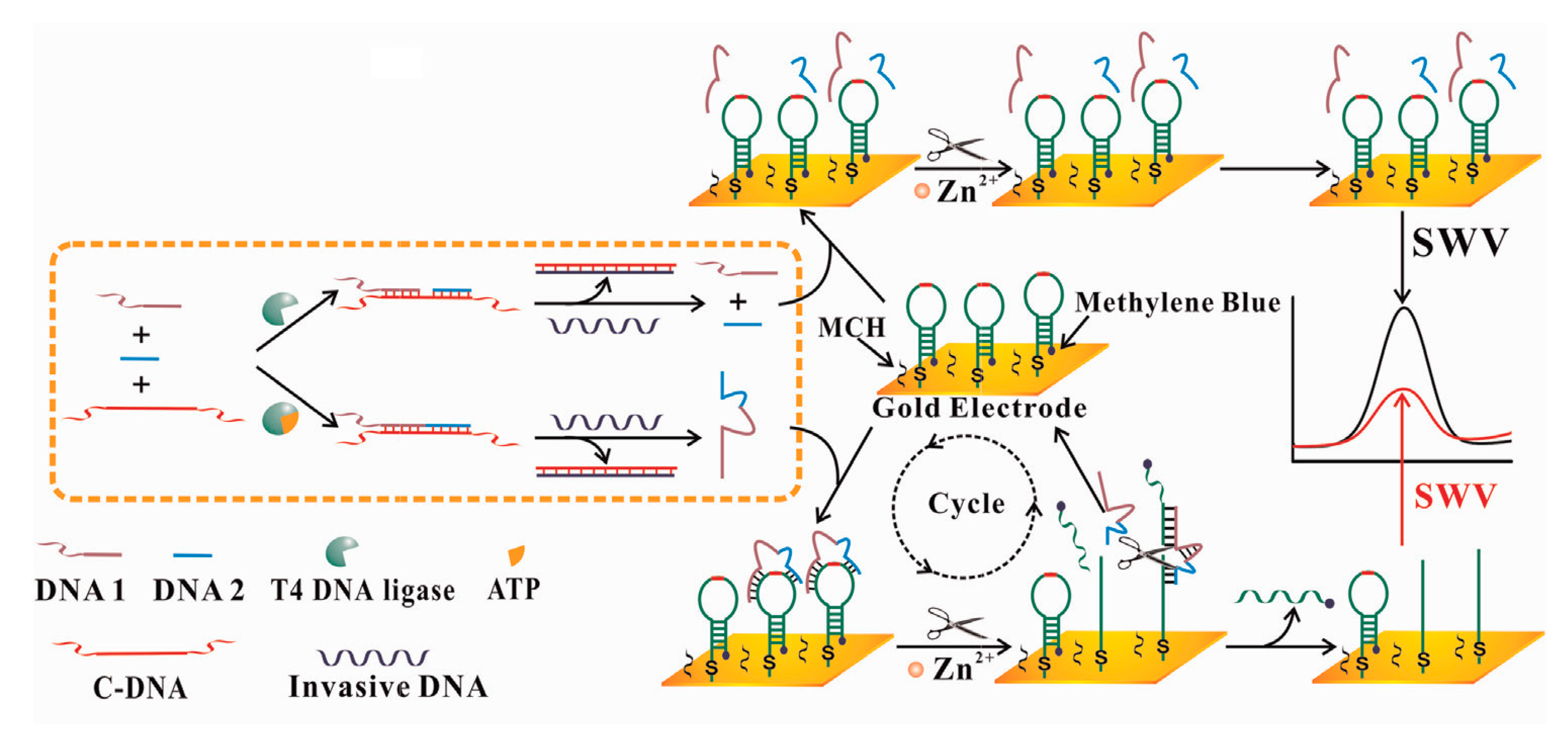
- Lu, L.; Si, J.C.; Gao, Z.F.; Zhang, Y.; Lei, J.L.; Luo, H.Q.; Li, N.B. Highly selective and sensitive electrochemical biosensor for ATP based on the dual strategy integrating the cofactor-dependent enzymatic ligation reaction with self-cleaving DNAzyme-amplified electrochemical detection. Biosens. Bioelectron. 2015, 63, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Çevik, E.; Bahar, Ö.; Şenel, M.; Abasıyanık, M.F. Construction of novel electrochemical immunosensor for detection of prostate specific antigen using ferrocene-PAMAM dendrimers. Biosens. Bioelectron. 2016, 86, 1074–1079. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-Y.; Wang, J.; Liu, G.; Wu, H.; Wai, C.M.; Lin, Y. A nanoparticle label/immunochromatographic electrochemical biosensor for rapid and sensitive detection of prostate-specific antigen. Biosens. Bioelectron. 2008, 23, 1659–1665. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.K.; Narayanan, J.; Pardasani, D.; Srivastava, D.N.; Upadhyay, S.; Goel, A.K. Ultrasensitive electrochemical immunoassay for surface array protein, a Bacillus anthracis biomarker using Au–Pd nanocrystals loaded on boron-nitride nanosheets as catalytic labels. Biosens. Bioelectron. 2016, 80, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Rahi, A.; Sattarahmady, N.; Heli, H. Label-free electrochemical aptasensing of the human prostate-specific antigen using gold nanospears. Talanta 2016, 156, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Teles, F.S.R.R. Biosensors and rapid diagnostic tests on the frontier between analytical and clinical chemistry for biomolecular diagnosis of dengue disease: A review. Anal. Chim. Acta 2011, 687, 28–42. [Google Scholar] [CrossRef] [PubMed]
