Spectral Similarity Assessment Based on a Spectrum Reflectance-Absorption Index and Simplified Curve Patterns for Hyperspectral Remote Sensing
Abstract
:1. Introduction
2. Related Similarities of Spectral Vectors
2.1. SID
2.2. SAM
3. Proposed Approach
3.1. SRAI
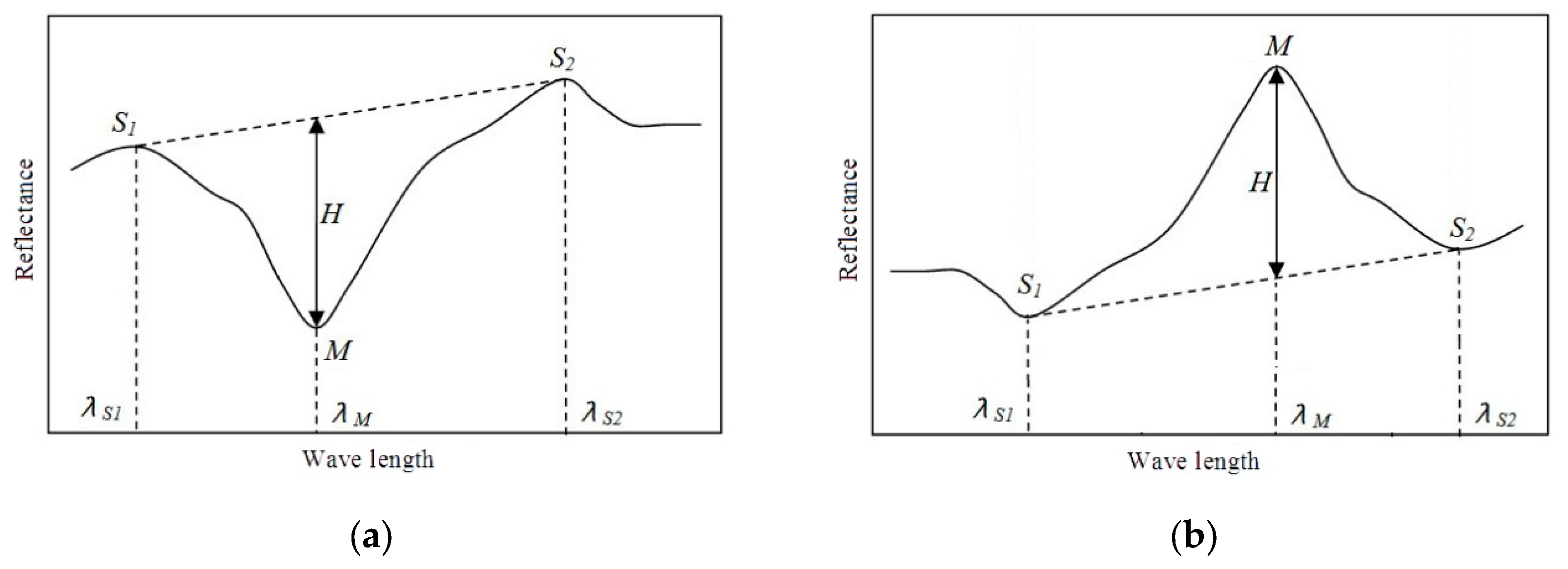
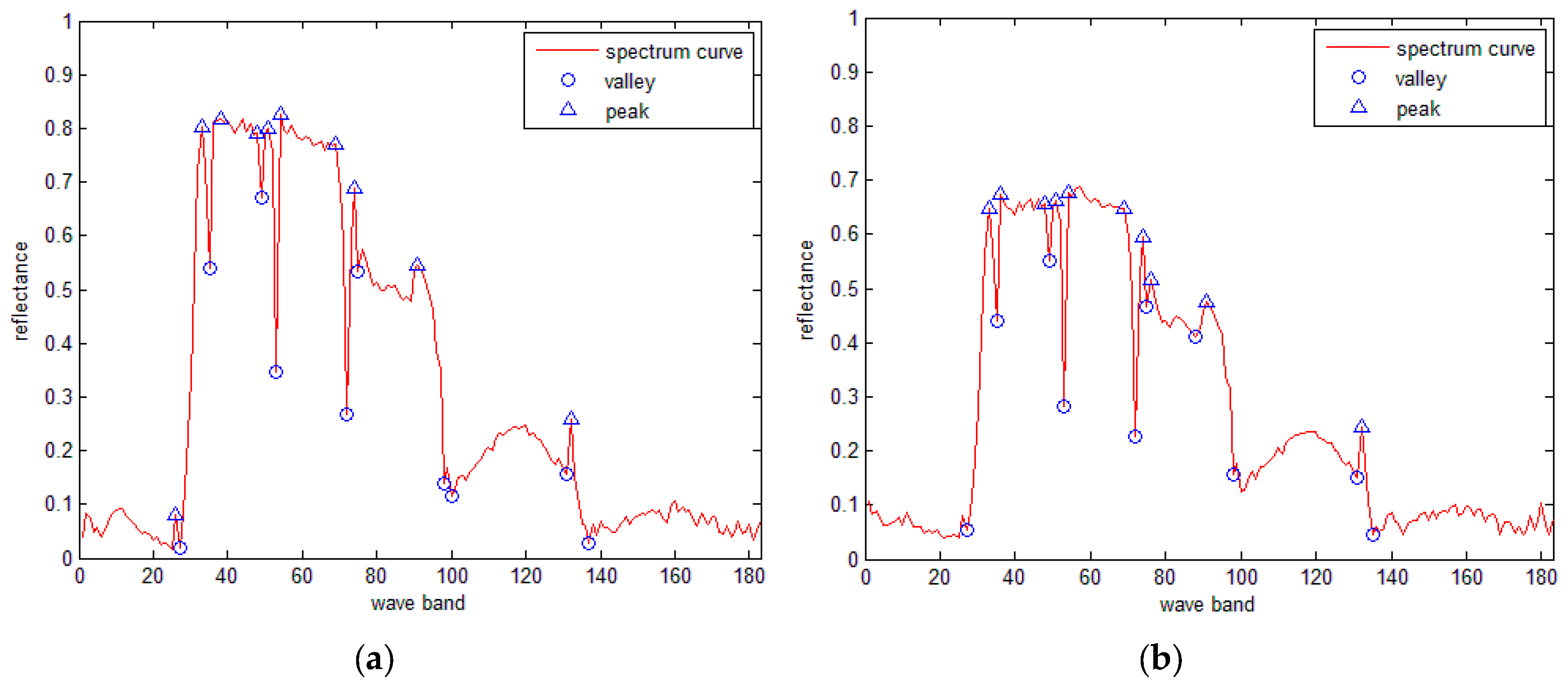
3.2. Classical DP Algorithm
- (1)
- Connect both ends of the curve with a straight line and calculate the distances (d) from all the points on curve to the line.
- (2)
- Compare the maximum distance (dmax) and threshold D; if dmax < D, then eliminate all points on the curve; otherwise, retain the point with the maximum distance (dmax) and split the curve into two parts;
- (3)
- Repeat steps 1 and 2 until dmax < D is true for all points on the curve, and all the retained points comprise the final simplified spectral curve.
- (1)
- The threshold must be specified as it can significantly affect the performance of curve simplification. On the one hand, if the threshold becomes higher, fewer points are retained, which may destroy the pattern. On the other hand, if the threshold becomes lower, more points are retained; therefore, less simplification is achieved.
- (2)
- Knowing how many points are left using a threshold before computing is impossible; in other words, setting a proper threshold to maintain the feature of a spectral curve using the necessary points is a difficult and complex task.
- (3)
- For the effective simplification of multiple curves, particularly for hyperspectral images, different thresholds are needed for various curves; moreover, to retain enough points, every curve should set different thresholds.
