A Review of Membrane-Based Biosensors for Pathogen Detection
Abstract
:1. Introduction
2. Membrane Materials and Fabrication
2.1. Inorganic Membranes
2.2. Organic Membranes
2.3. Hybrid Membranes
2.4. Composite Membranes
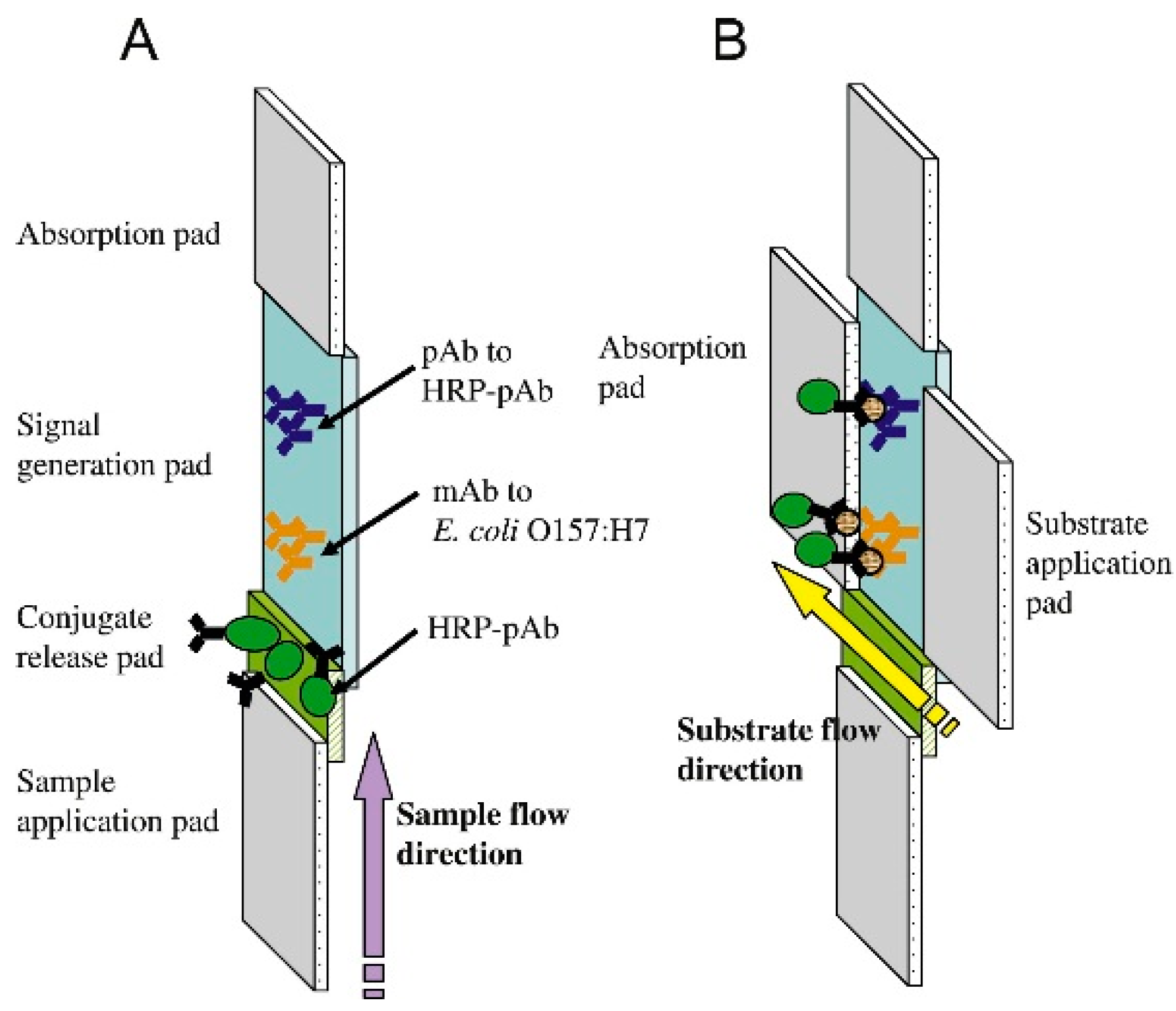
3. Molecular Probes
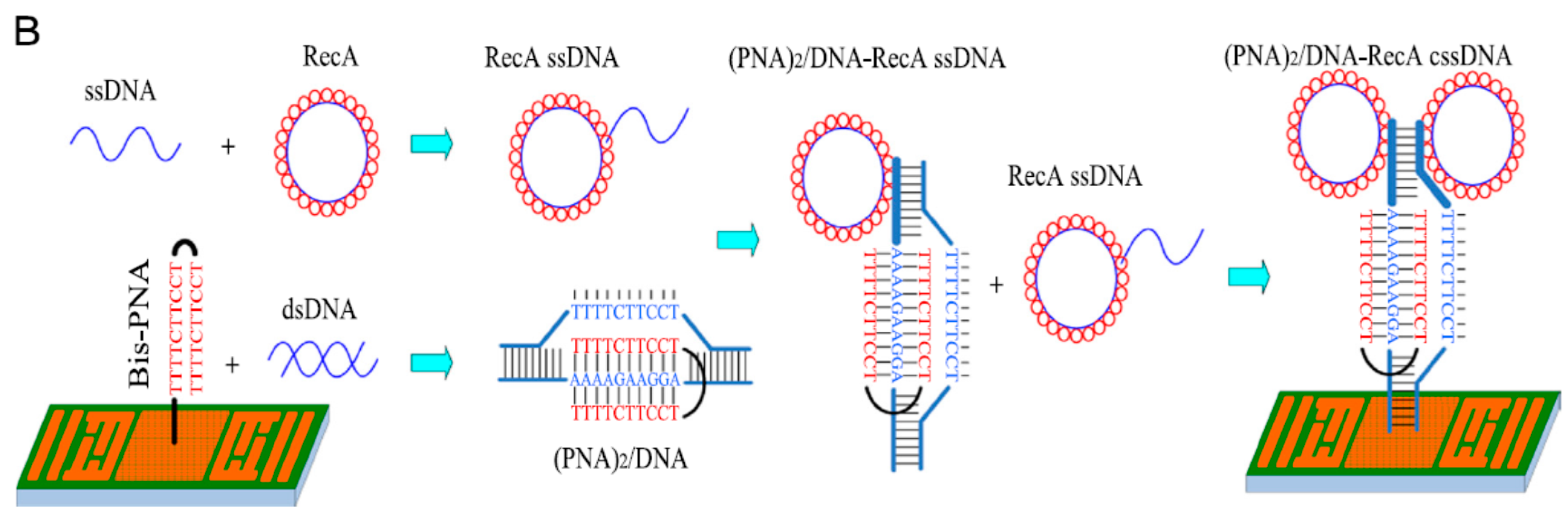
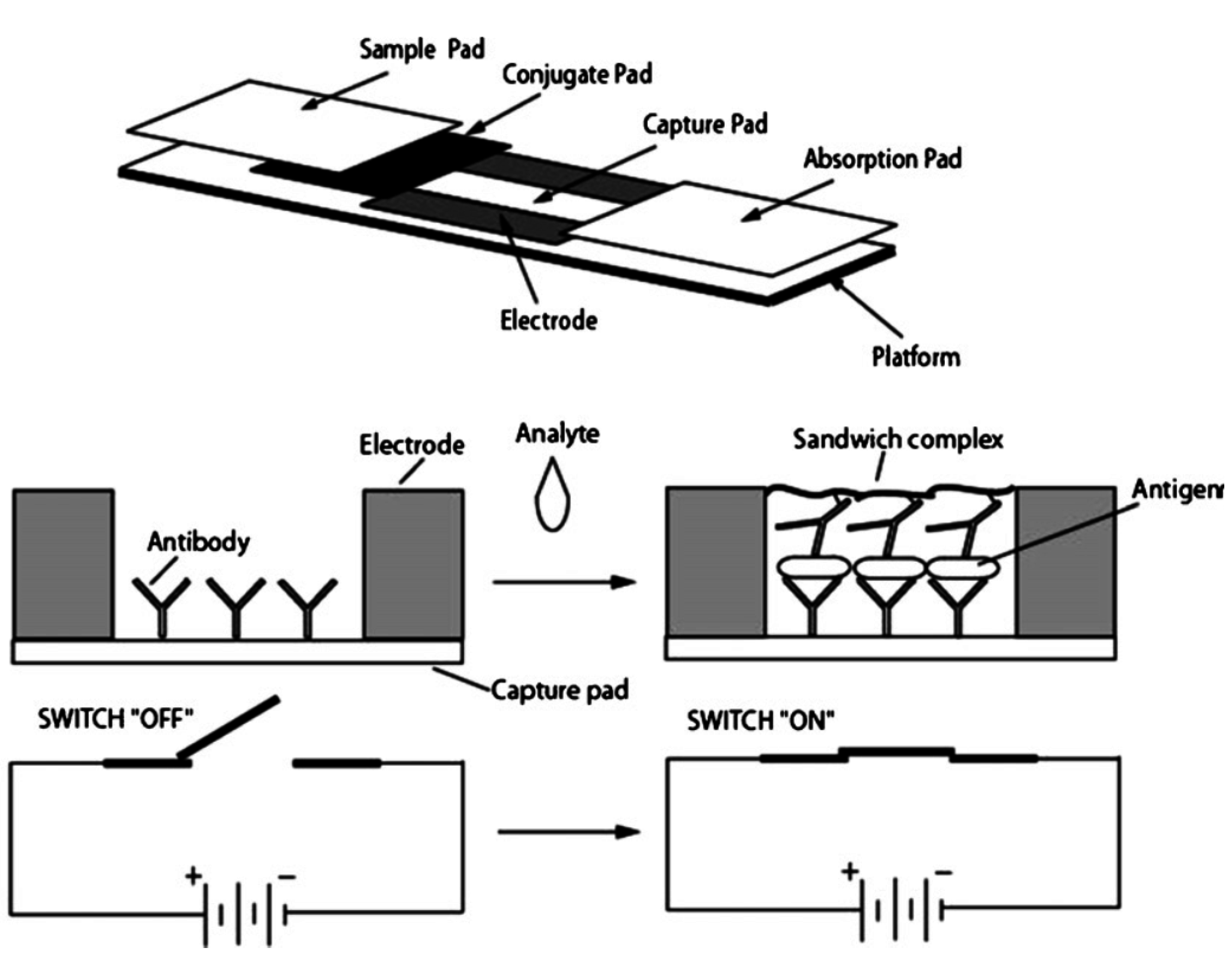
4. Linking Procedure
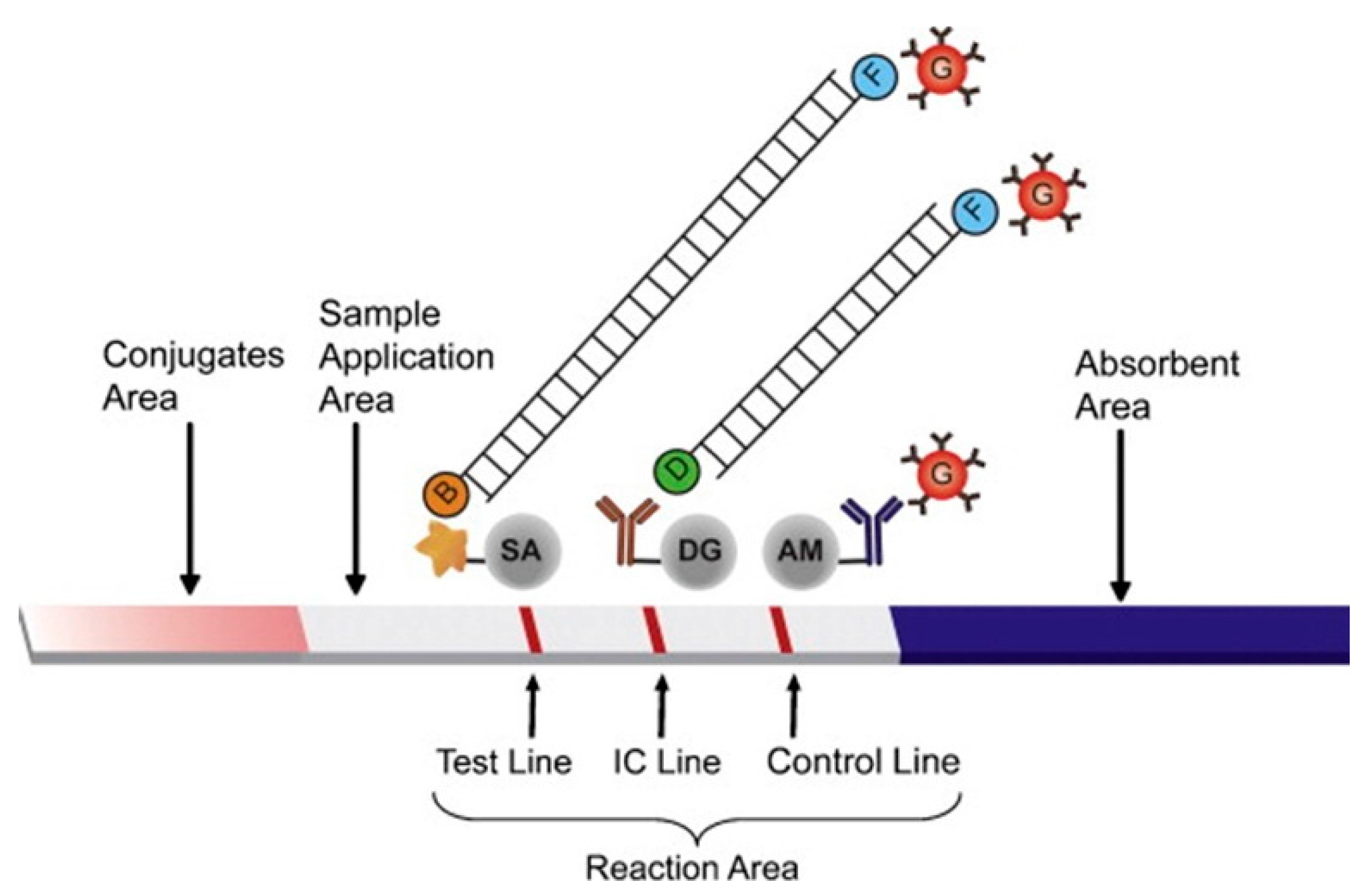

5. Transduction Systems
5.1. Electrical
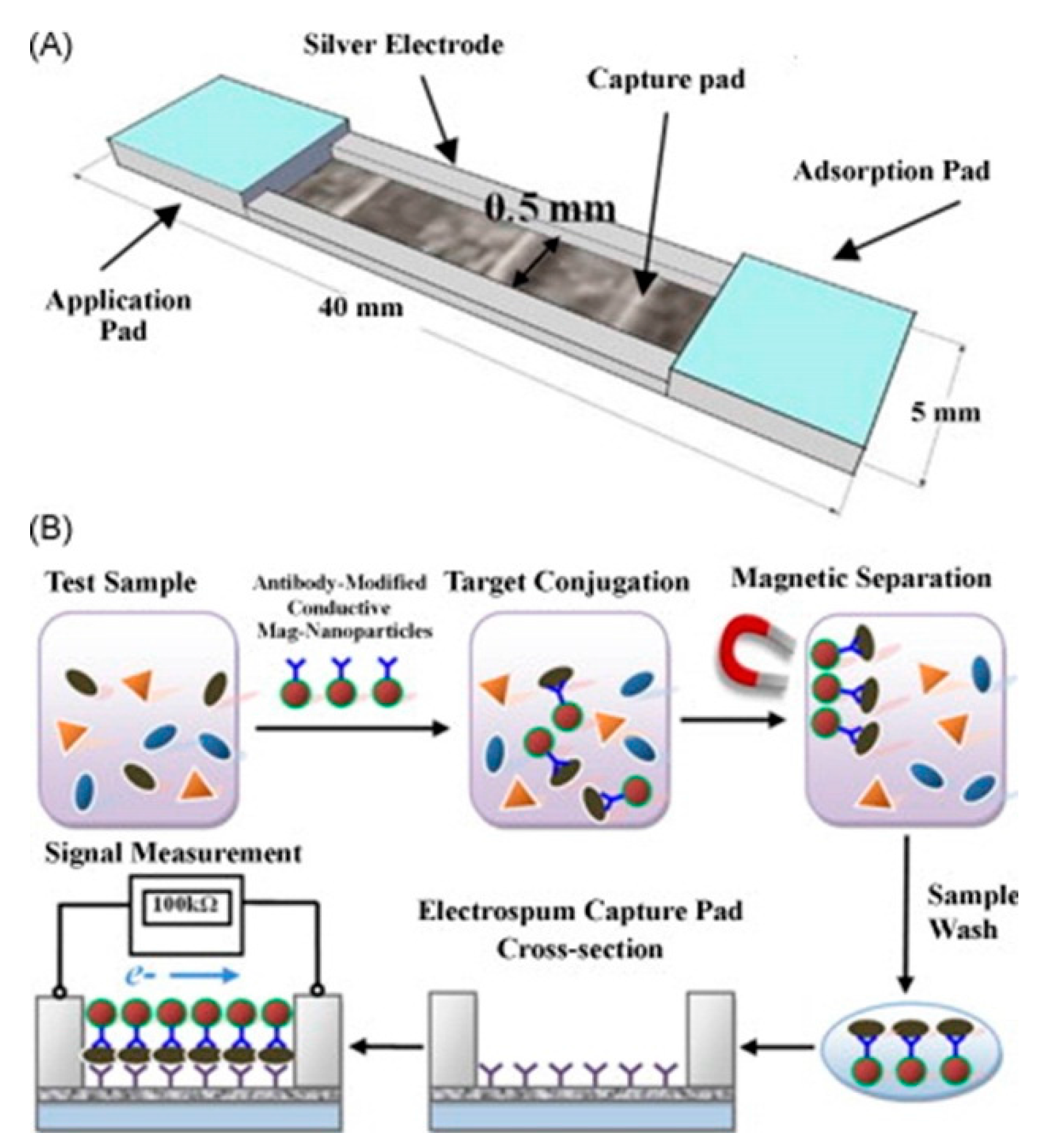
5.2. Optical
5.2.1. Color Change
5.2.2. Light Emission
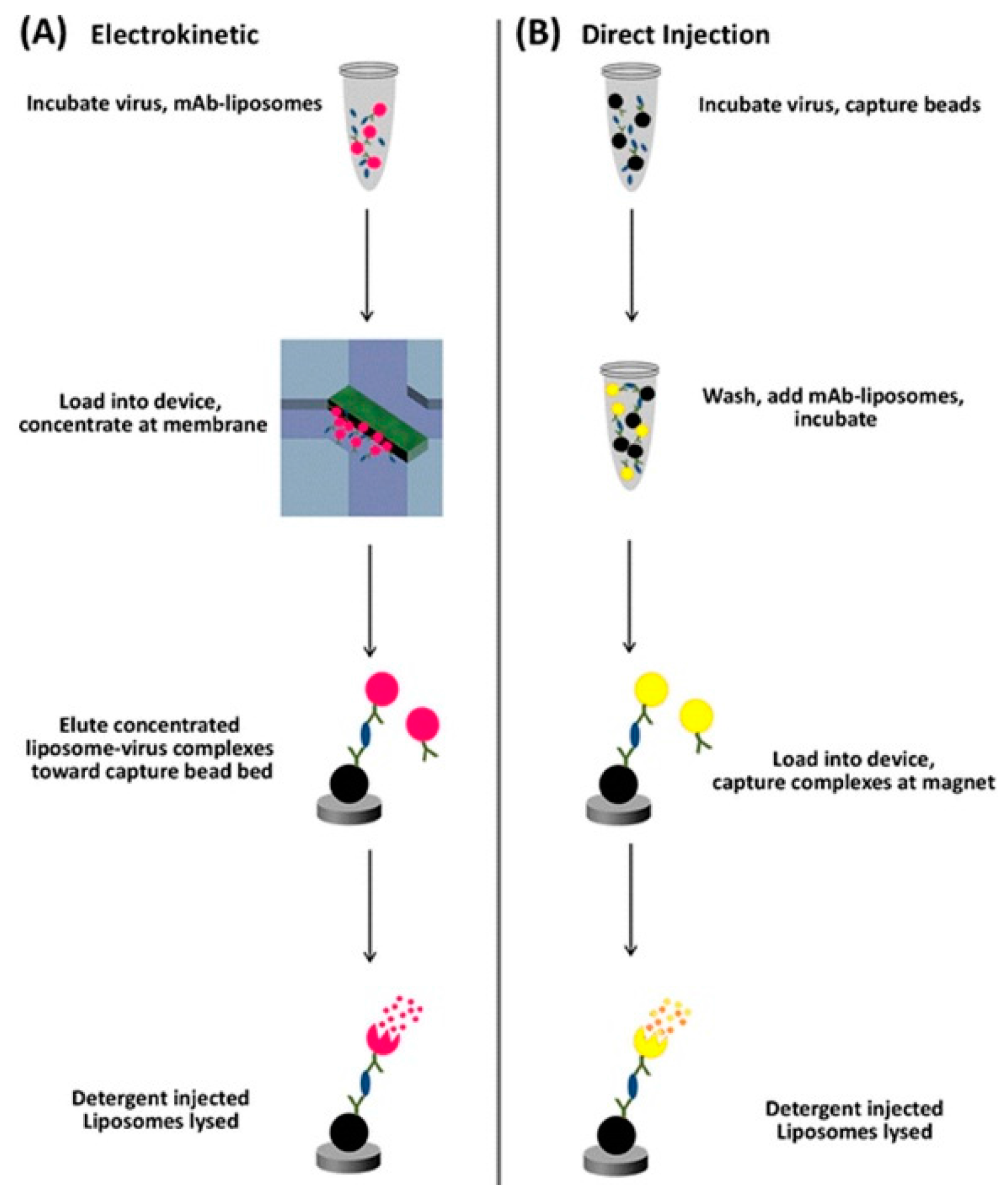
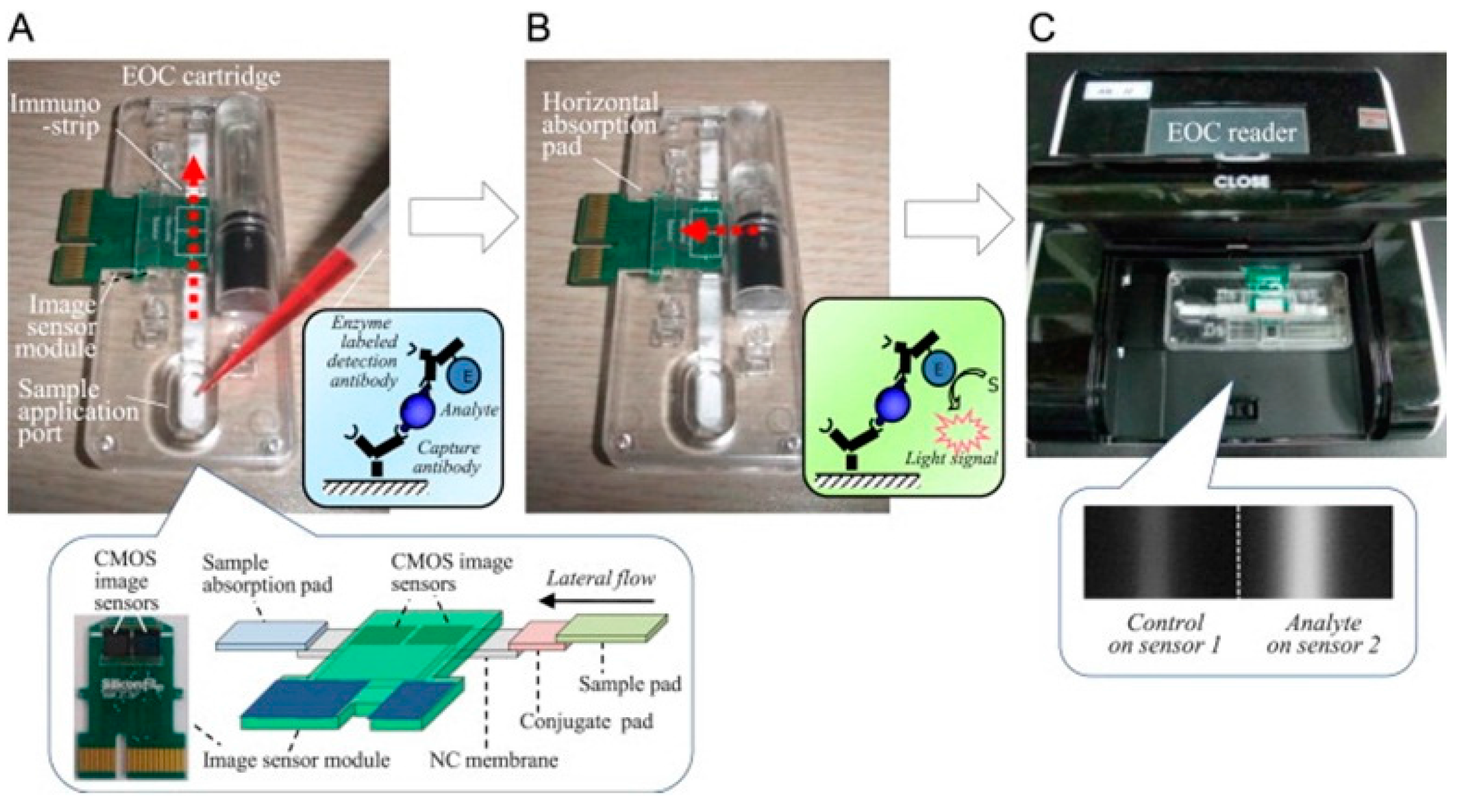
5.2.3. Spectroscopic/Interferometric
5.3. Other
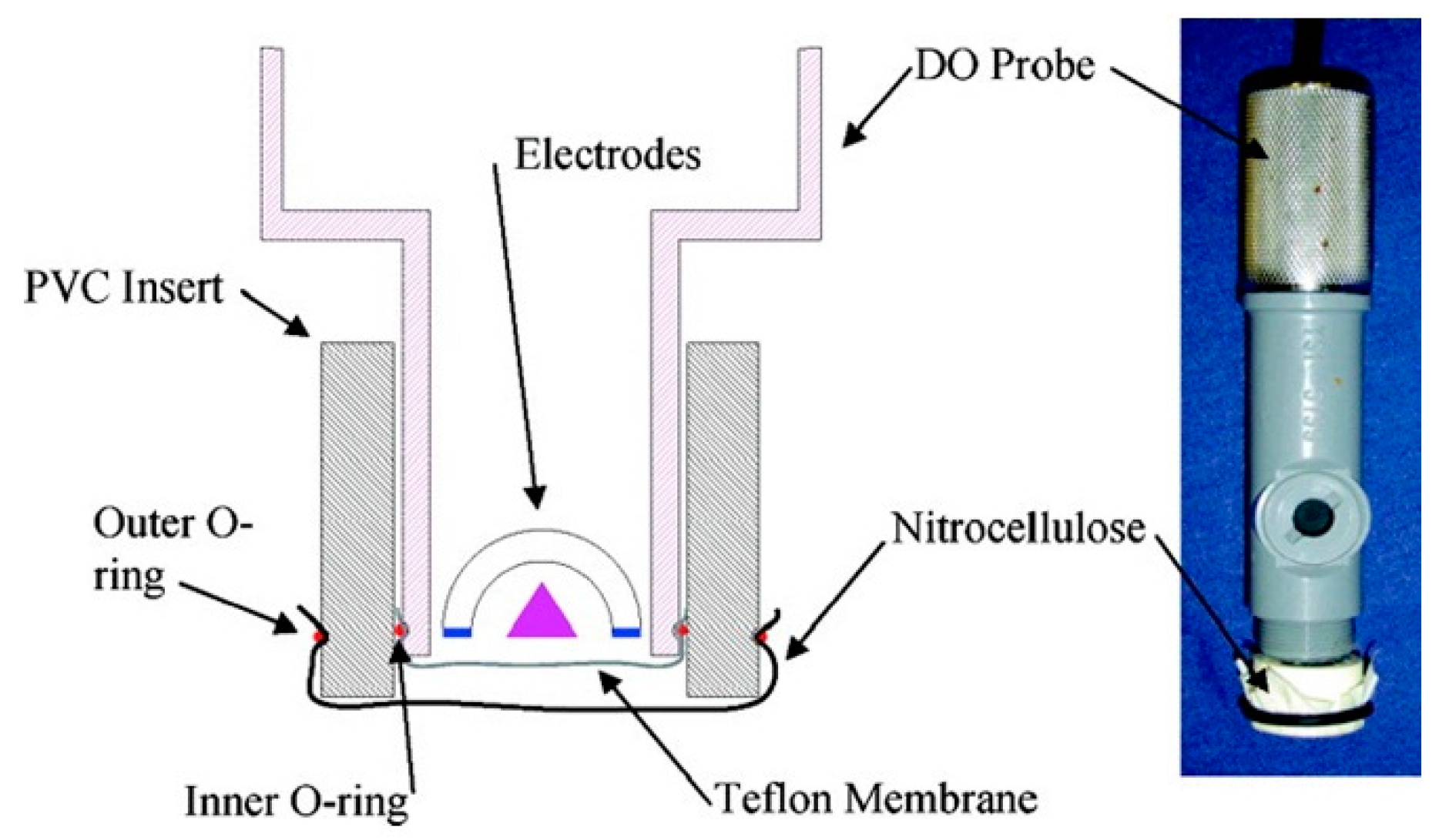
6. Detection Thresholds
| Pathogen | Detection Type | Membrane Sensor | Transduction Method | Detection Limit or Range |
|---|---|---|---|---|
| Bacillus Anthracis | RNA | Polyethersulfone membrane with linked ssDNA probe | Reflectometer-based detection of dye-filled liposome linked to reporter DNA probe | 1 nM [62] |
| RNA | Polyethersulfone membrane with linked ssDNA probe | Reflectometer-based detection of dye-filled lyposome linked to reporter probe | 1.5 fmol [64] | |
| Baccilus Cereus | whole bacteria | Immunodyne ABC membranes with various linked lectins | Chemometric data analysis of pathogen binding chronocoulometry results were used to distinguish between different pathogens | not given [80] |
| whole bacteria | Composite sensor composed of glass fiber, cellulose and nitrocellulose membranes with linked capture antibodies | Detection of antigen with conductive polyaniline nanowire-conjugated antibodies and quantification via change in conductance | 10 CFU/mL [91] | |
| Bovine viral diarrhea virus | virus particle | Nanofiber nitrocellulose membranes with linked antibodies | Pathogens coated by conductive nanoparticle-conjugated antibodies were immobilized on the membrane and quantified via the change in resistance. | 103 CCID/mL [59] |
| Brucella | RNA | Nanomembrane composed of polystyrene-divinylbenzene particles with quaternary ammonium groups and polyamide/polyestertextile fiber embedded in polyethylene with linked oligonucleotide probe | Change in ion current with oligonucleotide hybridization | 1 pM [81] |
| Cherry leaf roll virus | virus particle | Cellular membrane of live bacterial cells with inserted antibodies | Change in Membrane Potential due to binding | 1 pg/mL [82] |
| Clavibacter | anion channel formation | Lipid membrane composed of octanethiol, 1,2-Dimyristoyl-sn-glycero-3-phosphocholine, spacerlipid A on a gold electrode which are then coated with phospholipid | Toxic inserted channel proteins were detection by impedance spectroscopy | not given [85] |
| Cronobacter spp. | RNA | Composite sensor composed of glass fiber, cellulose and nitrocellulose membranes with linked oligonucleotide sandwich | Visual colour change due to carbon nanoparticles bound to ssDNA | 8 ng or 3 µg/mL [55] |
| Cucumber mosaic virus | virus particle | Cellular membrane of live fibroblast cells with electroinserted antibodies | Antibody-antigen binding was quantified by the observed change in electric potential | 1 ng/mL [84] |
| virus particle | Cellular membrane of live mammalian cells with electroinserted antibodies | Antibody-antigen binding was quantified by the observed change in electric potential | 1 ng/mL [83] | |
| Cyanobacteria | microcystin MC-LR protein | Ultrabind polyethersulfone membranes with linked protein phosphatase | Microcystin inhibits PP activity, reducing production of yellow pNP from colorless pNPP substrate | 0.30 µg/mL [67] |
| microcystin MC-RR protein | Ultrabind polyethersulfone membranes with linked protein phosphatase | Microcystin inhibits PP activity, reducing production of yellow pNP from colorless pNPP substrate | 0.52 µg/mL [67] | |
| Dengue virus | virus particle | Polyethersulfone membrane with linked DNA capture probe | Reflectometer-based detection of dye-filled liposomes linked to reported probes | serotype 2–50 molecules [65] |
| RNA | Nanoporous alumina membrane with linked ssDNA probe | Change in ionic conductivity due to oligonucleotide hybridization in pores was recorded by cyclic voltammetry and DPV | 9.55 × 10−12 M [40] | |
| glycoproteins | Lipid membrane modified by Concanavalin A on and gold electrode | Binding of Dengue virus particles was observed using cyclic voltammetry and electrochemical impedance techniques | not given [90] | |