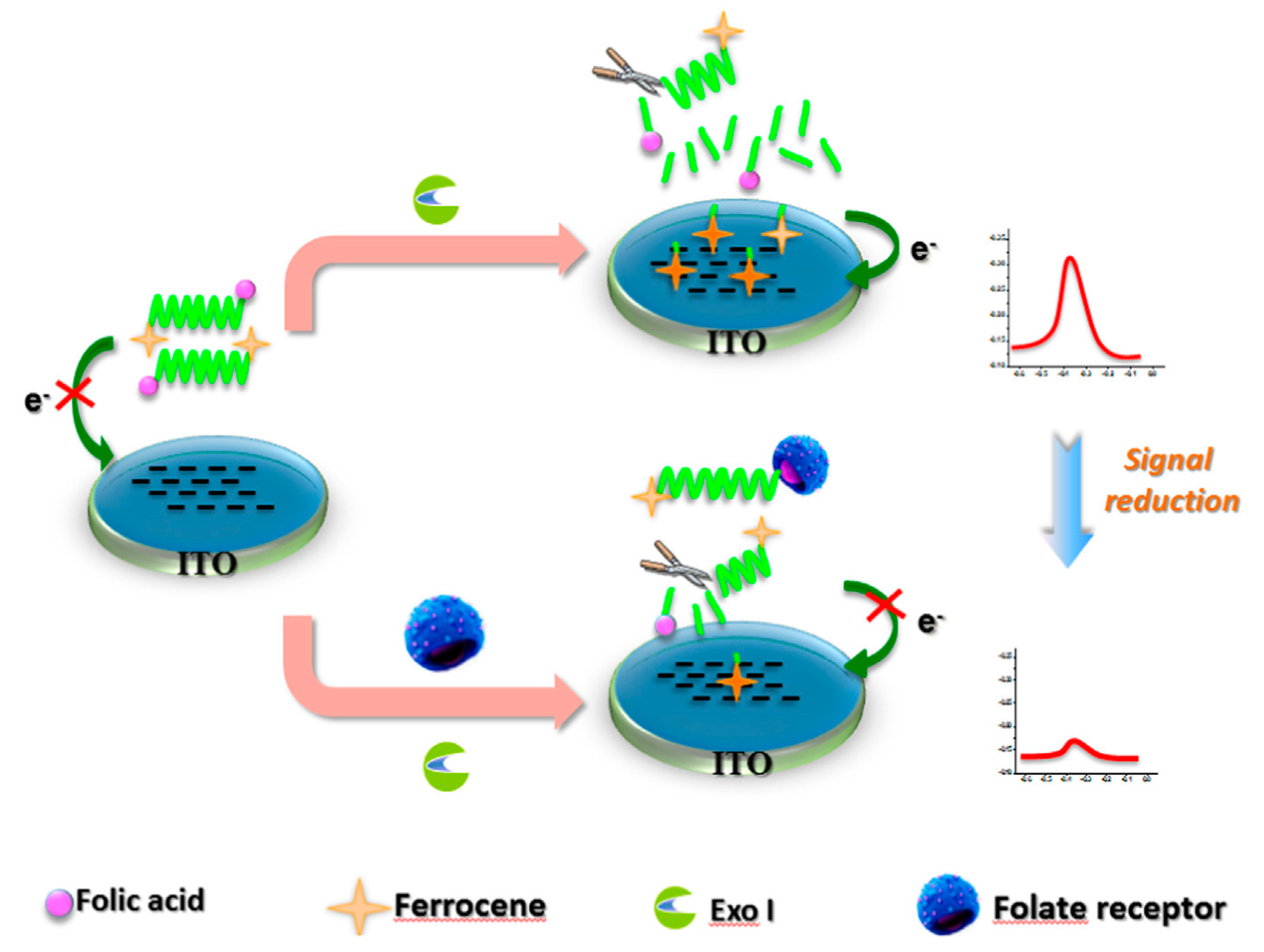
- Ni, J.; Wang, Q.; Yang, W.; Zhao, M.; Zhang, Y.; Guo, L.; Qiu, B.; Lin, Z.; Yang, H.-H. Immobilization free electrochemical biosensor for folate receptor in cancer cells based on terminal protection. Biosens. Bioelectron. 2016, 86, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.Y.; Wang, Z.; Li, P.; Lian, H.Z.; Chen, H.Y. Aptamer-based thrombin assay on microfluidic platform. Electrophoresis 2013, 34, 3260–3266. [Google Scholar] [CrossRef] [PubMed]
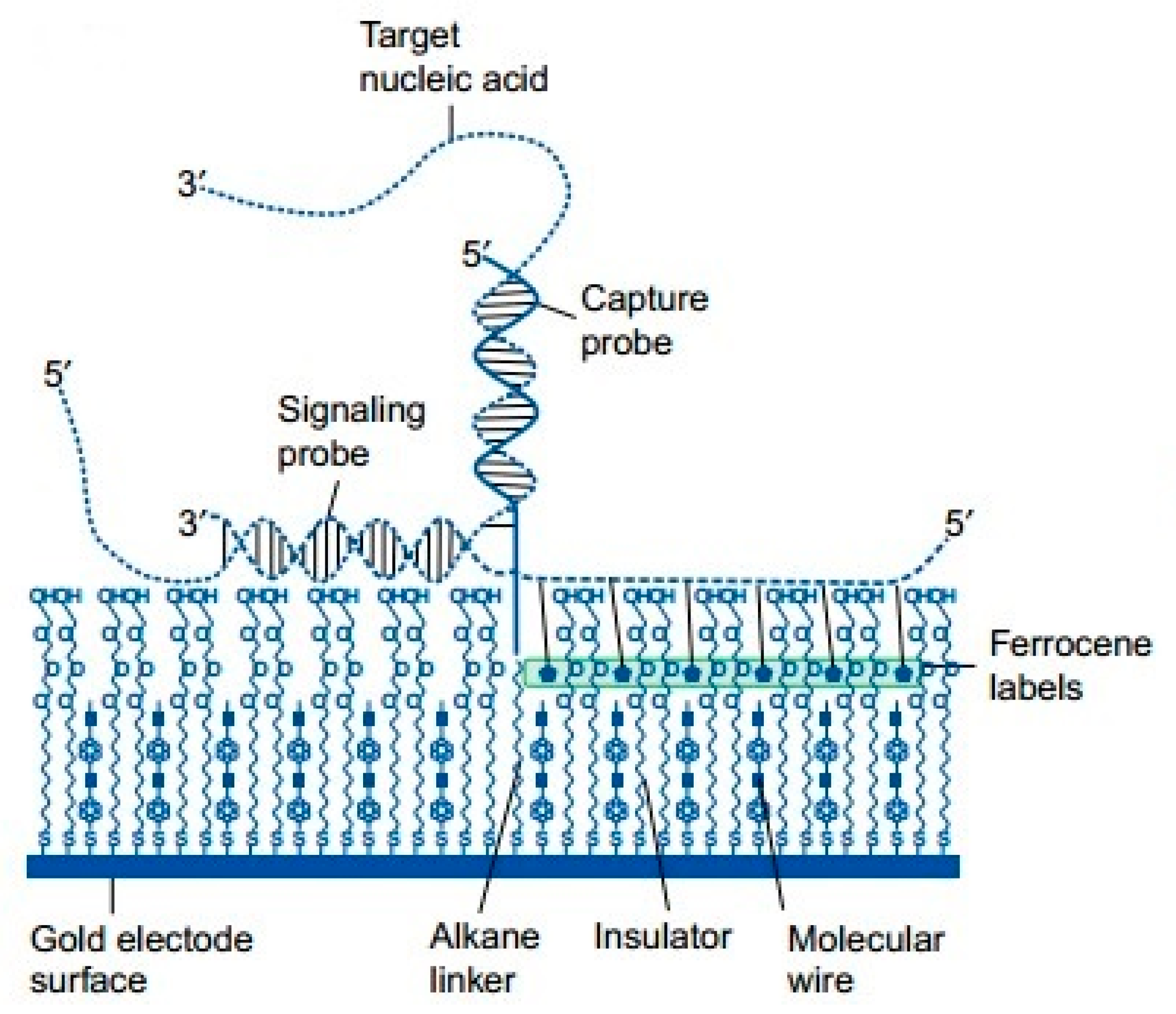
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA Sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Kerman, K.; Morita, Y.; Takamura, Y.; Tamiya, E. Label-free electrochemical detection of DNA hybridization on gold electrode. Electrochem. Commun. 2003, 5, 887–891. [Google Scholar] [CrossRef]
- Berney, H.; West, J.; Haefele, E.; Alderman, J.; Lane, W.; Collins, J. A DNA diagnostic biosensor: Development, characterisation and performance. Sens. Actuators B Chem. 2000, 68, 100–108. [Google Scholar] [CrossRef]
- Vestergaard, M.D.; Kerman, K.; Saito, M.; Nagatani, N.; Takamura, Y.; Tamiya, E. A rapid label-free electrochemical detection and kinetic study of Alzheimer’s amyloid beta aggregation. J. Am. Chem. Soc. 2005, 127, 11892–11893. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, B.; Wei, H.; Wang, Y.; Wang, E. Multifunctional label-free electrochemical biosensor based on an integrated aptamer. Anal. Chem. 2008, 80, 5110–5117. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Wei, X.; Zhao, M.; Qiu, B.; Guo, L.; Lin, Z.; Yang, H.