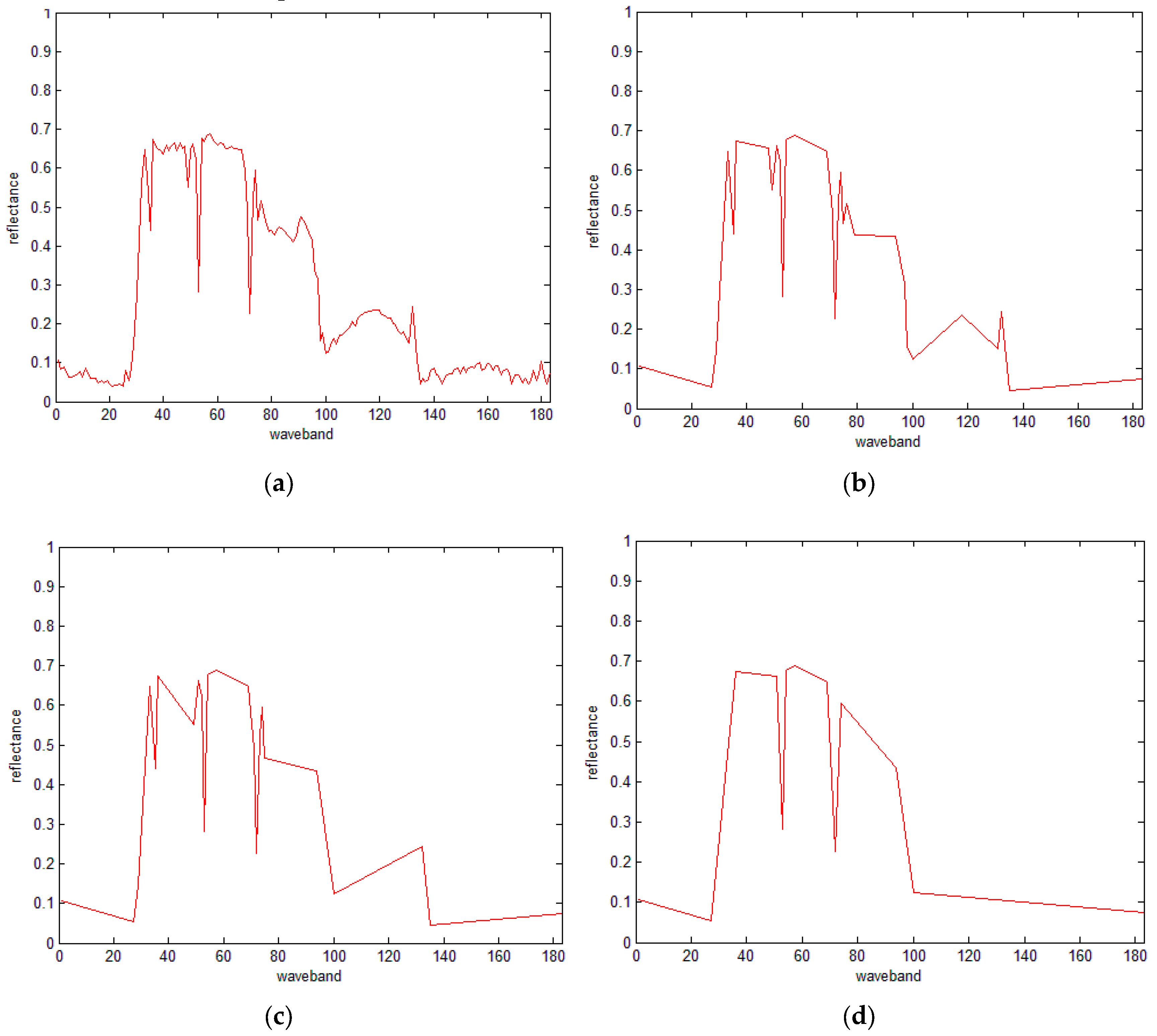
3.3. IDP Algorithm
- (1)
- Connect the starting point S and ending point E, calculate the distance from each point to segment SE, and retain the point M, which has the maximum distance; this is the same as step one in the traditional DP algorithm.
- (2)
- Connect SM and ME, calculate the distance from each point to segment SM and ME, and retain the point N, which has the maximum distance.
- (3)
- Divide the curve into three parts using points M and N. Repeat step (2) until it meets the point number retaining requirement.
- (4)
- If the distances are equal in step (2), calculate the ratio of the distance to the line sector and the length of sector, the point with the lower ratio is then retained.
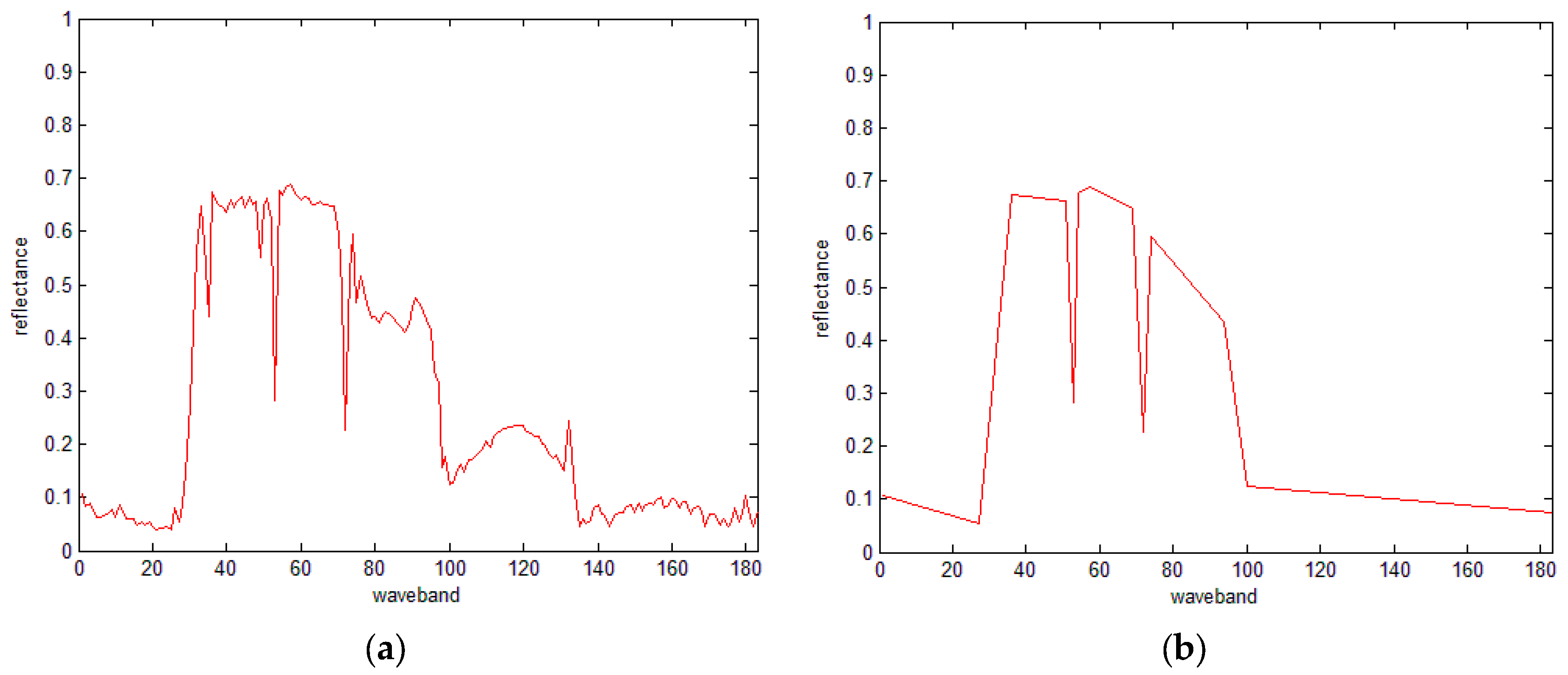
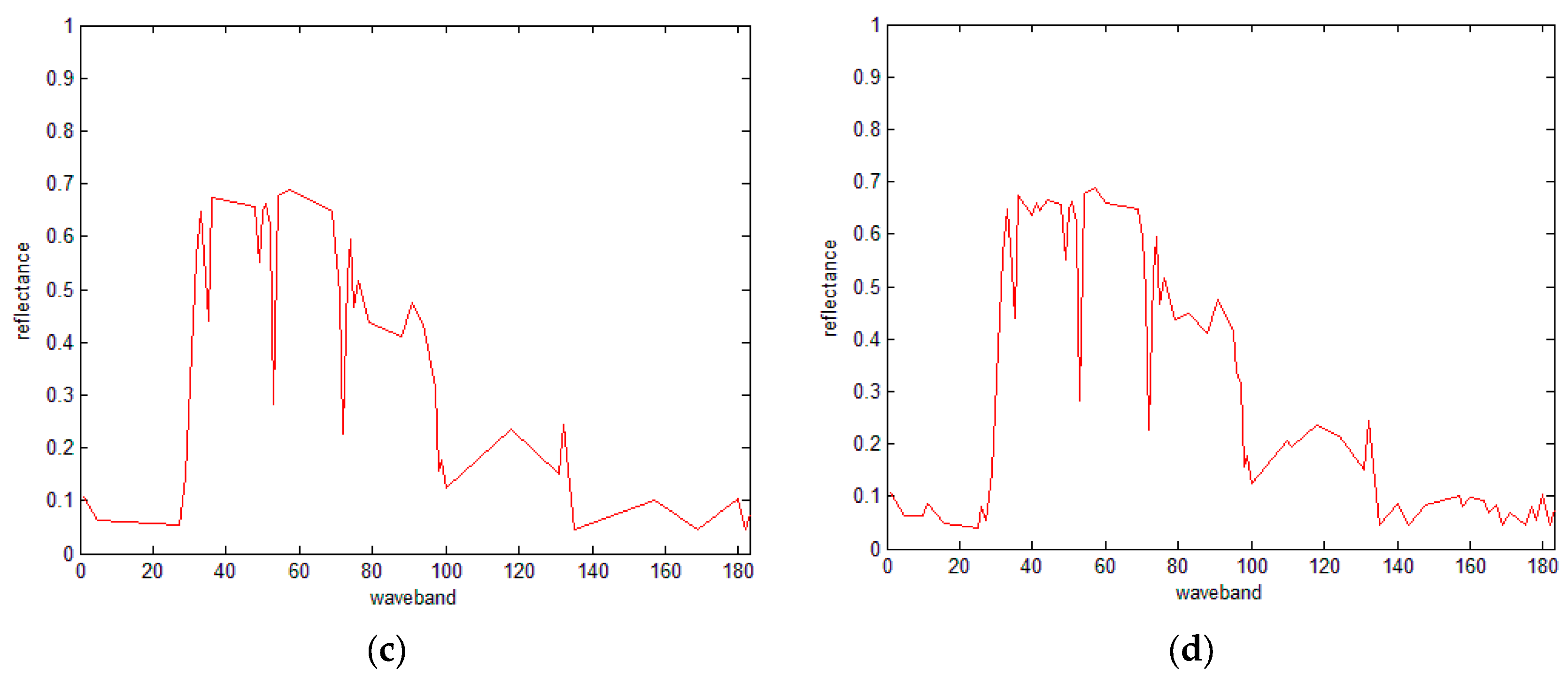
3.4. The IDP Algorithm under SRAI-Restriction
- (1)
- Specify the number of SRAI points and then calculate the SRAI feature points.
- (2)
- Specify the number of all the retained points and then set the SRAI points as the initial points of the IDP algorithm. Finally, run the IDP algorithm to determine the remaining points.
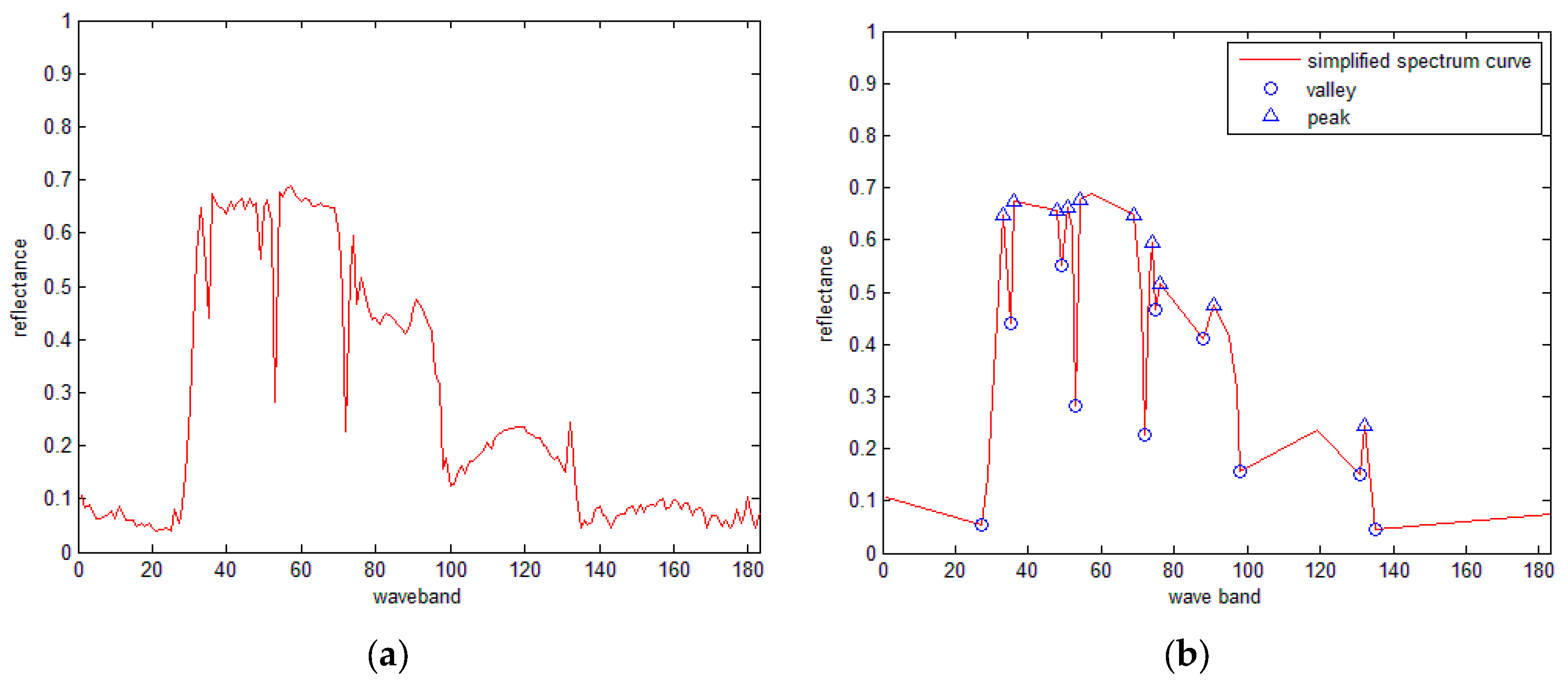
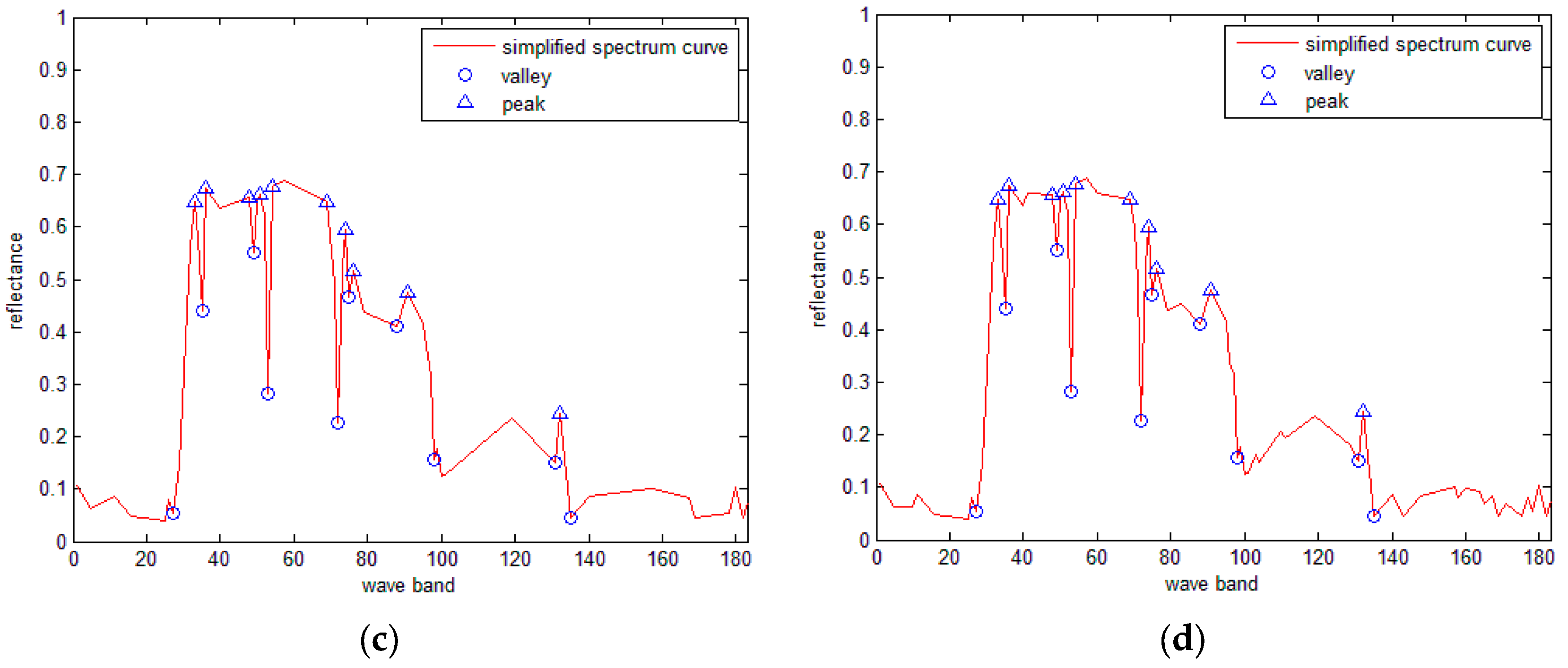
3.5. SRAI for the Hyperspectral Curves
- (1)
- For the simplified spectral curves A and B, for a point Ai on curve A, if a point Bj on curve B has the same band number as Ai, then Bj is matched with Ai. Repeat this procedure until all points on curve A have been processed. Let variable N denote the total number of matched points.
- (2)
- For every matched point, calculate the ED between the reflectance of every matched point, and obtain the sum of all the distances as the final distance.
4. Experiment and Analysis
4.1. Airborne Visible Infrared Imaging Spectrometer (AVIRIS) Dataset

| CC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 1 | 0.9980 | 0.9982 | 0.9979 | 0.9977 | 0.9967 | 0.9964 | 0.9969 | 0.9968 | A1, A3, A2 |
| A2 | 0.9980 | 1 | 0.9960 | 0.9983 | 0.9972 | 0.9962 | 0.9969 | 0.9976 | 0.9968 | A2, B1, A1 |
| A3 | 0.9982 | 0.9960 | 1 | 0.9967 | 0.9977 | 0.9973 | 0.9963 | 0.9968 | 0.9968 | A3, A1, B2 |
| B1 | 0.9979 | 0.9983 | 0.9967 | 1 | 0.9983 | 0.9968 | 0.9982 | 0.9983 | 0.9982 | B1, B2, C1/C2 |
| B2 | 0.9977 | 0.9972 | 0.9977 | 0.9983 | 1 | 0.9982 | 0.9971 | 0.9978 | 0.9978 | B2, B1, B3 |
| B3 | 0.9967 | 0.9962 | 0.9973 | 0.9968 | 0.9982 | 1 | 0.9962 | 0.9969 | 0.9965 | B3, B2, A3 |
| C1 | 0.9964 | 0.9969 | 0.9963 | 0.9982 | 0.9971 | 0.9962 | 1 | 0.9994 | 0.9993 | C1, C2, C3 |
| C2 | 0.9969 | 0.9976 | 0.9968 | 0.9983 | 0.9978 | 0.9969 | 0.9994 | 1 | 0.9993 | C2, C1, C3 |
| C3 | 0.9968 | 0.9968 | 0.9968 | 0.9982 | 0.9978 | 0.9965 | 0.9993 | 0.9993 | 1 | C3, C1, C2 |
| ED | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0 | 0.3739 | 0.3280 | 0.9241 | 0.8188 | 1.0386 | 1.3415 | 1.3367 | 1.2500 | A1,A3,A2 |