| RNA | Polyethersulfone membrane with linked DNA capture probe | Reflectometer-based detection of dye-filled liposomes linked to ssDNA reported probes | Roughly 10 PFU/mL [63] | |
| RNA | Nanomembrane composed of polystyrene-divinylbenzene particles with quaternary ammonium groups and polyamide/polyestertextile fiber embedded in polyethylene with linked oligonucleotide probe | Change in ion current with oligonucleotide hybridization | 1 pM [81] | |
| DNA sensing for pathogen detection | DNA | Nanoporous alumina membrane with linked ssDNA probe | EIS-based detection of DNA hybridization in the pores | 50 pM [35] |
| Enterobacter aerogenes | whole bacteria | Immunodyne ABC membranes with various linked lectins | Chemometric data analysis of pathogen binding chronocoulometry results were used to distinguish between different pathogens | Not given [80] |
| Escherichia coli | whole bacteria | ImmunodyneABC Nylon membranes coated with 10 different lectins | Detection of pathogen through chronocoulometric results and factor analysis for identification of 4 E. coli subspecies. | 1.8 × 107 CFU/mL [72] |
| whole bacteria | Nylon membrane used to prevent fouling of graphite–Teflon–peroxidase–ferrocene composite electrode | Change in current, due to presence or absence of catalase- based decomposition of hydrogen peroxide, was recorded by the electrode | 2 × 106 CFU/mL [69] | |
| RNA | Nanomembrane composed of polystyrene-divinylbenzene particles with quaternary ammonium groups and polyamide/polyestertextile fiber embedded in polyethylene with linked oligonucleotide probe | Change in ion current with oligonucleotide hybridization | 1 pM [81] | |
| whole bacteria | Nanoporous alumina membrane with linked antibodies | Antibody-antigen binding was quantified by impedance amplitude changes | ~1000 CFU/mL [39] | whole bacteria |
| RNA | Polyethersulfone membrane with linked ssDNA capture probe | Reflectometer-based detection of dye-filled liposomes linked to ssDNA reported probes | 5 fmol [66] | |
| virulence factors | Membranes were composed of either 2,3-di-O-phytanylglycerol-1-tetraethylene glycol-d,l-lipoic acid ester lipid, 2,3-di-Ophytanyl-sn-glycerol-1-tetra-ethylene glycol-(3-tryethoxysilane) ether lipid, or cholesterolpentaethyleneglycol and 1,2-di-O-phytanoyl-sn-glycero-3 phosphocholine or cholesterol | Bacterial toxins were detected through change in impedance caused by pore formation in the lipid bilayer | not given [51] | |
| Gold coated PDMS membrane with linked thiols | Stress-based membrane deflection detected by white light and fiber optic interferometers | Distinguish between living and dead cells [44] | whole bacteria | |
| whole bacteria | Immunodyne ABC membranes with various linked lectins | Chemometric data analysis of pathogen binding chronocoulometry results were used to distinguish between different pathogens | not given [80] | |
| whole bacteria | Vesicles formed from TRCDA and DMPC | TRCDA vesicles change colour when exposed to lipopolysaccharides from pathogens | ~108 CFU [89] | |
| Escherichia coli DH1 | DNA | Nitrocellulose membranes coated with the contents of lysed E. coli cells | PCR was performed and radiolabeled DNA probes were added to bind to the DNA from the lysed cells. The autoradiography was recorded using autoradiography film. | not given [58] |
| Escherichia coli O157:H7 | whole bacteria | Nanoporous alumina membrane with linked antibodies | Change in impedance due to antibody-antigen binding was recorded by an electrochemical analyzer | 102 CFU/mL [37] |
| whole bacteria | Nitrocellulose membrane with linked anti-E. coli O157:H7 antibody conjugated to HRP placed over oxygen probe membrane | On pathogen binding, decrease in HRP activity is recorded by a Clark-type oxygen electrode probe | 50 cells/mL [57] | |
| whole bacteria | Polypropylene microfiber membrane coated with conductive polypyrrole and linked with antibodies | Change in resistance due to antibody-antigen binding | log 0–9 CFU/mL [73] | |
| whole bacteria | Nanoporous nylon membrane with linked antibodies | Pathogen detected by photoluminescent CdSe/ZnS core/shell dendron nanocrystal-conjugated antibodies | 2.3 CFU/mL [70] | |
| whole bacteria | Nylon membrane with linked capture antibody | Sandwich ELISA with NaI, ortho-phenylenediamine and hydrogen peroxide substrates which were measured amperometrically | 100 cells/mL [71] | |