-H. Ultraselective homogeneous electrochemical biosensor for DNA species related to oral cancer based on nicking endonuclease assisted target recycling amplification. Anal. Chem. 2015, 87, 9204–9208. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Electrochemical nucleic acid biosensors. Anal. Chim. Acta 2002, 469, 63–71. [Google Scholar] [CrossRef]
- Marrazza, G.; Chiti, G.; Mascini, M.; Anichini, M. Detection of human apolipoprotein E genotypes by DNA electrochemical biosensor coupled with PCR. Clin. Chem. 2000, 46, 31–37. [Google Scholar] [PubMed]
- Ahmed, M.U.; Idegami, K.; Chikae, M.; Kerman, K.; Chaumpluk, P.; Yamamura, S.; Tamiya, E. Electrochemical DNA biosensor using a disposable electrochemical printed (DEP) chip for the detection of SNPs from unpurified PCR amplicons. Analyst 2007, 132, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Umek, R.M.; Lin, S.W.; Vielmetter, J.; Terbrueggen, R.H.; Irvine, B.; Yu, C.; Kayyem, J.F.; Yowanto, H.; Blackburn, G.F.; Farkas, D.H. Electronic detection of nucleic acids: A versatile platform for molecular diagnostics. J. Mol. Diagn. 2001, 3, 74–84. [Google Scholar] [CrossRef]
- Pournaghi-Azar, M.; Ahour, F.; Hejazi, M. Direct detection and discrimination of double-stranded oligonucleotide corresponding to hepatitis C virus genotype 3a using an electrochemical DNA biosensor based on peptide nucleic acid and double-stranded DNA hybridization. Anal. Bioanal. Chem. 2010, 397, 3581–3587. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.-J.; Kim, K.-C.; Choi, S.-H. Electrochemical DNA biosensor based on avidin–biotin conjugation for influenza virus (type A) detection. Appl. Surf. Sci. 2011, 257, 9390–9396. [Google Scholar] [CrossRef]