| A2 | 0.3739 | 0 | 0.3088 | 0.6276 | 0.3471 | 0.8056 | 1.0458 | 1.0417 | 0.9627 | A2,A3,B2 |
| A3 | 0.3280 | 0.3088 | 0 | 0.6942 | 0.5721 | 0.7933 | 1.0870 | 1.0815 | 0.9946 | A3,A2,A1 |
| B1 | 0.9241 | 0.6276 | 0.6942 | 0 | 0.2499 | 0.4251 | 0.4784 | 0.4823 | 0.4068 | B1,B2,C3 |
| B2 | 0.8188 | 0.3471 | 0.5721 | 0.2499 | 0 | 0.4570 | 0.6084 | 0.5983 | 0.4155 | B2,B1,A2 |
| B3 | 1.0386 | 0.8056 | 0.7933 | 0.4251 | 0.4570 | 0 | 0.4460 | 0.5134 | 0.4949 | B3,B1,C1 |
| C1 | 1.3415 | 1.0458 | 1.0870 | 0.4784 | 0.6084 | 0.4460 | 0 | 0.1618 | 0.1800 | C1,C2,C3 |
| C2 | 1.3367 | 1.0417 | 1.0815 | 0.4823 | 0.5983 | 0.5134 | 0.1618 | 0 | 0.1874 | C2,C1,C3 |
| C3 | 1.2500 | 0.9627 | 0.9946 | 0.4068 | 0.4155 | 0.4949 | 0.1800 | 0.1874 | 0 | C3,C1,C2 |
| SID | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0 | 0.0087 | 0.0066 | 0.0154 | 0.0183 | 0.0101 | 0.0159 | 0.0109 | 0.0147 | A1,A3,A2 |
| A2 | 0.0087 | 0 | 0.0082 | 0.0085 | 0.0114 | 0.0141 | 0.0105 | 0.0064 | 0.0117 | A2,A3,C2 |
| A3 | 0.0066 | 0.0082 | 0 | 0.0110 | 0.0126 | 0.0117 | 0.0112 | 0.0083 | 0.0109 | A3,A1,A2 |
| B1 | 0.0154 | 0.0085 | 0.0110 | 0 | 0.0078 | 0.0210 | 0.0059 | 0.0069 | 0.0076 | B1,C1,C2 |
| B2 | 0.0183 | 0.0114 | 0.0126 | 0.0078 | 0 | 0.0214 | 0.0100 | 0.0085 | 0.0075 | B2,C3,B1 |
| B3 | 0.0101 | 0.0141 | 0.0117 | 0.0210 | 0.0214 | 0 | 0.0219 | 0.0150 | 0.0214 | B3,A1,A3 |
| C1 | 0.0159 | 0.0105 | 0.0112 | 0.0059 | 0.0100 | 0.0219 | 0 | 0.0071 | 0.0078 | C1,B1,C2 |
| C2 | 0.0109 | 0.0064 | 0.0083 | 0.0069 | 0.0085 | 0.0150 | 0.0071 | 0 | 0.0076 | C2,A2,B1 |
| C3 | 0.0147 | 0.0117 | 0.0109 | 0.0076 | 0.0075 | 0.0214 | 0.0078 | 0.0076 | 0 | C3,B2,B1/C2 |
| KL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0 | 0.2441 | 0.2080 | 0.8186 | 0.7634 | 1.3321 | 1.4262 | 1.4227 | 1.2981 | A1,A3,A2 |
| A2 | 0.2441 | 0 | 0.2109 | 0.4512 | 0.2323 | 1.1203 | 0.9707 | 0.9656 | 0.9177 | A2,A3,B2 |
| A3 | 0.2080 | 0.2109 | 0 | 0.4835 | 0.4373 | 0.9866 | 0.9297 | 0.9518 | 0.8453 | A3,A1,A2 |
| B1 | 0.8186 | 0.4512 | 0.4835 | 0 | 0.2207 | 0.7410 | 0.2934 | 0.3687 | 0.3044 | B1,B2,C1 |
| B2 | 0.7634 | 0.2323 | 0.4373 | 0.2207 | 0 | 0.8417 | 0.4877 | 0.5064 | 0.3786 | B2,B1,A2 |
| B3 | 1.3321 | 1.1203 | 0.9866 | 0.7410 | 0.8417 | 0 | 0.6529 | 0.4534 | 0.6356 | B3,C1,C2 |
| C1 | 1.4262 | 0.9707 | 0.9297 | 0.2934 | 0.4877 | 0.6529 | 0 | 0.1272 | 0.1388 | C1,C2,C3 |
| C2 | 1.4227 | 0.9656 | 0.9518 | 0.3687 | 0.5064 | 0.4534 | 0.1272 | 0 | 0.1473 | C2,C1,C3 |
| C3 | 1.2981 | 0.9177 | 0.8453 | 0.3044 | 0.3786 | 0.6356 | 0.1388 | 0.1473 | 0 | C3,C1,C2 |
| SAM | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 1 | 0.9988 | 0.9992 | 0.9981 | 0.9980 | 0.9986 | 0.9972 | 0.9979 | 0.9976 | A1,A3,A2 |
| A2 | 0.9988 | 1 | 0.9982 | 0.9991 | 0.9986 | 0.9977 | 0.9983 | 0.9989 | 0.9984 | A2,B1,C2 |
| A3 | 0.9992 | 0.9982 | 1 | 0.9980 | 0.9985 | 0.9986 | 0.9977 | 0.9982 | 0.9981 | A3,A1,B3 |