| whole bacteria | Nitrocellulose membrane with linked capture antibody | Sandwich ELISA with luminol-based chemiluminescent output | 105–106 CFU/mL [54] | |
| whole bacteria | Nanofiber nitrocellulose membranes with linked antibodies | Pathogens coated by conductive nanoparticle-conjugated antibodies were immobilized on the membrane and quantified via the change in resistance. | 61 CFU/mL [59] | |
| DNA | Aluminum anodized oxide membrane with linked | Change in ionic conductivity due to DNA hybridization in pores measured by cyclic voltammetry and impedance spectroscopy | 0.5 nM [36] | |
| whole bacteria | Composite sensor composed of glass fiber, cellulose and nitrocellulose membranes with linked capture antibodies | Visual output from sandwich ELISA using 3,3′,5,5′-tetramethylbenzidene and SuperSignal West Femto substrates | 1.8 × 103 to 1.8 × 108 CFU/mL [47] | |
| whole bacteria | Nylon membrane with linked capture antibody | Sandwich ELISA with NaI, ortho-phenylenediamine and hydrogen peroxide substrates which were measured amperometrically | 100 cells/mL [71] | |
| whole bacteria | Nitrocellulose membrane with linked capture antibody | Sandwich ELISA with luminol-based chemiluminescent output | 105–106 CFU/mL [54] | |
| whole bacteria | Nanofiber nitrocellulose membranes with linked antibodies | Pathogens coated by conductive nanoparticle-conjugated antibodies were immobilized on the membrane and quantified via the change in resistance. | 61 CFU/mL [59] | |
| DNA | Aluminum anodized oxide membrane with linked | Change in ionic conductivity due to DNA hybridization in pores measured by cyclic voltammetry and impedance spectroscopy | 0.5 nM [36] | |
| whole bacteria | Composite sensor composed of cellulose and nitrocellulose membranes with linked antibodies | Detection of antigen with conductive nanoparticle-conjugated antibodies and quantification via change in conductance | 67 CFU/mL [60] | |
| whole bacteria | Nanoporous alumina membrane with linked antibodies | Change in ionic impedance of electrolytes in nanopores due to antibody-antigen binding | 83.7 CFU/mL [42] | |
| Feline calicivirus | virus particle | Nanoporous polyacrylamide membrane used for pathogen concentration | Antibodies conjugated to fluorescent dye filled liposomes were used to quantify the pathogen | 1.6 × 105 PFU/mL [76] |
| Giardia lamblia | Giardia lamblia cysts | Gold-coated PCTE membrane filter | Immunogold labeled antigen quantified via Raman spectroscopy | 200 cysts/mL [41] |
| Hepatitis B virus | surface antigen | Nanoporous nylon membrane with linked antibodies | Pathogen detected by photoluminescent CdSe/ZnS core/shell dendron nanocrystal-conjugated antibodies | 5 ng/mL [70] |
| Human Papilloma virus | DNA | Gold membrane with linked bis-peptide nucleic acid probe | Surface acoustic wave based detection of DNA hybridization | 1.21 pg/L [43] |
| Influenza A virus | virus particle | Nitrocellulose membrane coated with antigen | Detection of antigen with magnetic bead-conjugated antibodies which were quantified with a magnetic reader | 1 to 250 ng/mL [53] |
| Legionella pneumophilla | DNA | Nanoporous alumina membrane with linked ssDNA probe | Change in ionic conductivity due to oligonucleotide hybridization in pores was recorded by cyclic voltammetry and DPV | 3.1 × 10−13 M [38] |
| Mycobacterium avium subspecies paratuberculosis | RNA | Polyethersulfone with linked oligonucleotide sandwich | Reflectometer-based detection of dye-filled liposomes linked to reported probes | 10 CFU [61] |
| whole bacteria | Composite sensor composed of glass fiber, cellulose and nitrocellulose membranes | A primary antibody and secondary conductive nanoparticle-conjugated antibody bind to the antigen, and the change in conductivity is recorded. | serum dilution of 1:80 [48] | |
| Mycobacterium parafortuitum | whole bacteria | HPC modified cellulose acetate ultrafiltration membrane with linked antibody | Fluorescently labeled secondary antibodies were used to detect the immobilized pathogen | not given [78] |
| Potato virus Y | virus particle | Cellular membrane of live mammalian cells with electroinserted antibodies | Antibody-antigen binding was quantified by the observed change in electric potential | minimum detection of 1 ng/mL [83] |
| Proteus vulgaris | whole bacteria | Immunodyne ABC membranes with various linked lectins | Chemometric data analysis of pathogen binding chronocoulometry results were used to distinguish between different pathogens | not given [80] |