- Liu, Y.; Tuleouva, N.; Ramanculov, E.; Revzin, A. Aptamer-based electrochemical biosensor for interferon gamma detection. Anal. Chem. 2010, 82, 8131–8136. [Google Scholar] [CrossRef] [PubMed]
- Mandong, G.; Yanqing, L.; Hongxia, G.; Xiaoqin, W.; Lifang, F. Electrochemical detection of short sequences related to the hepatitis B virus using MB on chitosan-modified CPE. Bioelectrochemistry 2007, 70, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Jiang, Y.; Jia, F.; Yu, Y.; Chen, J.; Wang, Z. An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol. Methods 2014, 98, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Guo, Y.; Wu, X.; Yang, W.; Xu, L.; Chen, J.; Fu, F. A novel electrically magnetic-controllable electrochemical biosensor for the ultra sensitive and specific detection of attomolar level oral cancer-related microRNA. Biosens. Bioelectron. 2013, 45, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Kwakye, S.; Goral, V.N.; Baeumner, A.J. Electrochemical microfluidic biosensor for nucleic acid detection with integrated minipotentiostat. Biosens. Bioelectron. 2006, 21, 2217–2223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, Y.; Chen, W.; Cheng, W.; Li, S.; Ding, X.; Li, D.; Wang, H.; Ju, H.; Ding, S. A simple electrochemical biosensor for highly sensitive and specific detection of microRNA based on mismatched catalytic hairpin assembly. Biosens. Bioelectron. 2015, 68, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Deng, H.; Shen, W.; Gao, Z. A highly sensitive and selective electrochemical biosensor for direct detection of microRNAs in serum. Anal. Chem. 2013, 85, 4784–4789. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Li, W.; Liu, X.; Li, F. Label-Free and Enzyme-Free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: A facile, sensitive, and highly specific microRNA assay. Anal. Chem. 2015, 87, 11368–11374. [Google Scholar] [CrossRef] [PubMed]
- Mandli, J.; Mohammadi, H.; Amine, A. Electrochemical DNA sandwich biosensor based on enzyme amplified microRNA-21 detection and gold nanoparticles. Bioelectrochemistry 2017, 116, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003, 125, 6642–6643. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Hoshino, S.; Hazemoto, N.; Haga, M.; Kato, Y.; Suzuki, Y. A Liposome Immunoassay Based on a Chemiluminescense Reaction. Chem. Pharm. Bull. 2008, 37, 1629–1631. [Google Scholar] [CrossRef]
- Cho, I.H.; Paek, E.H.; Kim, Y.K.; Kim, J.H.; Paek, S.H. Chemiluminometric enzyme-linked immunosorbent assays (ELISA)-on-a-chip biosensor based on cross-flow chromatography. Anal. Chim. Acta 2009, 632, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Grieshaber, D.; MacKenzie, R.; Voeroes, J.; Reimhult, E. Electrochemical biosensors-sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.S.; Hsu, W.L.; Chend, H.Y. Determination of glutamate pyruvate transaminase activity in clinical specimens using a biosensor composed of immobilized L-glutamate oxidase in a photo-crosslinkable polymer membrane on a palladium-deposited screen-printed carbon electrode. Anal. Chim. Acta 2003, 481, 199–208. [Google Scholar] [CrossRef]
