Predicting Gilthead Sea Bream (Sparus aurata) Freshness by a Novel Combined Technique of 3D Imaging and SW-NIR Spectral Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.1.1. Destructive Analyses
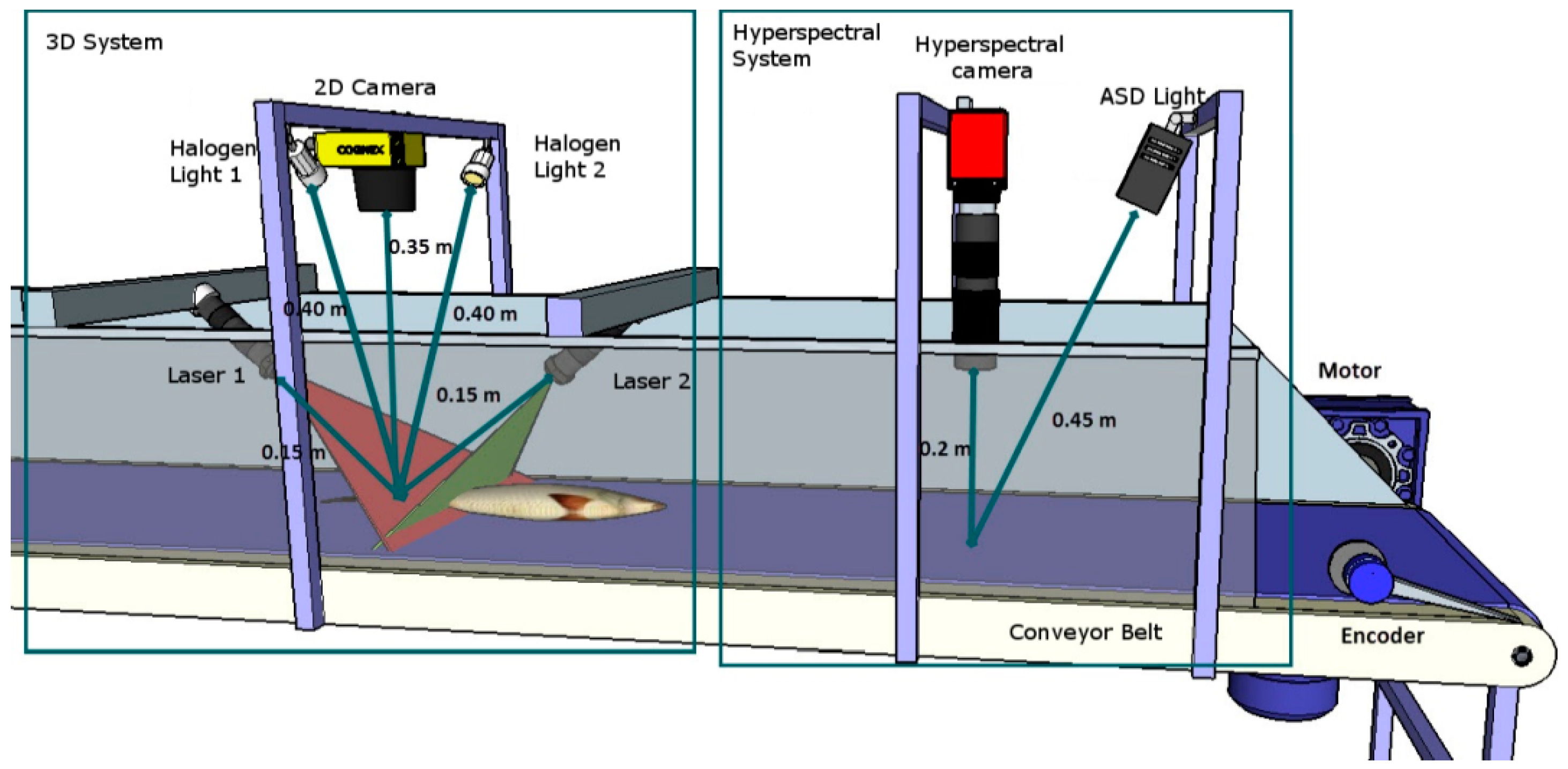
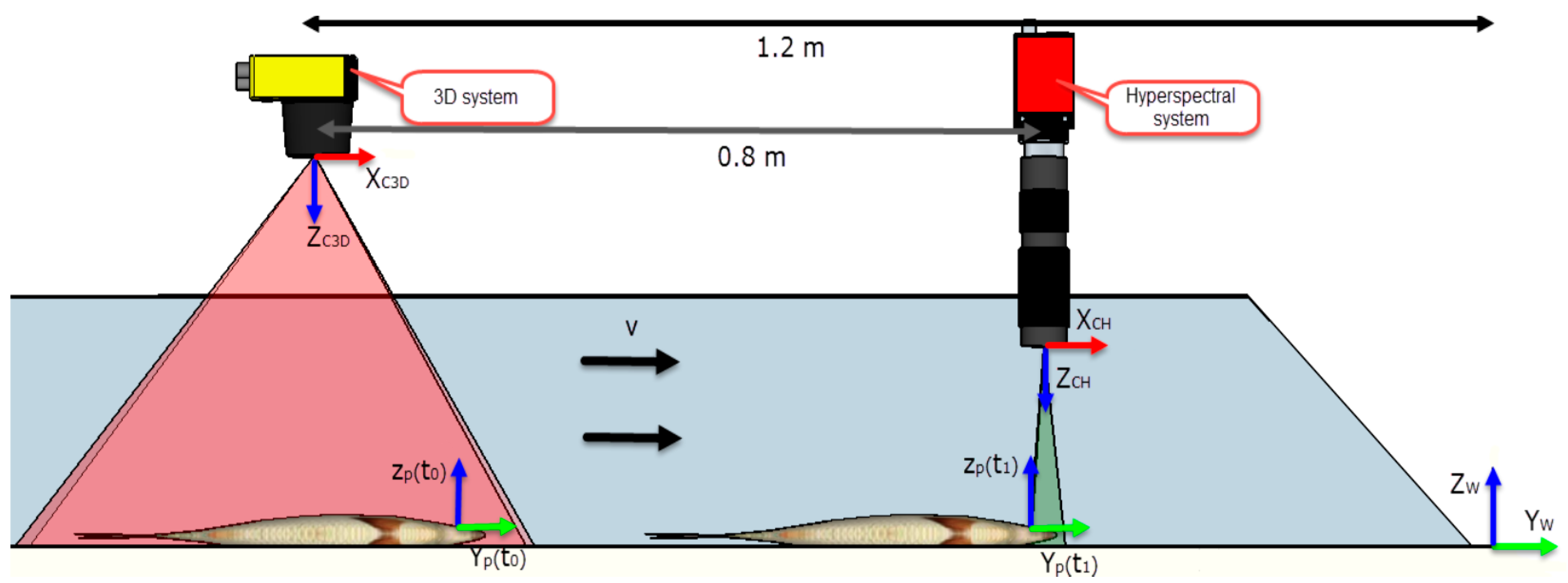