| B1 | 0.9981 | 0.9991 | 0.9980 | 1 | 0.9991 | 0.9975 | 0.9992 | 0.9993 | 0.9992 | B1,C2,C1/C3 |
| B2 | 0.9980 | 0.9986 | 0.9985 | 0.9991 | 1 | 0.9977 | 0.9987 | 0.9990 | 0.9990 | B2,B1,C2/C3 |
| B3 | 0.9986 | 0.9977 | 0.9986 | 0.9975 | 0.9977 | 1 | 0.9966 | 0.9975 | 0.9970 | B3,A1,A3 |
| C1 | 0.9972 | 0.9983 | 0.9977 | 0.9992 | 0.9987 | 0.9966 | 1 | 0.9990 | 0.9987 | C1,B1,C2 |
| C2 | 0.9979 | 0.9989 | 0.9982 | 0.9993 | 0.9990 | 0.9975 | 0.9990 | 1 | 0.9991 | C2,B1,C3 |
| C3 | 0.9976 | 0.9984 | 0.9981 | 0.9992 | 0.9990 | 0.9970 | 0.9987 | 0.9991 | 1 | C3,B1,C2 |
| Proposed | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0 | 0.0637 | 0.0748 | 0.1495 | 0.1960 | 0.2151 | 0.3167 | 0.2271 | 0.2295 | A1,A2,A3 |
| A2 | 0.0637 | 0 | 0.0568 | 0.1202 | 0.1343 | 0.1516 | 0.1829 | 0.1678 | 0.1838 | A2,A3,A1 |
| A3 | 0.0748 | 0.0568 | 0 | 0.1797 | 0.1412 | 0.2417 | 0.1819 | 0.2116 | 0.2304 | A3,A2,A1 |
| B1 | 0.1495 | 0.1202 | 0.1797 | 0 | 0.0518 | 0.0726 | 0.1101 | 0.1001 | 0.0934 | B1,B2,B3 |
| B2 | 0.1960 | 0.1343 | 0.1412 | 0.0518 | 0 | 0.0883 | 0.1509 | 0.1445 | 0.1443 | B2,B1,B3 |
| B3 | 0.2151 | 0.1516 | 0.2417 | 0.0726 | 0.0883 | 0 | 0.1122 | 0.1242 | 0.0966 | B3,B1,B2 |
| C1 | 0.3167 | 0.1829 | 0.1819 | 0.1101 | 0.1509 | 0.1122 | 0 | 0.0293 | 0.0412 | C1,C2,C3 |
| C2 | 0.2271 | 0.1678 | 0.2116 | 0.1001 | 0.1445 | 0.1242 | 0.0293 | 0 | 0.0461 | C2,C1,C3 |
| C3 | 0.2295 | 0.1838 | 0.2304 | 0.0934 | 0.1443 | 0.0966 | 0.0412 | 0.0461 | 0 | C3,C1,C2 |
4.2. Reflective Optics System Imaging Spectrometer (ROSIS) Dataset

| CC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 1.0000 | 0.9775 | 0.9798 | 0.9024 | 0.9156 | 0.9054 | 0.9441 | 0.9376 | 0.9437 | A1,A3,A2 |
| A2 | 0.9775 | 1.0000 | 0.9645 | 0.9570 | 0.9673 | 0.9579 | 0.9362 | 0.9327 | 0.9382 | A2,A1,B2 |
| A3 | 0.9798 | 0.9645 | 1.0000 | 0.9560 | 0.9643 | 0.9593 | 0.9392 | 0.9350 | 0.9406 | A3,A1,A2 |
| B1 | 0.9024 | 0.9570 | 0.9560 | 1.0000 | 0.9524 | 0.9961 | 0.9128 | 0.9152 | 0.9176 | B1,B3,A2 |
| B2 | 0.9156 | 0.9673 | 0.9643 | 0.9524 | 1.0000 | 0.9937 | 0.9243 | 0.9264 | 0.9278 | B2,B3,A2 |
| B3 | 0.9054 | 0.9579 | 0.9593 | 0.9961 | 0.9937 | 1.0000 | 0.9202 | 0.9223 | 0.9245 | B3,B1,B2 |
| C1 | 0.9441 | 0.9362 | 0.9392 | 0.9128 | 0.9243 | 0.9202 | 1.0000 | 0.9991 | 0.9994 | C1,C3,C2 |
| C2 | 0.9376 | 0.9327 | 0.9350 | 0.9152 | 0.9264 | 0.9223 | 0.9991 | 1.0000 | 0.9994 | C2,C3,C1 |
| C3 | 0.9437 | 0.9382 | 0.9406 | 0.9176 | 0.9278 | 0.9245 | 0.9994 | 0.9994 | 1.0000 | C3,C1,C2 |
| ED | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0.0000 | 0.1923 | 0.4887 | 0.3429 | 0.4139 | 0.4501 | 1.5770 | 2.0089 | 1.6241 | A1,A2,B1 |
| A2 | 0.1923 | 0.0000 | 0.0787 | 0.2705 | 0.3597 | 0.3867 | 1.4922 | 1.9360 | 1.5395 | A2,A3,A1 |
| A3 | 0.4887 | 0.0787 | 0.0000 | 0.3112 | 0.4091 | 0.4314 | 1.5348 | 1.9820 | 1.5831 | A3,A2,B1 |
| B1 | 0.3429 | 0.2705 | 0.3112 | 0.0000 | 0.3444 | 0.1491 | 1.3785 | 1.7978 | 1.4199 | B1,B3,A2 |