- He, Y.-N.; Chen, H.-Y. The kinetics-based electrochemical determination of serum glutamate pyruvate transaminase activity with a gold microelectrode. Anal. Chim. Acta 1997, 353, 319–323. [Google Scholar] [CrossRef]
- Jamal, M.; Worsfold, O.; McCormac, T.; Dempsey, E. A stable and selective electrochemical biosensor for the liver enzyme alanine aminotransferase (ALT). Biosens. Bioelectron. 2009, 24, 2926–2930. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Li, W.; Zhou, Y.; Liu, C.-C. A single-use, disposable iridium-modified electrochemical biosensor for fructosyl valine for the glycoslated hemoglobin detection. Sens. Actuators B Chem. 2009, 137, 235–238. [Google Scholar] [CrossRef]
- Jeppsson, J.-O.; Kobold, U.; Barr, J.; Finke, A.; Hoelzel, W.; Hoshino, T.; Miedema, K.; Mosca, A.; Mauri, P.; Paroni, R. Approved IFCC reference method for the measurement of HbA1c in human blood. Clin. Chem. Lab. Med. 2002, 40, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Wang, F.; Chen, Z. Aptamer-based electrochemical biosensor for label-free voltammetric detection of thrombin and adenosine. Sens. Actuators B Chem. 2011, 160, 1380–1385. [Google Scholar] [CrossRef]
- Liu, G.; Guo, W.; Song, D. A multianalyte electrochemical immunosensor based on patterned carbon nanotubes modified substrates for detection of pesticides. Biosens. Bioelectron. 2014, 52, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wen, W.; Wang, Q.; Xiong, H.; Zhang, X.; Gu, H.; Wang, S. Novel electrochemical aptamer biosensor based on an enzyme-gold nanoparticle dual label for the ultrasensitive detection of epithelial tumour marker MUC1. Biosens. Bioelectron. 2014, 53, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.A.; Wang, J.; Kawde, A.N.; Xiang, Y.; Gothelf, K.V.; Collins, G. Quantum-dot/aptamer-based ultrasensitive multi-analyte electrochemical biosensor. J. Am. Chem. Soc. 2006, 128, 2228–2229. [Google Scholar] [CrossRef] [PubMed]