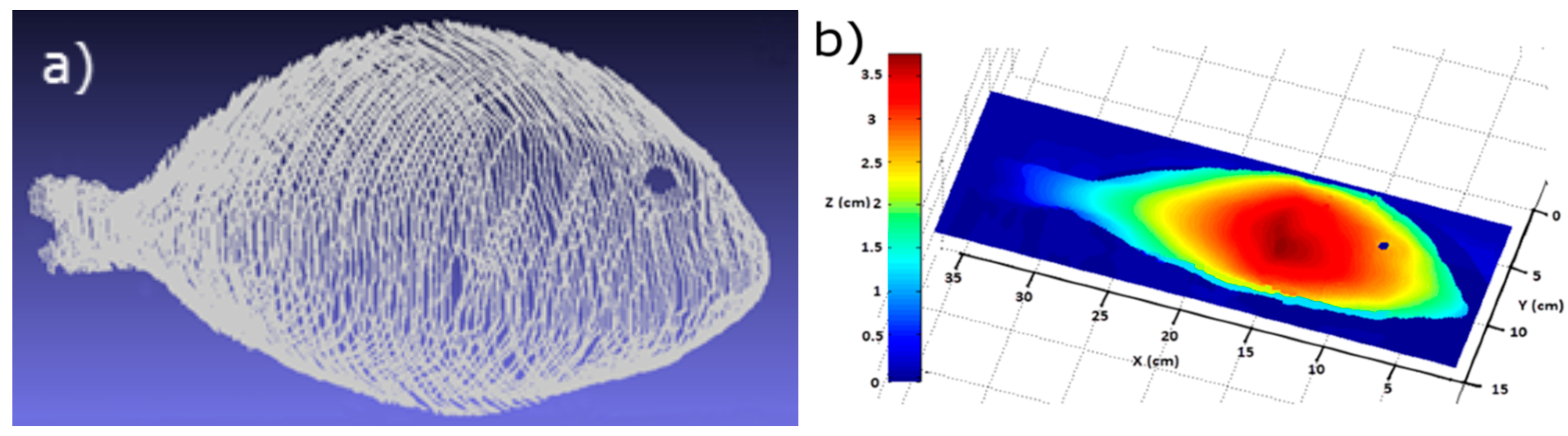
2.1.2. Imaging System
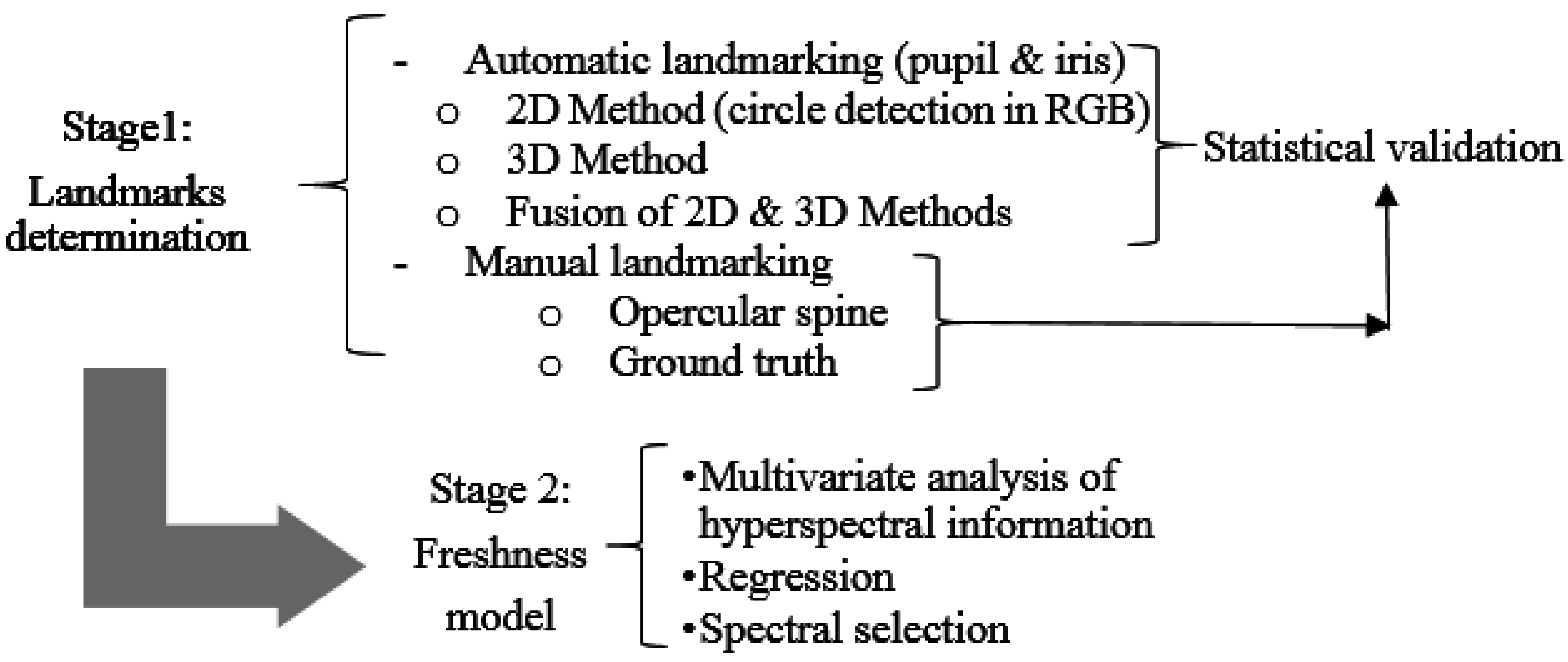
2.2. Data Processing
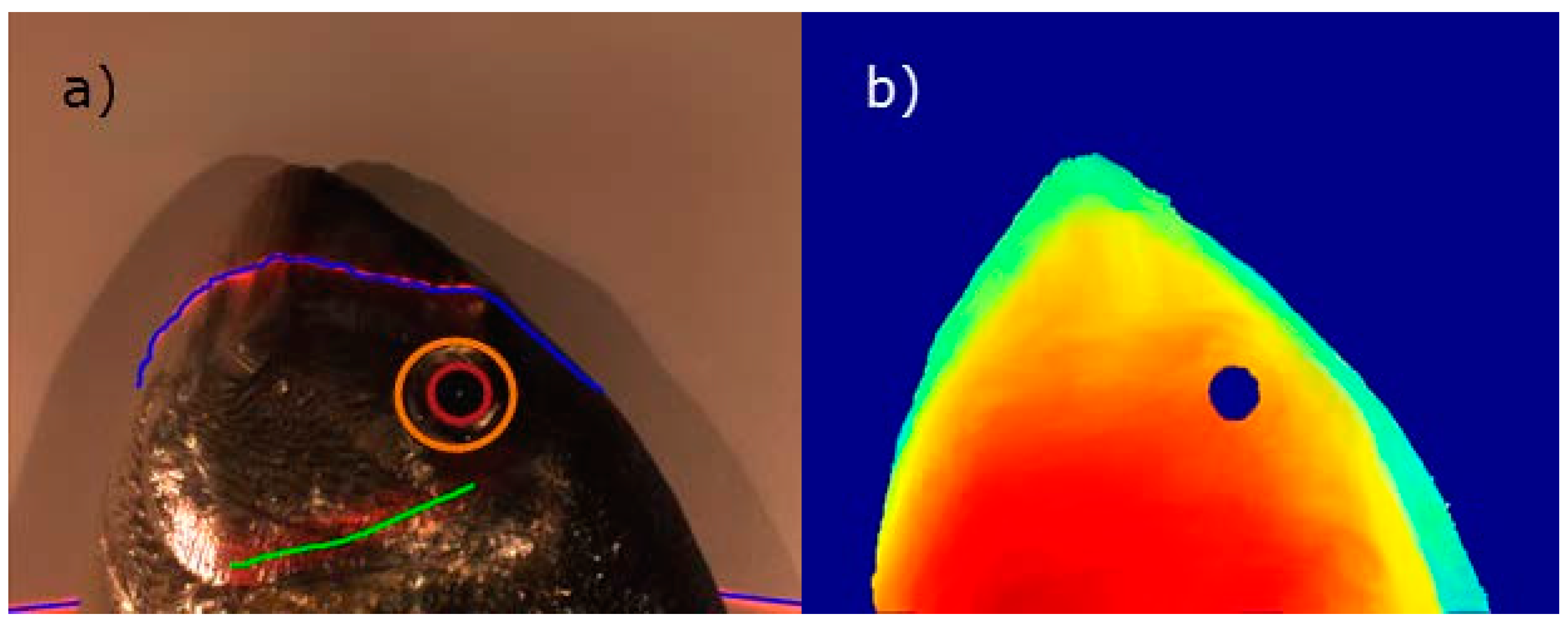