| Pseudomonas aeruginosa | DNA | TiO2 and TiO2-polyethylene glycol membranes on piezoelectric quartz with linked ssDNA probe | DNA hybridization detected by shift in resonant frequency | 10−4 g/L [45] |
| virulence factors | Membranes were composed of either 2,3-di-O-phytanylglycerol-1-tetraethylene glycol-D, l-lipoic acid ester lipid, 2,3-di-Ophytanyl-sn-glycerol-1-tetra-ethylene glycol-(3-tryethoxysilane) ether lipid, or cholesterolpentaethyleneglycol and 1,2-di-O-phytanoyl-sn-glycero-3 phosphocholine or cholesterol | Bacterial toxins were detected through change in impedance caused by pore formation in the lipid bilayer | not given [51] | |
| Saccharomyces cerevisiae | whole bacteria | Immunodyne ABC membranes with various linked lectins | Chemometric data analysis of pathogen binding chronocoulometry results were used to distinguish between different pathogens | not given [80] |
| Salmonella Newport | GIII bacteriophage | Polypyrrole modified microporous polycarbonate membrane | Pathogen cells drawn into membrane pores, GIII bacteriophage added to pathogen and change in impedance recorded | not given [79] |
| Salmonella spp. | whole bacteria | Nitrocellulose membrane with linked capture antibody | Sandwich ELISA with luminol-based chemiluminescent output | 106–107 CFU/mL [54] |
| Salmonella Typhi | whole bacteria | Polycarbonate membranes with linked antibodies | Sandwich ELISA with colourimetric output from 3,3',5,5' tetramethyl benzidine-hydrogen peroxide substrates | 2 × 103 cells/mL [75] |
| Salmonella typhimurium | whole bacteria | Vesicles formed from TRCDA and DMPC | TRCDA vesicles change colour when exposed to lipolysaccharides from pathogens | ~108 CFU [89] |
| whole bacteria | Nitrocellulose membrane coated with | Urease, linked to bacteria on the surface, converts urea to ammonia and CO2 which results in a pH change which is measured as a change in electric potential | 119 CFU [52] | |
| whole bacteria | Composite sensor composed of glass fiber, cellulose and nitrocellulose membranes with linked capture antibodies | Visual output from sandwich ELISA using chemiluminescent substrate solution quantified by CMOS image sensor | 4.22 × 103 CFU/mL and 1.1 × 102 CFU/mL with pre-separation and concentration [49] | |
| Shigella sonei | whole bacteria | Vesicles formed from TRCDA and DMPC | TRCDA vesicles change colour when exposed to lipolysaccharides from pathogens | ~108 CFU [89] |
| Stapholococcus aureus | whole bacteria | Polyethersulfone membrane | Pathogen cells were labeled with HRP conjugated antibodies, collected by the membrane and quantified by a luminol-based luminescent reaction | 3.8 × 104 CFU/mL [68] |
| DNA (enterotoxins B gene) | Membranes composed of egg phosphatidylcholine, cholesterol and hexadecylamine with linked ssDNA probes | DNA hybridization detected by change in current through the membrane | 20 ng/mL [87] | |
| whole bacteria | Immunodyne ABC membranes with various linked lectins | Chemometric data analysis of pathogen binding chronocoulometry results were used to distinguish between different pathogens | not given [80] | |
| virulence factors | Membranes were composed of either 2,3-di-O-phytanylglycerol-1-tetraethylene glycol-D,L-lipoic acid ester lipid, 2,3-di-Ophytanyl-sn-glycerol-1-tetra-ethylene glycol-(3-tryethoxysilane) ether lipid, or cholesterolpentaethyleneglycol and 1,2-di-O-phytanoyl-sn-glycero-3 phosphocholine or cholesterol | Bacterial toxins were detected through change in impedance caused by pore formation in the lipid bilayer | ~240 pM [51] | |
| whole bacteria | Nanoporous alumina membrane with linked antibodies | Antibody-antigen binding was quantified by impedance amplitude changes | ~1000 CFU/mL [39] | |
| whole bacteria | Celluloseacetate membrane filters | Pathogen-antibody/gold nanoparticle/magnetic nanoparticle complexes were filtered through the membrane and the colour change was quantified by the optical density. | 1.5 × 103 CFU for pure bacteria and 1.5 × 105 CFU in milk [77] | |
| whole bacteria | Nanoporous alumina membrane with linked antibodies | Change in impedance due to antibody-antigen binding was recorded by an electrochemical analyzer | 102 CFU/mL [37] | |
| Streptococcus pneumoniae | whole bacteria | Nylon membrane used to prevent fouling of graphite–Teflon–peroxidase–ferrocene composite electrode | Change in current, due to presence or absence of catalase- based decomposition of hydrogen peroxide, was recorded by the electrode | 2 × 105 cfu/mL [69] |