| B2 | 0.4139 | 0.3597 | 0.4091 | 0.3444 | 0.0000 | 0.0904 | 1.3043 | 1.7101 | 1.3443 | B2,B3,B1 |
| B3 | 0.4501 | 0.3867 | 0.4314 | 0.1491 | 0.0904 | 0.0000 | 1.2787 | 1.6847 | 1.3175 | B3,B2,B1 |
| C1 | 1.5770 | 1.4922 | 1.5348 | 1.3785 | 1.3043 | 1.2787 | 0.0000 | 0.4871 | 0.0842 | C1,C3,C2 |
| C2 | 2.0089 | 1.9360 | 1.9820 | 1.7978 | 1.7101 | 1.6847 | 0.4871 | 0.0000 | 0.4313 | C2,C3,C1 |
| C3 | 1.6241 | 1.5395 | 1.5831 | 1.4199 | 1.3443 | 1.3175 | 0.0842 | 0.4313 | 0.0000 | C3,C1,C2 |
| SID | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0.0000 | 0.0165 | 0.0190 | 0.0468 | 0.0456 | 0.0594 | 0.5079 | 0.5212 | 0.5029 | A1,A2,A3 |
| A2 | 0.0165 | 0.0000 | 0.0020 | 0.0166 | 0.0138 | 0.0224 | 0.3945 | 0.4035 | 0.3859 | A2,A3,B2 |
| A3 | 0.0190 | 0.0020 | 0.0000 | 0.0018 | 0.0164 | 0.0234 | 0.4069 | 0.4172 | 0.3992 | A3,A2,B1 |
| B1 | 0.0468 | 0.0166 | 0.0018 | 0.0000 | 0.0040 | 0.0028 | 0.3804 | 0.3839 | 0.3682 | B1,B3,B2 |
| B2 | 0.0456 | 0.0138 | 0.0164 | 0.0040 | 0.0000 | 0.0046 | 0.3675 | 0.3703 | 0.3568 | B2,A2,B1 |
| B3 | 0.0594 | 0.0224 | 0.0234 | 0.0028 | 0.0046 | 0.0000 | 0.3516 | 0.3542 | 0.3395 | B3,B2,B1 |
| C1 | 0.5079 | 0.3945 | 0.4069 | 0.3804 | 0.3675 | 0.3516 | 0.0000 | 0.0035 | 0.0028 | C1,C3,C2 |
| C2 | 0.5212 | 0.4035 | 0.4172 | 0.3839 | 0.3703 | 0.3542 | 0.0035 | 0.0000 | 0.0029 | C2,C3,C1 |
| C3 | 0.5029 | 0.3859 | 0.3992 | 0.3682 | 0.3568 | 0.3395 | 0.0028 | 0.0029 | 0.0000 | C3,C1,C2 |
| KL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0.0000 | 0.1542 | 0.1540 | 0.4020 | 0.4914 | 0.5996 | 4.7123 | 6.0548 | 4.8043 | A1,A3,A2 |
| A2 | 0.1542 | 0.0000 | 0.0203 | 0.3606 | 0.0828 | 0.4234 | 3.9258 | 5.3117 | 3.9991 | A2,A3,B1 |
| A3 | 0.1540 | 0.0203 | 0.0000 | 0.0955 | 0.4606 | 0.5081 | 4.1037 | 5.5636 | 4.1915 | A3,A2,A1 |
| B1 | 0.4020 | 0.3606 | 0.0955 | 0.0000 | 0.0916 | 0.0542 | 3.5761 | 4.6501 | 3.5883 | B1,B3,A2 |
| B2 | 0.4914 | 0.0828 | 0.4606 | 0.0916 | 0.0000 | 0.0411 | 3.4130 | 4.3212 | 3.4172 | B2,B3,B1 |
| B3 | 0.5996 | 0.4234 | 0.5081 | 0.0542 | 0.0411 | 0.0000 | 3.2701 | 4.1749 | 3.2621 | B3,B2,B1 |
| C1 | 4.7123 | 3.9258 | 4.1037 | 3.5761 | 3.4130 | 3.2701 | 0.0000 | 0.3048 | 0.0321 | C1,C3,C2 |
| C2 | 6.0548 | 5.3117 | 5.5636 | 4.6501 | 4.3212 | 4.1749 | 0.3048 | 0.0000 | 0.2392 | C2,C3,C1 |
| C3 | 4.8043 | 3.9991 | 4.1915 | 3.5883 | 3.4172 | 3.2621 | 0.0321 | 0.2392 | 0.0000 | C3,C1,C2 |
| SAM | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 1.0000 | 0.9932 | 0.9947 | 0.9841 | 0.9845 | 0.9805 | 0.8631 | 0.8601 | 0.8628 | A1,A3,A2 |
| A2 | 0.9932 | 1.0000 | 0.9993 | 0.9950 | 0.9954 | 0.9991 | 0.9009 | 0.8985 | 0.9011 | A2,A3,B3 |
| A3 | 0.9947 | 0.9993 | 1.0000 | 0.9940 | 0.9989 | 0.9930 | 0.8967 | 0.8941 | 0.8968 | A3,A2,B2 |
| B1 | 0.9841 | 0.9950 | 0.9940 | 1.0000 | 0.9948 | 0.9933 | 0.9014 | 0.9006 | 0.9025 | B1,A2,B2 |