2.2.1. Stage 1: Determining Landmarks
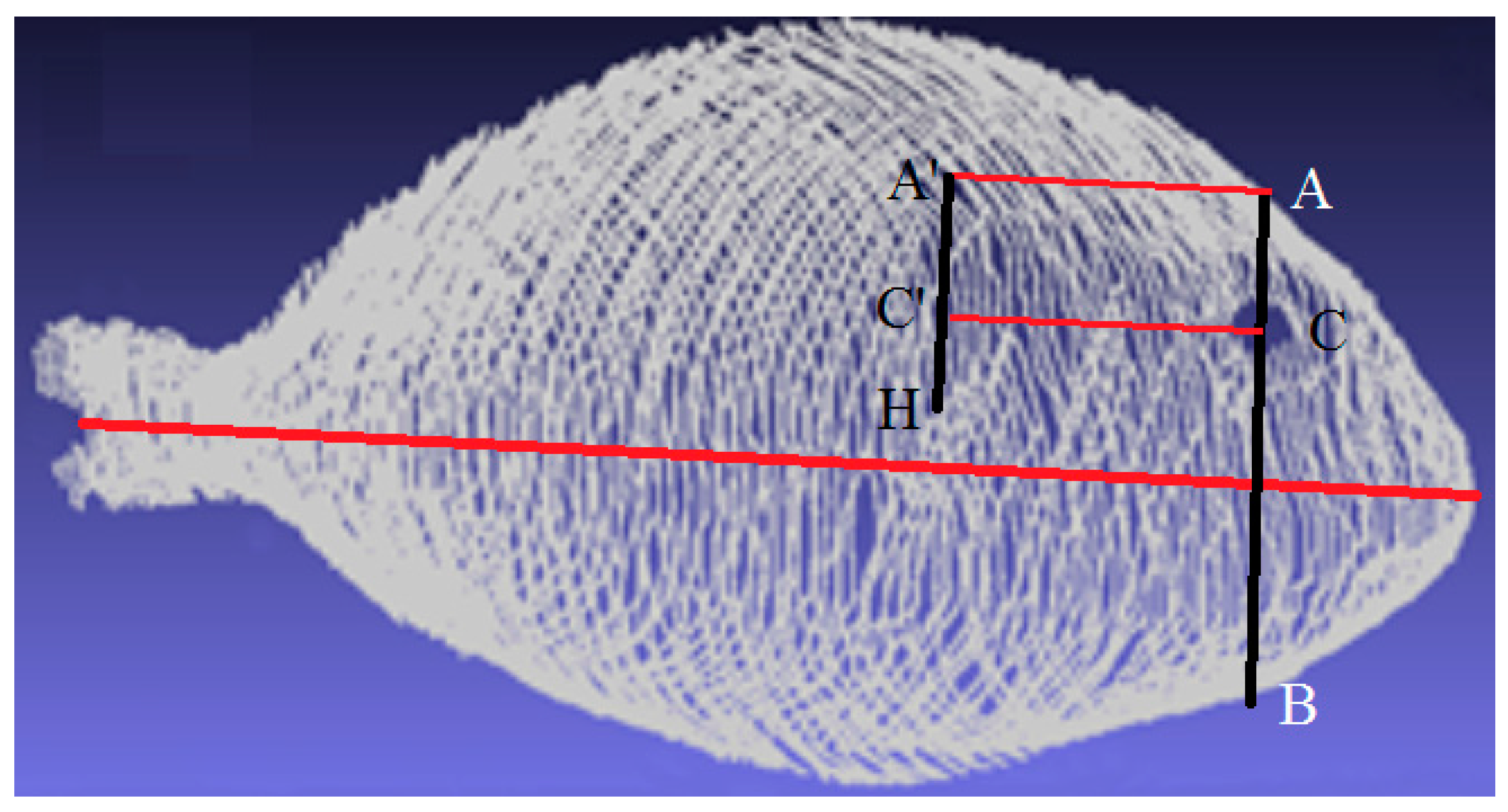
- The central axis is detected to take into account orientation changes.