| Tobacco mosaic virus | virus particle | Cellular membrane of live bacterial cells with electroinserted antibodies | Antibody-antigen binding was quantified by the observed change in electric potential | 1 pg/mL [82] |
| Tobacco rattle virus | virus particle | Cellular membrane of live mammalian cells with electroinserted antibodies | Antibody-antigen binding was quantified by the observed change in electric potential | 1 ng/mL [83] |
| Vibrio cholerae | DNA | Composite sensor composed of glass fiber and cellulose membranes with linked oligonucleotide sandwich | Visual colour change due to gold nanoparticles bound to ssDNA | 5 ng or 250 ng/mL [46] |
| cholera toxin protein complex | Lipid membrane composed of octanethiol on a gold electrode which was then coated with DPPC and GM1 | Cholera toxin induced liposome agglutination on the piezoelectric sensor was detected by the resonant frequency shift | 25 ng/mL [86] | |
| cholera toxin protein complex | Polydiacetylene liposomes with incorporated ganglioside, GM1 | Cholera toxin induces a change in the liposome light absorption | not given [51] | |
| Yersinia pestis | whole bacteria | Composite sensor composed of glass fiber, cellulose and nitrocellulose membranes with linked capture antibodies | Secondary antibodies conjugated to up-converting phosphor particles were excited, and the resultant luminescence was quantified by a photomultiplier tube | 104 CFU/mL [50] |
7. Conclusions
Conflicts of Interest
References
- Steffan, R.J.; Atlas, R.M. DNA amplification to enhance detection of genetically engineered bacteria in environmental-samples. Appl. Environ. Microb. 1988, 54, 2185–2191. [Google Scholar]
- Janyapoon, K.; Korbsrisate, S.; Thamapa, H.; Thongmin, S.; Kanjanahareutai, S.; Wongpredee, N.; Sarasombath, S. Rapid detection of salmonella enterica serovar choleraesuis in blood cultures by a dot blot enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 2000, 7, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Downes, F.P.; Green, J.H.; Greene, K.; Strockbine, N.; Wells, J.G.; Wachsmuth, I.K. Development and evaluation of enzyme-linked immunosorbent assays for detection of shiga-like toxin I and shiga-like toxin II. J. Clin. Microbiol. 1989, 27, 1292–1297. [Google Scholar] [PubMed]
- Basta, M.; Karmali, M.; Lingwood, C. Sensitive receptor-specified enzyme-linked immunosorbent assay for Escherichia coli verocytotoxin. J. Clin. Microbiol. 1989, 27, 1617–1622. [Google Scholar] [PubMed]
- Jones, M.E.; Fox, A.J.; Barnes, A.J.; Oppenheim, B.A.; Balagopal, P.; Morgenstern, G.R.; Scarffe, J.H. PCR-ELISA for the early diagnosis of invasive pulmonary aspergillus infection in neutropenic patients. J. Clin. Pathol. 1998, 51, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Berrang, M.E.; Liu, T.; Hofacre, C.L.; Sanchez, S.; Wang, L.; Maurer, J.J. Rapid detection of Campylobacter coli, C. jejuni, and Salmonella enterica on poultry carcasses by using PCR-enzyme-linked immunosorbent assay. Appl. Environ. Microbiol. 2003, 69, 3492–3499. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, K.; Shipley, M.; Stiles, B.; Hadfield, T.L.; Sofi Ibrahim, M. Identification of staphylococcus aureus enterotoxins a and b genes by PCR-ELISA. Mol. Cell. Probes 2000, 14, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Baez, L.A.; Juneja, V.K.; Sackitey, S.K. Chemiluminescent enzyme immunoassay for detection of PCR-amplified enterotoxin a from clostridium perfringens. Int. J. Food Microbiol. 1996, 32, 145–158. [Google Scholar] [CrossRef]
- Abdel-Hamid, I.; Atanasov, P.; Ghindilis, A.L.; Wilkins, E. Development of a flow-through immunoassay system. Sens. Actuators B Chem. 1998, 49, 202–210. [Google Scholar] [CrossRef]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Fung, Y.S.; Wong, Y.Y. Self-assembled monolayers as the coating in a quartz piezoelectric crystal immunosensor to detect salmonella in aqueous solution. Anal. Chem. 2001, 73, 5302–5309. [Google Scholar] [CrossRef] [PubMed]
- Ilic, B.; Czaplewski, D.; Zalalutdinov, M.; Craighead, H.G.; Neuzi, P.; Campagnolo, C.; Batt, C. Single cell detection with micromechanical oscillators. J. Vac. Sci. Technol. B 2001, 19, 2825–2828. [Google Scholar] [CrossRef]