| B2 | 0.9845 | 0.9954 | 0.9989 | 0.9948 | 1.0000 | 0.9989 | 0.9063 | 0.9054 | 0.9070 | B2,B3,A3 |
| B3 | 0.9805 | 0.9991 | 0.9930 | 0.9933 | 0.9989 | 1.0000 | 0.9114 | 0.9106 | 0.9124 | B3,A2,B2 |
| C1 | 0.8631 | 0.9009 | 0.8967 | 0.9014 | 0.9063 | 0.9114 | 1.0000 | 0.9996 | 0.9997 | C1,C3,C2 |
| C2 | 0.8601 | 0.8985 | 0.8941 | 0.9006 | 0.9054 | 0.9106 | 0.9996 | 1.0000 | 0.9997 | C2,C3,C1 |
| C3 | 0.8628 | 0.9011 | 0.8968 | 0.9025 | 0.9070 | 0.9124 | 0.9997 | 0.9997 | 1.0000 | C3,C1,C2 |
| Proposed | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | Rank | |
| A1 | 0.0000 | 0.0829 | 0.0622 | 0.1473 | 0.1192 | 0.2132 | 0.5461 | 0.9955 | 0.6399 | A1,A3,A2 |
| A2 | 0.0829 | 0.0000 | 0.0339 | 0.1180 | 0.1333 | 0.1645 | 0.4875 | 1.0929 | 0.5742 | A2,A3,A1 |
| A3 | 0.0622 | 0.0339 | 0.0000 | 0.1149 | 0.1514 | 0.1349 | 0.6716 | 0.6859 | 0.4885 | A3,A2,A1 |
| B1 | 0.1473 | 0.1180 | 0.1149 | 0.0000 | 0.0510 | 0.0736 | 0.6502 | 0.7601 | 0.4397 | B1,B2,B3 |
| B2 | 0.1192 | 0.1333 | 0.1514 | 0.0510 | 0.0000 | 0.0501 | 0.5167 | 0.5091 | 0.5821 | B2,B3,B1 |
| B3 | 0.2132 | 0.1645 | 0.1349 | 0.0736 | 0.0501 | 0.0000 | 0.4990 | 0.5677 | 0.3716 | B3,B2,B1 |
| C1 | 0.5461 | 0.4875 | 0.6716 | 0.6502 | 0.5167 | 0.4990 | 0.0000 | 0.1852 | 0.0376 | C1,C3,C2 |
| C2 | 0.9955 | 1.0929 | 0.6859 | 0.7601 | 0.5091 | 0.5677 | 0.1852 | 0.0000 | 0.1232 | C2,C3,C1 |
| C3 | 0.6399 | 0.5742 | 0.4885 | 0.4397 | 0.5821 | 0.3716 | 0.0376 | 0.1232 | 0.0000 | C3,C1,C2 |
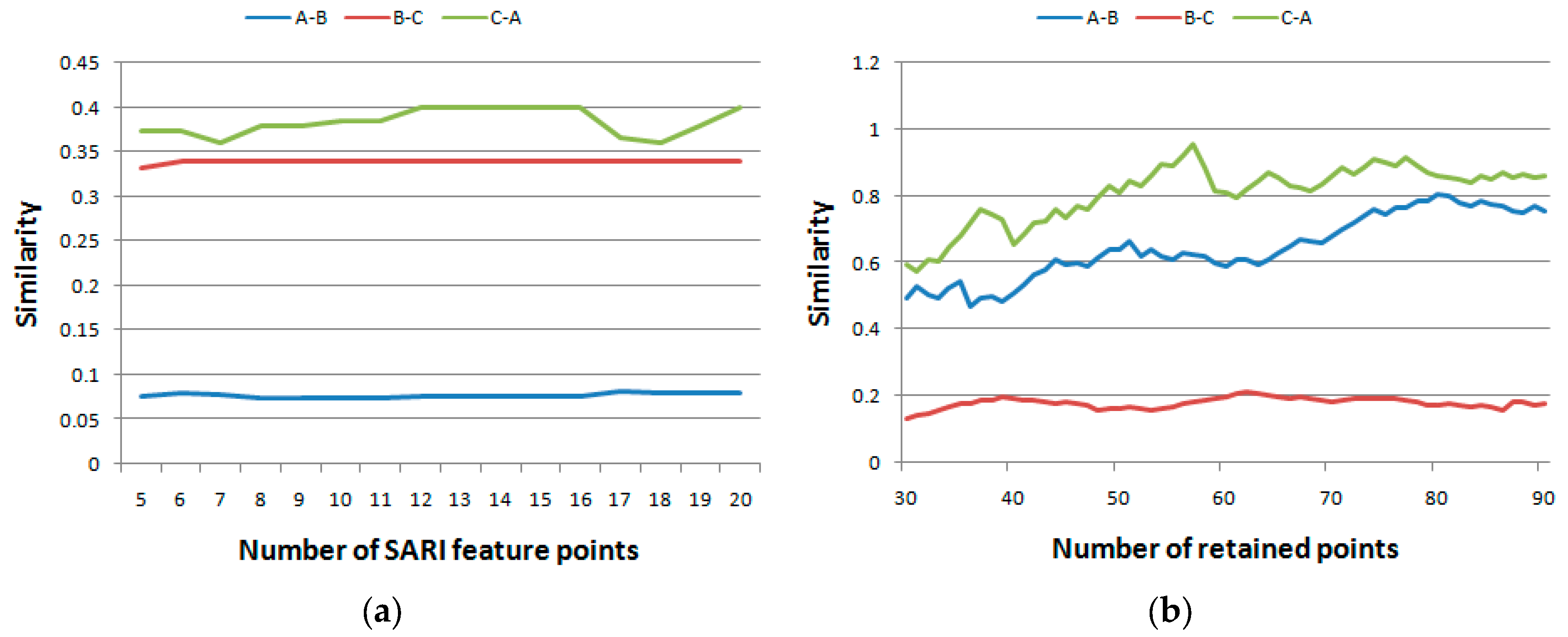
4.3.The Impacts of the Parameters
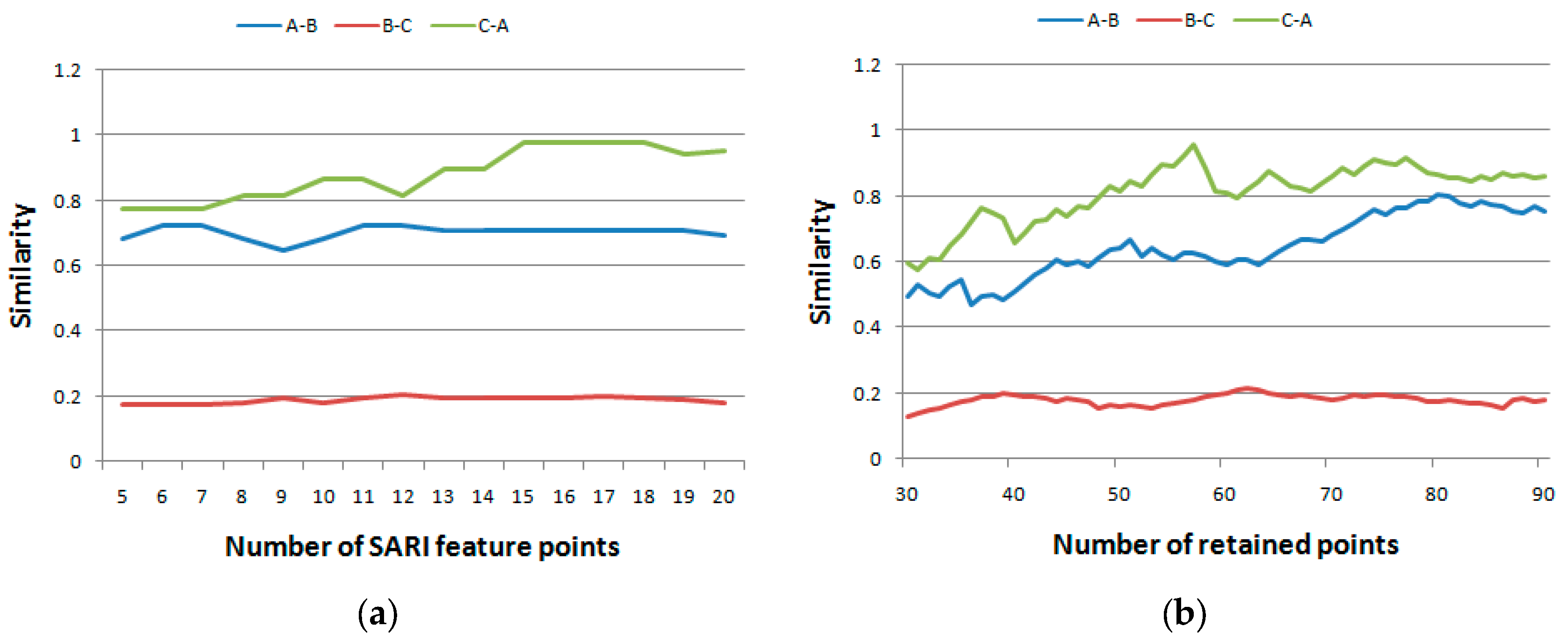
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van der Woerd, H.J.; Wernand, M.R. True Colour Classification of Natural Waters with Medium-Spectral Resolution Satellites: SeaWiFS, MODIS, MERIS and OLCI. Sensors 2015, 15, 25663–25680. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Hong, H.M.; You, Z.H.; Cheng, F. Spectral and Image Integrated Analysis of Hyperspectral Data for Waxy Corn Seed Variety Classification. Sensors 2015, 15, 15578–15594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Liu, F.; He, Y.; Li, X.L. Application of Hyperspectral Imaging and Chemometric Calibrations for Variety Discrimination of Maize Seeds. Sensors 2012, 12, 17234–17246. [Google Scholar] [CrossRef] [PubMed]
- Stratoulias, D.; Balzter, H.; Sykioti, O.; Zlinszky, A.; Tóth, V.R. Evaluating Sentinel-2 for Lakeshore Habitat Mapping Based on Airborne Hyperspectral Data. Sensors 2015, 15, 22956–22969. [Google Scholar] [CrossRef] [PubMed]
- Gmur, S.; Vogt, D.; Zabowski, D.; Moskal, L.M. Hyperspectral Analysis of Soil Nitrogen, Carbon, Carbonate, and Organic Matter Using Regression Trees. Sensors 2012, 12, 10639–10658. [Google Scholar] [CrossRef] [PubMed]
- Kaarna, A.; Zemcik, P.; Kalviainen, H. Compression of multispectral remote sensing images using clustering and spectral reduction. IEEE Trans. Geosci. Remote Sens. 2000, 38, 1588–1592. [Google Scholar] [CrossRef]