- The center of the eye is located by the previously explained techniques.
- An imaginary profile AB, perpendicular to the central axis, is calculated that crosses the fish, as depicted in Figure 4.
- The AC profile is found because AC is shorter than CB.
- Highest point H is found.
- The AC profile is projected in the direction of the central axis as a distance defined by H to obtain the A’C’ profile.
- A region around the A’C’ profile is the opercular spine region.
2.2.2. Stage 2: Freshness Model
2.3. Statistical Validation
3. Results and Discussion
3.1. Chemical and Microbiological Results
3.2. Image Analysis
3.2.1. Stage 1: Landmarks Determination
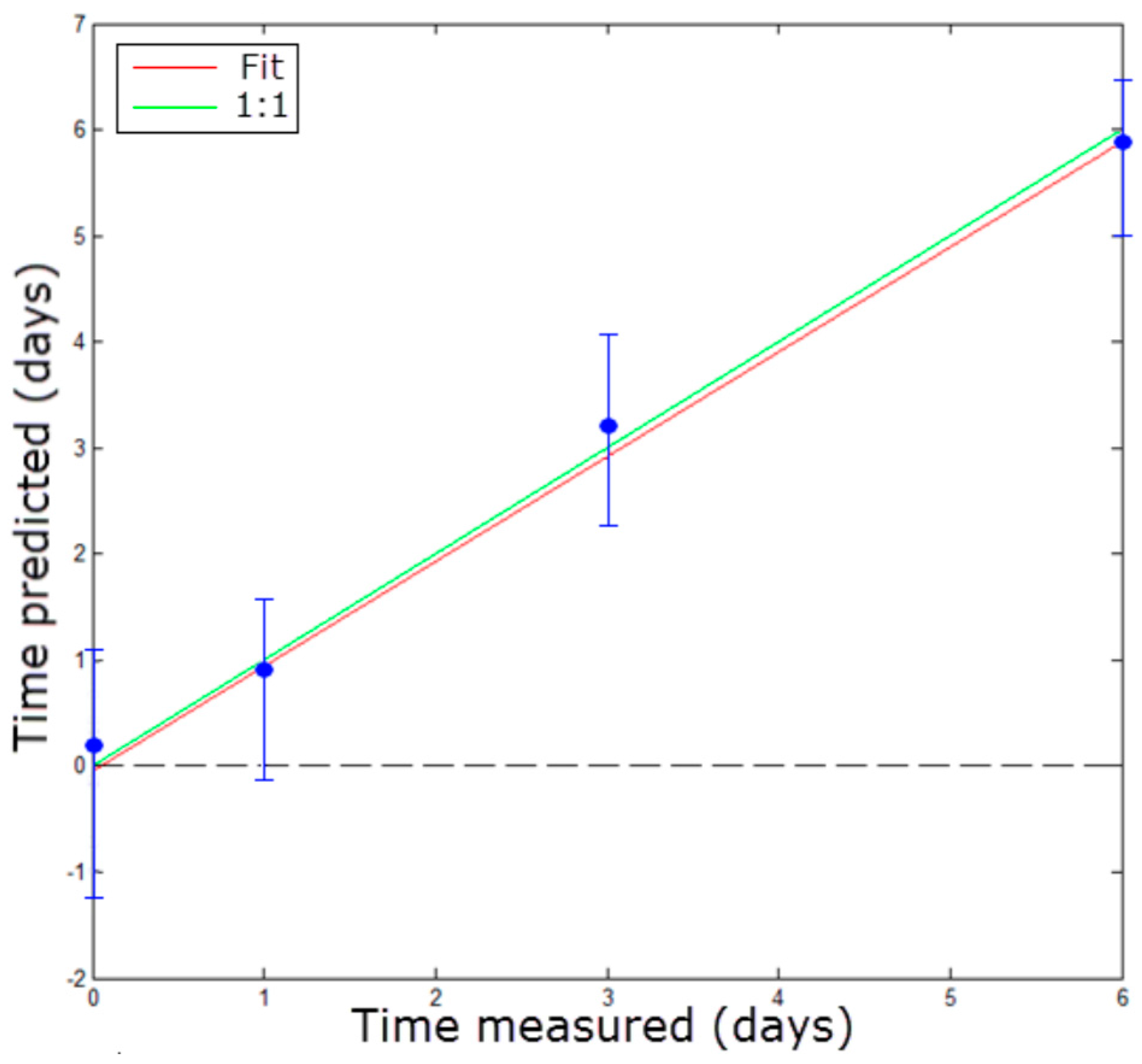
3.2.2. Stage 2: Freshness Evaluation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- FAO Fisheries and Aquaculture Department. Global Aquaculture Production Statistics for the Year; FAO Fisheries and Aquaculture Department: Rome, Italy, 2013. [Google Scholar]
- Olafsdattir, G.; Martinsdattir, E.; Oehlenschlager, J.; Dalgaard, P.; Jensen, B.; Undeland, I.; Mackie, I.M.; Henehan, G.; Nielsen, J.; Nilsen, H. Methods to evaluate fish freshness in research and industry. Trends Food Sci. Technol. 1997, 8, 258–265. [Google Scholar] [CrossRef]
- Rehbein, H.; Oehlenschlger, J. Fishery Products; Wiley-Blackwell: Oxford, UK, 2009. [Google Scholar]
- Pérez-Esteve, E.; Fuentes, A.; Grau, R.; Fernández-Segovia, I.; Masot, R.; Alcañiz, M.; Barat, J.M. Use of impedance spectroscopy for predicting freshness of sea bream (Sparus aurata). Food Control 2014, 35, 360–365. [Google Scholar] [CrossRef]
- Zaragozá, P.; Fuentes, A.; Ruiz-Rico, M.; Vivancos, J.-L.; Fernández-Segovia, I.; Ros-Lis, J.V.; Barat, J.M.; Martínez-Máñez, R. Development of a colorimetric sensor array for squid spoilage assessment. Food Chem. 2015, 175, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Federation European Aquaculture Producers. European Aquaculture Production Report; Federation European Aquaculture Producers: Liege, Belgium, 2014. [Google Scholar]
- Alasalvar, C.; Taylor, K.D.A.; Öksüz, A.; Garthwaite, T.; Alexis, M.N.; Grigorakis, K. Freshness assessment of cultured sea bream (Sparus aurata) by chemical, physical and sensory methods. Food Chem. 2001, 72, 33–40. [Google Scholar] [CrossRef]
- Barat, J.M.; Gil, L.; Garcia-Breijo, E.; Aristoy, M.C.; Toldrá, F.; Martínez-Máñez, R.; Soto, J. Freshness monitoring of sea bream (Sparus aurata) with a potentiometric sensor. Food Chem. 2008, 108, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Lougovois, V.P.; Kyranas, E.R.; Kyrana, V.R. Comparison of selected methods of assessing freshness quality and remaining storage life of iced gilthead sea bream (Sparus aurata). Food Res. Int. 2003, 36, 551–560. [Google Scholar] [CrossRef]