- Ilic, B.; Yang, Y.; Craighead, H.G. Virus detection using nanoelectromechanical devices. Appl. Phys. Lett. 2004, 85, 2604–2606. [Google Scholar] [CrossRef]
- Poshtiban, S.; Singh, A.; Fitzpatrick, G.; Evoy, S. Bacteriophage tail-spike protein derivitized microresonator arrays for specific detection of pathogenic bacteria. Sens. Actuators B Chem. 2013, 181, 410–416. [Google Scholar] [CrossRef]
- Campbell, G.A.; Medina, M.B.; Mutharasan, R. Detection of staphylococcus enterotoxin B at picogram levels using piezoelectric-excited millimeter-sized cantilever sensors. Sens. Actuators B Chem. 2007, 126, 354–360. [Google Scholar] [CrossRef]
- Fischer, L.M.; Wright, V.A.; Guthy, C.; Yang, N.; McDermott, M.T.; Buriak, J.M.; Evoy, S. Specific detection of proteins using nanomechanical resonators. Sens. Actuators B Chem. 2008, 134, 613–617. [Google Scholar] [CrossRef]
- Mu, C.J.; Zhang, Z.Y.; Lin, M.; Du, Y.; Cao, X.D. Detecting low concentration bacterial cells in complex media using a microchip-based flow cytometer. Sens. Actuators B Chem. 2014, 202, 1051–1057. [Google Scholar] [CrossRef]
- Gau, J.J.; Lan, E.H.; Dunn, B.; Ho, C.M.; Woo, J.C. A MEMS based amperometric detector for E. coli bacteria using self-assembled monolayers. Biosens. Bioelectron. 2001, 16, 745–755. [Google Scholar] [CrossRef]
- Gervals, L.; Gel, M.; Allain, B.; Tolba, M.; Brovko, L.; Zourob, M.; Mandeville, R.; Griffiths, M.; Evoy, S. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens. Actuators B Chem. 2007, 125, 615–621. [Google Scholar] [CrossRef]
- Taylor, A.D.; Yu, Q.; Chen, S.; Homola, J.; Jiang, S. Comparison of E. coli O157: H7 preparation methods used for detection with surface plasmon resonance sensor. Sens. Actuators B Chem. 2005, 107, 202–208. [Google Scholar] [CrossRef]
- Foudeh, A.M.; Daoud, J.T.; Faucher, S.P.; Veres, T.; Tabrizian, M. Sub-femtomole detection of 16s rRNA from legionella pneumophila using surface plasmon resonance imaging. Biosens. Bioelectron. 2014, 52, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Singh, A.; Naidoo, R.; Wu, P.; McDermott, M.T.; Evoy, S. Chemically immobilized t4-bacteriophage for specific escherichia coli detection using surface plasmon resonance. Analyst 2011, 136, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arya, S.K.; Glass, N.; Hanifi-Moghaddam, P.; Naidoo, R.; Szymanski, C.M.; Tanha, J.; Evoy, S. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 2010, 26, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.C.; Ho, J.A.A. Attomole DNA electrochemical sensor for the detection of Escherichia coli O157. Anal. Chem. 2009, 81, 2470–2476. [Google Scholar] [CrossRef] [PubMed]
- Joung, H.A.; Lee, N.R.; Lee, S.K.; Ahn, J.; Shin, Y.B.; Choi, H.S.; Lee, C.S.; Kim, S.; Kim, M.G. High sensitivity detection of 16s rRNA using peptide nucleic acid probes and a surface plasmon resonance biosensor. Anal. Chim. Acta 2008, 630, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Ohk, S.H.; Koo, O.K.; Sen, T.; Yamamoto, C.M.; Bhunia, A.K. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of listeria monocytogenes from food. J. Appl. Microbiol. 2010, 109, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.K.; Lee, W.; Chun, B.S.; Bae, Y.M.; Lee, W.H.; Choi, J.W. The fabrication of protein chip based on surface plasmon resonance for detection of pathogens. Biosens. Bioelectron. 2005, 20, 1847–1850. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.D.; Ladd, J.; Yu, Q.M.; Chen, S.F.; Homola, J.; Jiang, S.Y. Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens. Bioelectron. 2006, 22, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Sorokulova, I.B.; Vodyanoy, V.J.; Simonian, A.L. Lytic phage as a specific and selective probe for detection of staphylococcus aureus—A surface plasmon resonance spectroscopic study. Biosens. Bioelectron. 2007, 22, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.H.; Johnson, M.L.; Guntupalli, R.; Petrenko, V.A.; Chin, B.A. Detection of bacillus anthracis spores in liquid using phage-based magnetoelastic micro-resonators. Sens. Actuators B Chem. 2007, 127, 559–566. [Google Scholar] [CrossRef]