- Du, Q.; Fowler, J.E. Low-complexity principal component analysis for hyperspectral image compression. Int. J. High Perform. Comput. Appl. 2008, 22, 438–448. [Google Scholar] [CrossRef]
- Wang, J.; Chang, C.I. Independent component analysis-based dimensionality reduction with applications in hyperspectral image analysis. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1586–1600. [Google Scholar] [CrossRef]
- Guo, B.; Gunn, S.R.; Damper, R.I. Band selection for hyperspectral image classification using mutual information. IEEE Geosci. Remote Sens. Lett. 2006, 3, 522–526. [Google Scholar] [CrossRef]
- Jimenez, L.O.; Landgrebe, D.A. Hyperspectral data analysis and supervised feature reduction via projection pursuit. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2653–2667. [Google Scholar] [CrossRef]
- Banskota, A.; Wynne, R.H.; Kayastha, N. Improving within-genus tree species discrimination using the discrete wavelet transform applied to airborne hyperspectral data. Int. J. Remote Sens. 2011, 32, 3551–3563. [Google Scholar] [CrossRef]
- Amato, U.; Cavalli, R.M.; Palombo, A. Experimental approach to the selection of the components in the minimum noise fraction. IEEE Trans. Geosci. Remote Sens. 2009, 47, 153–160. [Google Scholar] [CrossRef]
- Licciardi, G.A.; Frate, F.D. A comparison of feature extraction methodologies applied on hyperspectral data. In Proceedings of ESA Hyperspectral 2010 Workshop, Frascati, Italy, 17–19 March 2010.
- Jia, X.; Riehards, J.A. Segmented prineipal components transformation for efficient hyperspectral remote sensing image display and classification. IEEE Trans. Geosci. Remote Sens. 1999, 37, 538–540. [Google Scholar]
- Maghsoudi, Y.; Zoej, M.J.V.; Collins, M. Using class-based feature selection for the classification of hyperspectral data. Int. J. Remote Sens. 2011, 32, 4311–4326. [Google Scholar] [CrossRef]
- Goel, P.K.; Prasher, S.O.; Patel, R.M. Classification of hyperspectral data by decision trees and artificial neural networks to identify weed stress and nitrogen status of corn. Comput. Electron. Agric. 2003, 39, 67–93. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Fauvel, M.; Chanussot, J. SVM- and MRF-based method for accurate classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2010, 7, 736–740. [Google Scholar] [CrossRef]
- Kruse, F.A.; Lefkoff, A.B.; Boardman, J.W. The spectral image processing system (SIPS)–interactive visualization and analysis of imaging spectrometer data. Remote Sens. Environ. 1993, 44, 145–163. [Google Scholar] [CrossRef]
- Liu, W.; Chang, Q.R.; Guo, M. Extraction of first derivative spectrum features ofsoil organic matter via wavelet de-noising. Spectrosc. Spectr. Anal. 2011, 31, 100–104. [Google Scholar]