- Zaragozá, P.; Fuentes, A.; Fernández-Segovia, I.; Vivancos, J.-L.; Rizo, A.; Ros-Lis, J.V.; Barat, J.M.; Martínez-Máñez, R. Evaluation of sea bream (Sparus aurata) shelf life using an optoelectronic nose. Food Chem. 2013, 138, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Dowlati, M.; Mohtasebi, S.S.; Omid, M.; Razavi, S.H.; Jamzad, M.; de la Guardia, M. Freshness assessment of gilthead sea bream (Sparus aurata) by machine vision based on gill and eye color changes. J. Food Eng. 2013, 119, 277–287. [Google Scholar] [CrossRef]
- Menesatti, P.; Costa, C.; Aguzzi, J. Quality Evaluation of Fish by Hyperspectral Imaging. In Hyperspectral Imaging for Food Quality Analysis and Control; Academic Press: San Diego, CA, USA, 2010; pp. 273–294. [Google Scholar]
- Cheng, J.-H.; Sun, D.-W. Hyperspectral imaging as an effective tool for quality analysis and control of fish and other seafoods: Current research and potential applications. Trends Food Sci. Technol. 2014, 37, 78–91. [Google Scholar] [CrossRef]
- Cheng, J.-H.; Sun, D.-W.; Pu, H.-B.; Wang, Q.-J.; Chen, Y.-N. Suitability of hyperspectral imaging for rapid evaluation of thiobarbituric acid (TBA) value in grass carp (Ctenopharyngodon idella) fillet. Food Chem. 2015, 171, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-H.; Sun, D.-W.; Qu, J.-H.; Pu, H.-B.; Zhang, X.-C.; Song, Z.; Chen, X.; Zhang, H. Developing a multispectral imaging for simultaneous prediction of freshness indicators during chemical spoilage of grass carp fish fillet. J. Food Eng. 2016, 182, 9–17. [Google Scholar] [CrossRef]
- Ivorra, E.; Girón, J.; Sánchez, A.J.; Verdú, S.; Barat, J.M.; Grau, R. Detection of expired vacuum-packed smoked salmon based on PLS-DA method using hyperspectral images. J. Food Eng. 2013, 117, 342–349. [Google Scholar] [CrossRef]
- Sivertsen, A.H.; Kimiya, T.; Heia, K. Automatic freshness assessment of cod (Gadus morhua) fillets by Vis/Nir spectroscopy. J. Food Eng. 2011, 103, 317–323. [Google Scholar] [CrossRef]
- Udomkun, P.; Nagle, M.; Mahayothee, B.; Müller, J. Laser-based imaging system for non-invasive monitoring of quality changes of papaya during drying. Food Control 2014, 42, 225–233. [Google Scholar] [CrossRef]
- Verdú, S.; Ivorra, E.; Sánchez, A.J.; Girón, J.; Barat, J.M.; Grau, R. Comparison of TOF and SL techniques for in-line measurement of food item volume using animal and vegetable tissues. Food Control 2013, 33, 221–226. [Google Scholar] [CrossRef]
- Marchant, J. Fusing 3D information for crop/weeds classification. In Proceedings of the 15th International Conference on Pattern Recognition, Barcelona, Spain, 3–7 September 2000; pp. 295–298.