- Singh, A.; Arutyunov, D.; Szymanski, C.M.; Evoy, S. Bacteriophage based probes for pathogen detection. Analyst 2012, 137, 3405–3421. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arutyunov, D.; McDermott, M.T.; Szymanski, C.M.; Evoy, S. Specific detection of campylobacter jejuni using the bacteriophage NCTC 12673 receptor binding protein as a probe. Analyst 2011, 136, 4780–4786. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. Aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Chibli, H.; Ghali, H.; Park, S.; Peter, Y.A.; Nadeau, J.L. Immobilized phage proteins for specific detection of staphylococci. Analyst 2014, 139, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.W.; Shi, J.Y.; Chan, C.Y.; Zhang, Y.; Yang, M. A nanoporous membrane based impedance sensing platform for DNA sensing with gold nanoparticle amplification. Sens. Actuators B Chem. 2014, 193, 877–882. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Q.; Hu, Z.; Zhang, Y.; Wu, C.; Yang, M.; Wang, P. A novel electrochemical biosensor based on dynamic polymerase-extending hybridization for E. coli O157: H7 DNA detection. Talanta 2009, 78, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Leung, P.H.M.; Liu, Z.B.; Zhang, Y.; Xiao, L.; Ye, W.; Zhang, X.; Yi, L.; Yang, M. A PDMS microfluidic impedance immunosensor for E. coli O157: H7 and staphylococcus aureus detection via antibody-immobilized nanoporous membrane. Sens. Actuators B Chem. 2011, 159, 328–335. [Google Scholar] [CrossRef]
- Rai, V.; Deng, J.; Toh, C.S. Electrochemical nanoporous alumina membrane-based label-free DNA biosensor for the detection of legionella sp. Talanta 2012, 98, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Tan, F. Foodborne Pathogens Detection with Nanoporous Anodic Aluminum Oxide Membrane Based Biosensor; Hong Kong Polytechnic University: Hong Kong, China, 2012. [Google Scholar]
- Rai, V.; Hapuarachchi, H.C.; Ng, L.C.; Soh, S.H.; Leo, Y.S.; Toh, C.S. Ultrasensitive cDNA detection of dengue virus RNA using electrochemical nanoporous membrane-based biosensor. PLoS ONE 2012, 7, e42346. [Google Scholar] [CrossRef] [PubMed]
- Wigginton, K.R.; Vikesland, P.J. Gold-coated polycarbonate membrane filter for pathogen concentration and sers-based detection. Analyst 2010, 135, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Joung, C.K.; Kim, H.N.; Lim, M.C.; Jeon, T.J.; Kim, H.Y.; Kim, Y.R. A nanoporous membrane-based impedimetric immunosensor for label-free detection of pathogenic bacteria in whole milk. Biosens. Bioelectron. 2013, 44, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Chen, M.; Luo, Y.; Deng, K.; Chen, D.; Fu, W. A new system for the amplification of biological signals: Reca and complimentary single strand DNA probes on a leaky surface acoustic wave biosensor. Biosens. Bioelectron. 2014, 60, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Witte, H. A novel PDMS micro membrane biosensor based on the analysis of surface stress. Biosens. Bioelectron. 2010, 25, 2420–2424. [Google Scholar] [CrossRef] [PubMed]
- He, F.J.; Liu, S.Q. Detection of P. aeruginosa using nano-structured electrode-separated piezoelectric DNA biosensor. Talanta 2004, 62, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Chua, A.L.; Yean, C.Y.; Ravichandran, M.; Lim, B.; Lalitha, P. A rapid DNA biosensor for the molecular diagnosis of infectious disease. Biosens. Bioelectron. 2011, 26, 3825–3831. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.; Paek, S.H.; Hong, J.W.; Kim, Y.K. Enzyme-linked immuno-strip biosensor to detect Escherichia coli O157: H7. Ultramicroscopy 2008, 108, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Karthik, K.; Das, P.; Murugan, M.S.; Singh, P. Evaluation of bioelectronics sensor compared to other diagnostic test in diagnosis of johneʼs disease in goats. Small Rumin. Res. 2013, 109, 56–63. [Google Scholar] [CrossRef]
- Jeon, J.W.; Kim, J.H.; Lee, J.M.; Lee, W.H.; Lee, D.Y.; Paek, S.H. Rapid immuno-analytical system physically integrated with lens-free CMOS image sensor for food-borne pathogens. Biosens. Bioelectron. 2014, 52, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Zhou, L.; Zhao, Y.; Wang, J.; Huang, L.; Hu, K.; Liu, H.; Wang, H.; Guo, Z.; Song, Y.; et al. Rapid quantitative detection of yersinia pestis by lateral-flow immunoassay and up-converting phosphor technology-based biosensor. Sens. Actuators B Chem. 2006, 119, 656–663. [Google Scholar] [CrossRef]
- Thet, N.T. Modified Tethered Bilayer Lipid Membranes for Detection of Pathogenic Bacterial Toxins and Characterization of Ion Channels. Ph.D. Thesis, University of Bath, Bath, UK, 2010. [Google Scholar]
- Dill, K.; Stanker, L.H.; Young, C.R. Detection of salmonella in poultry using a silicon chip-based biosensor. J. Biochem. Biophys. Methods 1999, 41, 61–67. [Google Scholar] [CrossRef]