- Mohan, R.; Mach, K.E.; Bercovici, M.; Pan, Y.; Dhulipala, L.; Wong, P.K.; Liao, J.C. Clinical Validation of Integrated Nucleic Acid and Protein Detection on an Electrochemical Biosensor Array for Urinary Tract Infection Diagnosis. J. Urol. 2013, 189, 152–153. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Sonn, G.A.; Sin, M.L.Y.; Mach, K.E.; Shih, M.C.; Gau, V.; Wong, P.K.; Liao, J.C. Electrochemical immunosensor detection of urinary lactoferrin in clinical samples for urinary tract infection diagnosis. Biosens. Bioelectron. 2010, 26, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.T.C.; Dutra, R.A.F.; Noronha, J.P.C.; Sales, M.G.F. Electrochemical biosensor based on biomimetic material for myoglobin detection. Electrochim. Acta 2013, 107, 481–487. [Google Scholar] [CrossRef]
- Hoeben, A.; Landuyt, B.; Highley, M.S.; Wildiers, H.; van Oosterom, A.T.; De Bruijn, E.A. Vascular endothelial growth factor and angiogenesis. Pharmacol. Rev. 2004, 56, 549–580. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yang, W.; Lai, R.Y. A folding-based electrochemical aptasensor for detection of vascular endothelial growth factor in human whole blood. Biosens. Bioelectron. 2011, 26, 2442–2447. [Google Scholar] [CrossRef] [PubMed]
- Devadoss, A.; Burgess, J.D. Steady-state detection of cholesterol contained in the plasma membrane of a single cell using lipid bilayer-modified microelectrodes incorporating cholesterol oxidase. J. Am. Chem. Soc. 2004, 126, 10214–10215. [Google Scholar] [CrossRef] [PubMed]