- Mundt, J.T.; Streutker, D.R.; Glenn, N.F. Mapping Sagebrush Distribution Using Fusion of Hyperspectral and Lidar Classifications. Photogramm. Eng. Remote Sens. 2006, 72, 47–54. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Kurz, T.; Buckley, S.; Howell, J.; Schneider, D. Geological outcrop modelling and interpretation using ground based hyperspectral and laser scanning data fusion. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 37, B5. [Google Scholar]
- Buckley, S.J.; Kurz, T.H.; Howell, J.A.; Schneider, D. Terrestrial lidar and hyperspectral data fusion products for geological outcrop analysis. Comput. Geosci. 2013, 54, 249–258. [Google Scholar] [CrossRef]
- Taoukis, P.S.; Koutsoumanis, K.; Nychas, G.J.E. Use of time–temperature integrators and predictive modelling for shelf life control of chilled fish under dynamic storage conditions. Int. J. Food Microbiol. 1999, 53, 21–31. [Google Scholar] [CrossRef]
- Boran, G.; Karaçam, H.; Boran, M. Changes in the quality of fish oils due to storage temperature and time. Food Chem. 2006, 98, 693–698. [Google Scholar] [CrossRef]
- Fuentes, A.; Barat, J.M.; Fernández-Segovia, I.; Serra, J.A. Study of sea bass (Dicentrarchus labrax L.) salting process: Kinetic and thermodynamic control. Food Control 2008, 19, 757–763. [Google Scholar] [CrossRef]
- Malle, P.; Tao, S.H. Rapid quantitative determination of trimethylamine using steam distillation. J. Food Prot. 1987, 50, 756–760. [Google Scholar]
- Kiertzmann, H.J.; Wegner, K.; Priebe, U.; Rakow, D. Freshness test of fish using measurement of refractive index of eye and mussel fluids of fish. Zentralbratt Veterinär-Medezin 1964, 11, 551–560. [Google Scholar]
- Trobina, M. Error Model of a Coded-Light Range Sensor. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.34.6434&rep=rep1&type=pdf (accessed on 17 October 2016).
- Zhang, Z. A flexible new technique for camera calibration. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1330–1334. [Google Scholar] [CrossRef]
- Ivorra, E.; Amat, S.V.; Sánchez, A.J.; Barat, J.M.; Grau, R. Continuous monitoring of bread dough fermentation using a 3D vision Structured Light technique. J. Food Eng. 2014, 130, 8–13. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Wold, S.; Antti, H.; Lindgren, F.; Öhman, J. Orthogonal signal correction of near-infrared spectra. Chemom. Intell. Lab. Syst. 1998, 44, 175–185. [Google Scholar] [CrossRef]
- Horaud, R.; Mohr, R.; Lorecki, B. Linear Camera Calibration. In Proceedings of the 1992 IEEE International Conference on Robotics and Automation, Nice, France, 12–14 May 1992; pp. 1539–1544.