- Pluim, J.P.W.; Maintz, J.B.A.; Viergever, M. Mutual information based registration of medical images: A survey. IEEE Trans. Med. Imag. 2003, 22, 986–1004. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.I. An information-theoretic approach to spectral variability, similarity, and discrimination for hyperspectral image analysis. IEEE Trans. Inf. Theory 2000, 46, 1927–1932. [Google Scholar] [CrossRef]
- Li, Q.L.; Xue, Y.Q.; Wang, J.Y. Automated tongue segmentation algorithm based on hyperspectral image. J. Infrared Millim. Waves 2007, 26, 77–80. [Google Scholar]
- Du, Y.Z.; Chang, C.; Ren, H. New hyperspectral discrimination measure for spectral characterization. Opt. Eng. 2004, 43, 1777–1786. [Google Scholar]
- Kumar, M.N.; Seshasai, M.V.R.; Prasad, K.S.V. A new hybrid spectral similarity measure for discrimination among Vigna species. Int. J. Remote Sens. 2011, 32, 4041–4053. [Google Scholar] [CrossRef]
- Kong, X.B.; Shu, N.; Tao, J.B. A new spectral similarity measure based on multiple features integration. Spectrosc. Spectr. Anal. 2011, 31, 2166–2170. [Google Scholar]
- Du, P.J.; Chen, Y.H.; Fang, T. Spectral feature-based hyperspectral RS image retrieval. Spectrosc. Spectr. Anal. 2005, 25, 1171–1175. [Google Scholar]
- Sun, H.; Li, M.Z.; Zhang, Y.E. Spectral characteristics of corn under different nitrogen treatments. Spectrosc. Spectr. Anal. 2010, 30, 715–719. [Google Scholar]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Bakker, W.H.; Schmidt, K.S. Hyperspectral edge filtering for measuring homo-geneity of surface cover types. ISPRS J. Photogramm. Remote Sens. 2002, 56, 246–256. [Google Scholar] [CrossRef]
- Du, Y.; Ives, R.; Etter, D.; Welch, T.; Chang, C.-I. A one-dimensional approach for iris recognition. In Proceedings of the SPIE Biometric Technology for Human Identification, Orlando, FL, USA, 12–16 April 2004; pp. 237–247.
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, D.; Liu, J.; Huang, J.; Li, H.; Liu, P.; Chen, H.; Qian, J. Spectral Similarity Assessment Based on a Spectrum Reflectance-Absorption Index and Simplified Curve Patterns for Hyperspectral Remote Sensing. Sensors 2016, 16, 152. https://doi.org/10.3390/s16020152
Ma D, Liu J, Huang J, Li H, Liu P, Chen H, Qian J. Spectral Similarity Assessment Based on a Spectrum Reflectance-Absorption Index and Simplified Curve Patterns for Hyperspectral Remote Sensing. Sensors. 2016; 16(2):152. https://doi.org/10.3390/s16020152
Chicago/Turabian StyleMa, Dan, Jun Liu, Junyi Huang, Huali Li, Ping Liu, Huijuan Chen, and Jing Qian. 2016. "Spectral Similarity Assessment Based on a Spectrum Reflectance-Absorption Index and Simplified Curve Patterns for Hyperspectral Remote Sensing" Sensors 16, no. 2: 152. https://doi.org/10.3390/s16020152
APA StyleMa, D., Liu, J., Huang, J., Li, H., Liu, P., Chen, H., & Qian, J. (2016). Spectral Similarity Assessment Based on a Spectrum Reflectance-Absorption Index and Simplified Curve Patterns for Hyperspectral Remote Sensing. Sensors, 16(2), 152. https://doi.org/10.3390/s16020152