- Aravamudhan, S.; Ramgir, N.S.; Bhansali, S. Electrochemical biosensor for targeted detection in blood using aligned Au nanowires. Sens. Actuators B Chem. 2007, 127, 29–35. [Google Scholar] [CrossRef]
- Yadav, S.K.; Agrawal, B.; Chandra, P.; Goyal, R.N. In vitro chloramphenicol detection in a Haemophilus influenza model using an aptamer-polymer based electrochemical biosensor. Biosens. Bioelectron. 2014, 55, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Chen, J.; Nie, L.; Nie, Z.; Yao, S. Sensitive bifunctional aptamer-based electrochemical biosensor for small molecules and protein. Anal. Chem. 2009, 81, 9972–9978. [Google Scholar] [CrossRef] [PubMed]
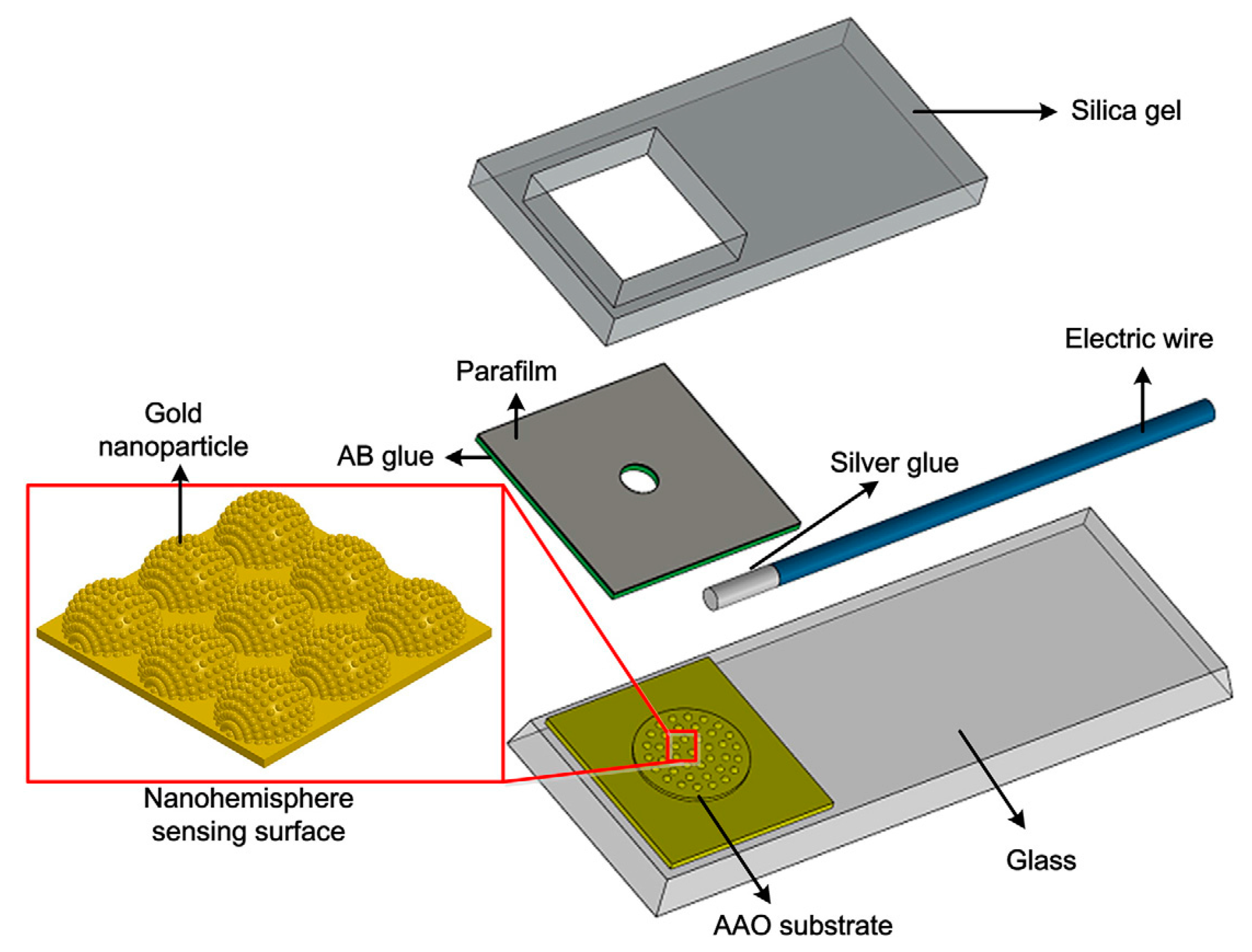
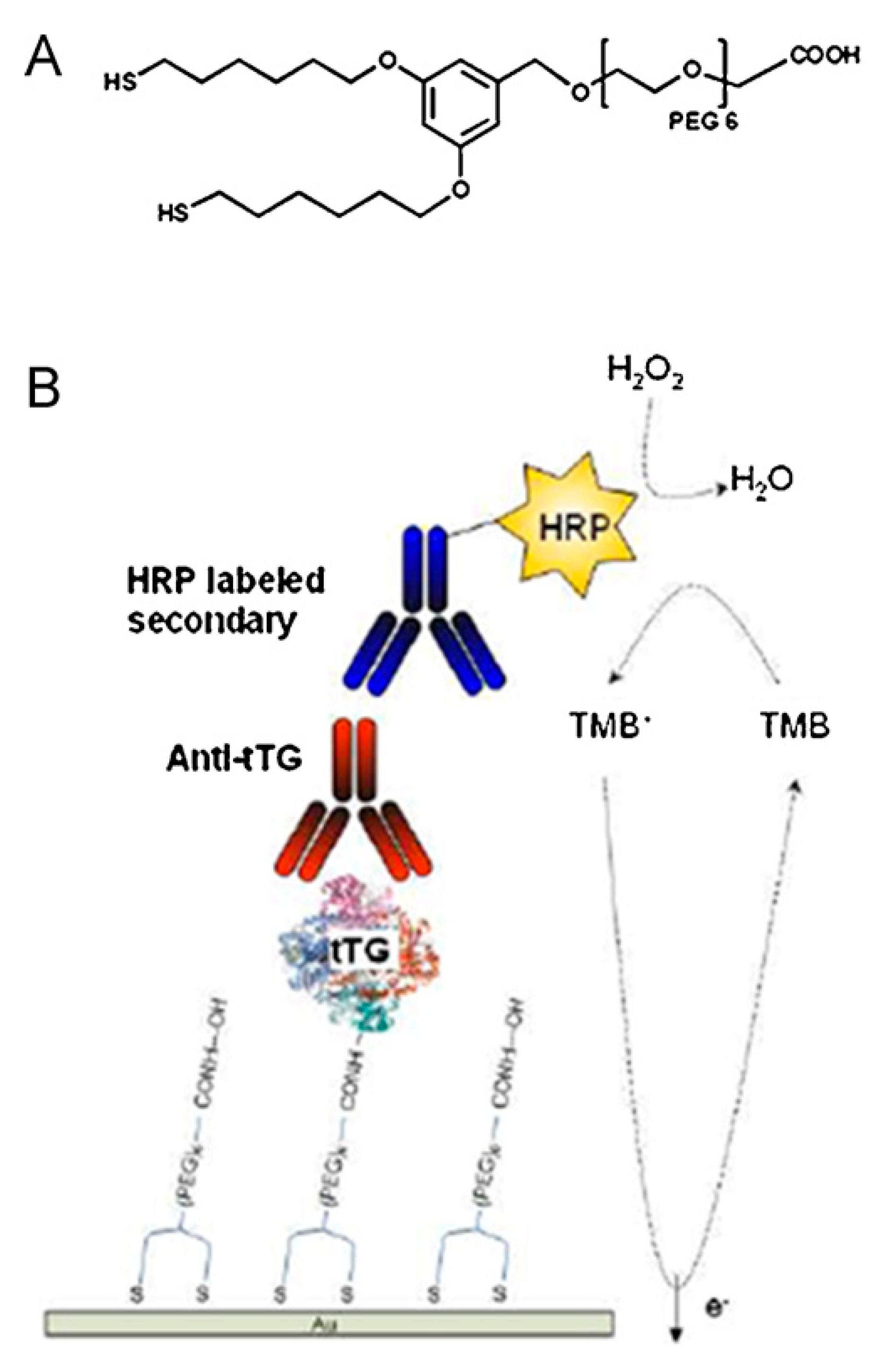
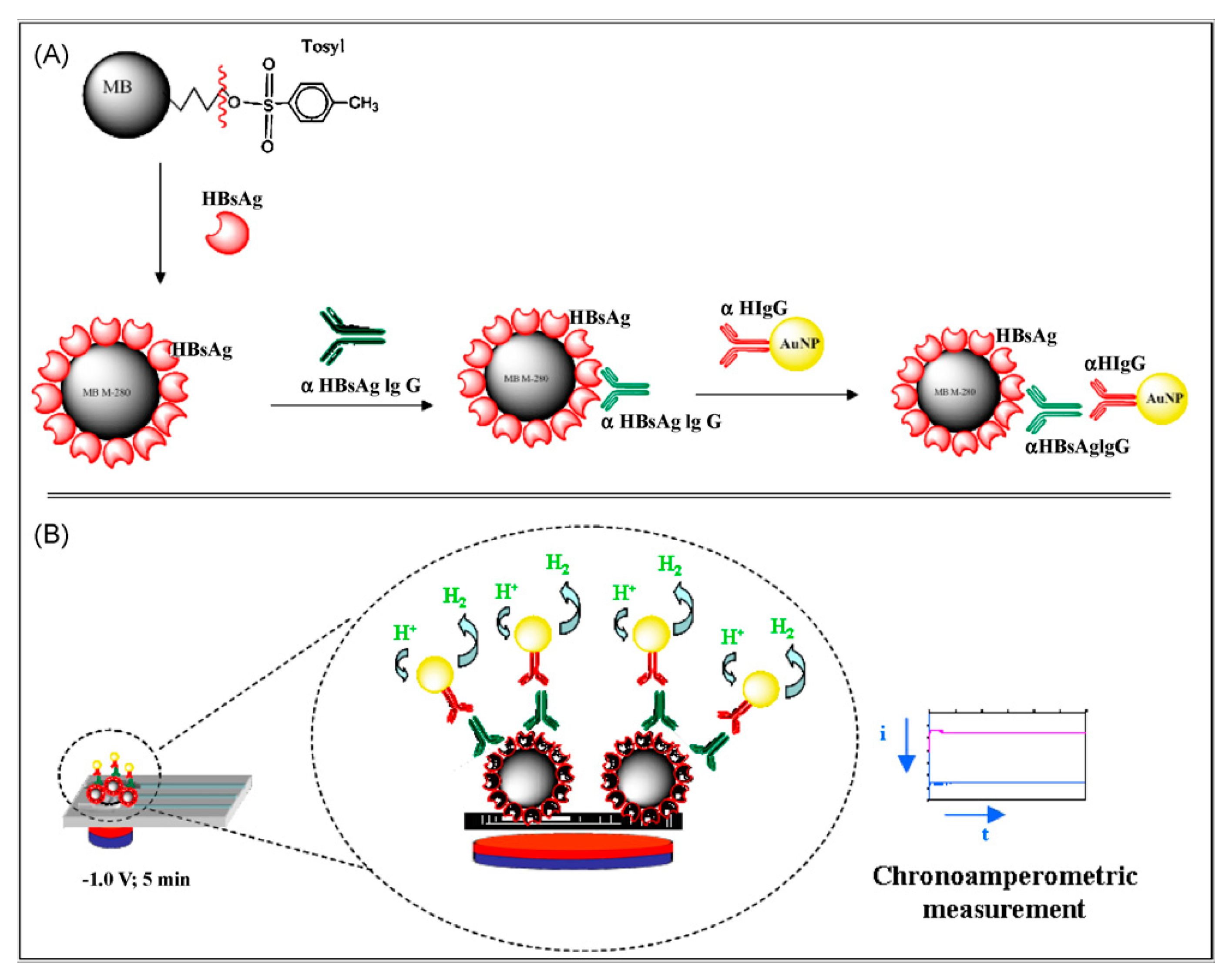
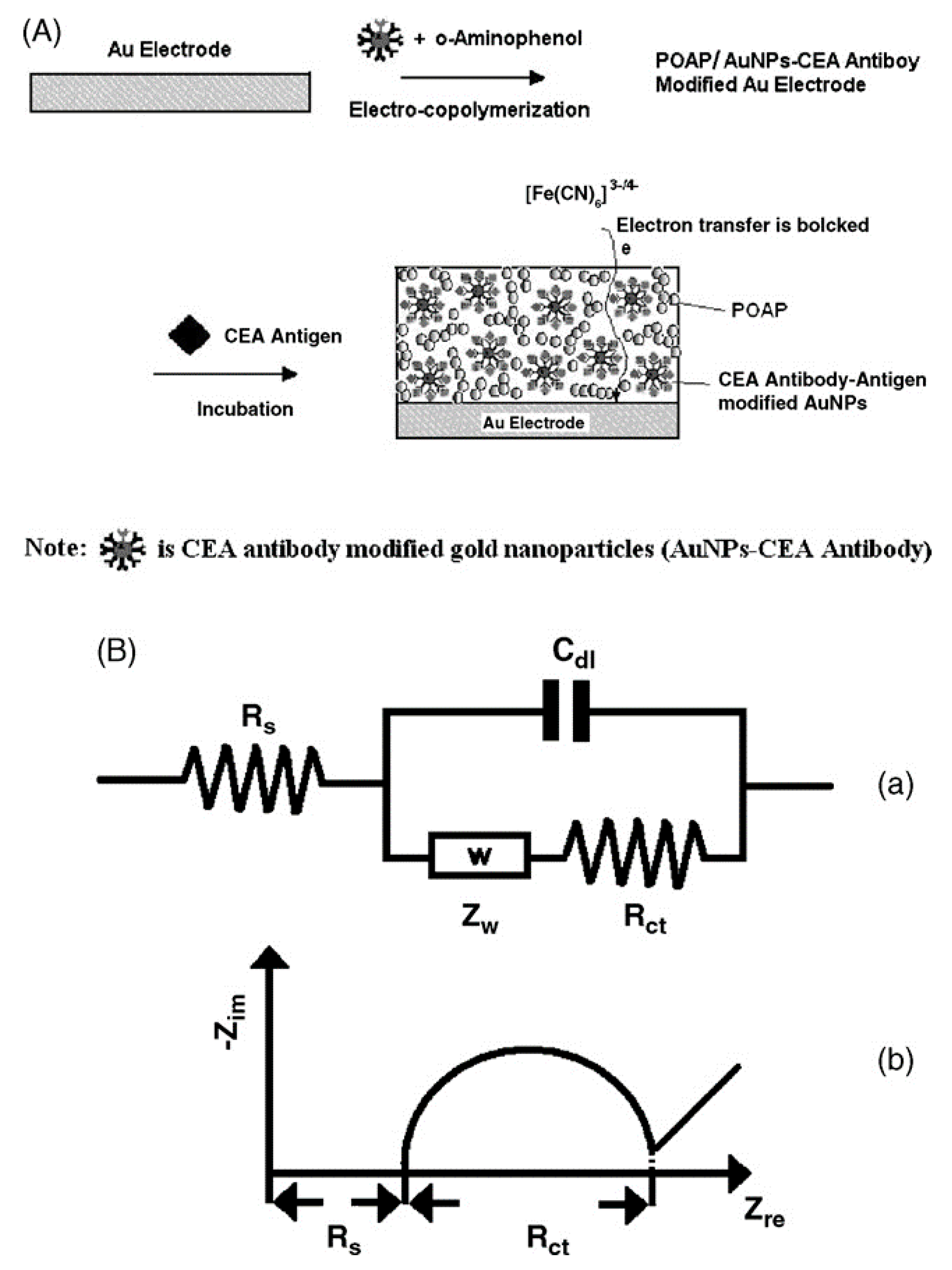


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Xu, J.; Liu, J.; Wang, X.; Chen, B. Disease-Related Detection with Electrochemical Biosensors: A Review. Sensors 2017, 17, 2375. https://doi.org/10.3390/s17102375
Huang Y, Xu J, Liu J, Wang X, Chen B. Disease-Related Detection with Electrochemical Biosensors: A Review. Sensors. 2017; 17(10):2375. https://doi.org/10.3390/s17102375
Chicago/Turabian StyleHuang, Ying, Jin Xu, Junjie Liu, Xiangyang Wang, and Bin Chen. 2017. "Disease-Related Detection with Electrochemical Biosensors: A Review" Sensors 17, no. 10: 2375. https://doi.org/10.3390/s17102375
APA StyleHuang, Y., Xu, J., Liu, J., Wang, X., & Chen, B. (2017). Disease-Related Detection with Electrochemical Biosensors: A Review. Sensors, 17(10), 2375. https://doi.org/10.3390/s17102375