- Wold, S.; Geladi, P.; Esbensen, K.; Öhman, J. Multi-way principal components-and PLS-analysis. J. Chemom. 1987, 1, 41–56. [Google Scholar] [CrossRef]
- De Jong, S. SIMPLS: An alternative approach to partial least squares regression. Chemom. Intell. Lab. Syst. 1993, 18, 251–263. [Google Scholar] [CrossRef]
- Holland, J.H. Adaptation in Natural and Artificial Systems; University of Michigan Press: Ann Arbor, MI, USA, 1975. [Google Scholar]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.-J.E. Data mining derived from food analyses using non-invasive/non-destructive analytical techniques; determination of food authenticity, quality & safety in tandem with computer science disciplines. Trends Food Sci. Technol. 2016, 50, 11–25. [Google Scholar]
- Carrascosa, C.; Millán, R.; Saavedra, P.; Jaber, J.R.; Raposo, A.; Pérez, E.; Montenegro, T.; Sanjuán, E. Microbiological evolution of gilthead sea bream (Sparus aurata) in Canary Islands during ice storage. J. Food Sci. Technol. 2013, 52, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Arashisar, Ş.; Hisar, O.; Kaya, M.; Yanik, T. Effects of modified atmosphere and vacuum packaging on microbiological and chemical properties of rainbow trout (Oncorynchus mykiss) fillets. Int. J. Food Microbiol. 2004, 97, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Özogul, F.; Polat, A.; Özogul, Y. The effects of modified atmosphere packaging and vacuum packaging on chemical, sensory and microbiological changes of sardines (Sardina pilchardus). Food Chem. 2004, 85, 49–57. [Google Scholar] [CrossRef]
- Pascual, M.; Calderón, V.; Pascual, M.A. Metodología Analítica Para Alimentos y Bebidas; Díaz de Santos: A Coruña, Spain, 1999. [Google Scholar]
- Ingram, M. Sampling for Microbiological Analysis: Principles and Specific Applications; University of Toronto Press: Toronto, ON, Canada, 1978. [Google Scholar]
- Costa, C.; Negretti, P.; Vandeputte, M.; Pallottino, F.; Antonucci, F.; Aguzzi, J.; Bianconi, G.; Menesatti, P. Innovative Automated Landmark Detection for Food Processing: The Backwarping Approach. Food Bioprocess Technol. 2014, 7, 2291–2298. [Google Scholar] [CrossRef]
- Masniyom, P. Deterioration and shelf-life extension of fish and fishery products by modified atmosphere packaging. Sonklanakarin J. Sci. Technol. 2011, 33, 181. [Google Scholar]
- James, R.A.; Hoadley, P.A.; Sampson, B.G. Determination of Postmortem Interval by Sampling Vitreous Humour. Am. J. Forensic Med. Pathol. 1997, 18, 158–162. [Google Scholar] [CrossRef] [PubMed]
| Storage Time (Days) | RI | TVB-N | pH | Enterobacteriaceae (log cfu) | Mesophilic (log cfu) |
|---|---|---|---|---|---|
| 0 | 1.3348 ± 0.0003 a | 18.14 ± 2.25 a | 6.28 ± 0.18 a | <1 | 0.37 ± 0.74 |
| 1 | 1.3350 ± 0.0002 a | 18.57 ± 0.61 a | 6.11 ± 0.04 ab | ||
| 3 | 1.3352 ± 0.0006 a | 19.47 ± 1.79 a | 6.22 ± 0.108 ab | ||
| 6 | 1.3371 ± 0.0010 b | 28.06 ± 1.29 b | 6.23 ± 0.09 b | 3.30 ± 0.22 | 5.02 ± 0.24 |
| Iris | Pupil | Pupil & Iris | Opercular Spine | |||
|---|---|---|---|---|---|---|
| Whole Spectrum | I-PLS Selection | Whole Spectrum | I-PLS Selection | Whole Spectrum | Whole Spectrum | |
| Num. LVs | 7 | 3 | 7 | 3 | 5 | 6 |
| RMSEC (days) | 0.805 | 1.041 | 0.604 | 0.908 | 1.174 | 0.662 |
| RMSECV (days) | 0.968 | 1.071 | 0.712 | 0.941 | 1.212 | 0.803 |
| RMSEPred (days) | 0.882 | 0.971 | 0.651 | 0.846 | 1.253 | 0.783 |
| R2 Cal | 0.87 | 0.79 | 0.93 | 0.84 | 0.74 | 0.566 |
| R2 CV | 0.82 | 0.77 | 0.90 | 0.82 | 0.72 | 0.391 |
| R2 Pred | 0.86 | 0.83 | 0.92 | 0.87 | 0.7 | 0.413 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivorra, E.; Verdu, S.; Sánchez, A.J.; Grau, R.; Barat, J.M. Predicting Gilthead Sea Bream (Sparus aurata) Freshness by a Novel Combined Technique of 3D Imaging and SW-NIR Spectral Analysis. Sensors 2016, 16, 1735. https://doi.org/10.3390/s16101735
Ivorra E, Verdu S, Sánchez AJ, Grau R, Barat JM. Predicting Gilthead Sea Bream (Sparus aurata) Freshness by a Novel Combined Technique of 3D Imaging and SW-NIR Spectral Analysis. Sensors. 2016; 16(10):1735. https://doi.org/10.3390/s16101735
Chicago/Turabian StyleIvorra, Eugenio, Samuel Verdu, Antonio J. Sánchez, Raúl Grau, and José M. Barat. 2016. "Predicting Gilthead Sea Bream (Sparus aurata) Freshness by a Novel Combined Technique of 3D Imaging and SW-NIR Spectral Analysis" Sensors 16, no. 10: 1735. https://doi.org/10.3390/s16101735
APA StyleIvorra, E., Verdu, S., Sánchez, A. J., Grau, R., & Barat, J. M. (2016). Predicting Gilthead Sea Bream (Sparus aurata) Freshness by a Novel Combined Technique of 3D Imaging and SW-NIR Spectral Analysis. Sensors, 16(10), 1735. https://doi.org/10.3390/s16101735