- Hong, H.B.; Krause, H.J.; Song, K.B.; Choi, C.J.; Chung, M.A.; Son, S.W.; Offenhaeusser, A. Detection of two different influenza a viruses using a nitrocellulose membrane and a magnetic biosensor. J. Immunol. Methods 2011, 365, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Karoonuthaisiri, N.; Charlermroj, R.; Uawisetwathana, U.; Luxananil, P.; Kirtikara, K.; Gajanandana, O. Development of antibody array for simultaneous detection of foodborne pathogens. Biosens. Bioelectron. 2009, 24, 1641–1648. [Google Scholar] [CrossRef] [PubMed]
- Blazkova, M.; Javurkova, B.; Fukal, L.; Rauch, P. Immunochromatographic strip test for detection of genus cronobacter. Biosens. Bioelectron. 2011, 26, 2828–2834. [Google Scholar] [CrossRef] [PubMed]
- Low, S.C.; Ahmad, A.L.; Ideris, N.; Ng, Q.H. Interaction of isothermal phase inversion and membrane formulation for pathogens detection in water. Bioresour. Technol. 2012, 113, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Theegala, C.S.; Small, D.D.; Monroe, W.T. Oxygen electrode-based single antibody amperometric biosensor for qualitative detection of E. coli and bacteria in water. J. Environ. Sci. Health Part A 2008, 43, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Toranzos, G.A.; Alvarez, A.J. Solid-phase polymerase chain-reaction-applications for direct detection of enteric pathogens in waters. Can. J. Microbiol. 1992, 38, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Nartker, S.; Miller, H.; Hochhalter, D.; Wiederoder, M.; Wiederoder, S.; Setterington, E.; Drzal, L.T.; Alocilja, E.C. Surface functionalization of electrospun nanofibers for detecting E. coli O157: H7 and BVDV cells in a direct-charge transfer biosensor. Biosens. Bioelectron. 2010, 26, 1612–1617. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Nartker, S.; Wiederoder, M.; Miller, H.; Hochhalter, D.; Drzal, L.T.; Alocilja, E.C. Novel biosensor based on electrospun nanofiber and magnetic nanoparticles for the detection of E. coli O157: H7. IEEE Trans. Nanotechnol. 2012, 11, 676–681. [Google Scholar] [CrossRef]
- Kumanan, V.; Nugen, S.R.; Baeumner, A.J.; Chang, Y.F. A biosensor assay for the detection of mycobacterium avium subsp paratuberculosis in fecal samples. J. Vet. Sci. 2009, 10, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Baeumner, A.J.; Leonard, B.; McElwee, J.; Montagna, R.A. A rapid biosensor for viable B-anthracis spores. Anal. Bioanal. Chem. 2004, 380, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Baeumner, A.J.; Schlesinger, N.A.; Slutzki, N.S.; Romano, J.; Lee, E.M.; Montagna, R.A. Biosensor for dengue virus detection: Sensitive, rapid, and serotype specific. Anal. Chem. 2002, 74, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Hartley, H.A.; Baeumner, A.J. Biosensor for the specific detection of a single viable B-anthracis spore. Anal. Bioanal. Chem. 2003, 376, 319–327. [Google Scholar] [PubMed]
- Zaytseva, N.V.; Montagna, R.A.; Lee, E.M.; Baeumner, A.J. Multi-analyte single-membrane biosensor for the serotype-specific detection of dengue virus. Anal. Bioanal. Chem. 2004, 380, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Baeumner, A.J.; Cohen, R.N.; Miksic, V.; Min, J.H. RNA biosensor for the rapid detection of viable Escherichia coli in drinking water. Biosens. Bioelectron. 2003, 18, 405–413. [Google Scholar] [CrossRef]
- Campas, M.; Szydlowska, D.; Trojanowicz, M.; Marty, J.L. Towards the protein phosphatase-based biosensor for microcystin detection. Biosens. Bioelectron. 2005, 20, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.M.; Pivarnik, P.E.; Senecal, A.G.; Rand, A.G. Rapid detection of staphylococcus aureus using a membrane fiber optic biosensor. In Pathogen Detection and Remediation for Safe Eating; Chen, Y.R., Ed.; SPIE: Bellingham, WA, USA, 1999; pp. 2–9. [Google Scholar]
- Serra, B.; Zhang, J.; Morales, M.D.; Guzman-Vazquez de Prada, A.; Reviejo, A.J.; Pingarron, J.M. A rapid method for detection of catalase-positive and catalase-negative bacteria based on monitoring of hydrogen peroxide evolution at a composite peroxidase biosensor. Talanta 2008, 75, 1134–1139. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Brandon, R.; Cate, M.; Peng, X.; Stony, R.; Johnson, M. Detection of pathogens using luminescent CdSe/ZnS dendron nanocrystals and a porous membrane immunofilter. Anal. Chem. 2007, 79, 8796–8802. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamid, I.; Ivnitski, D.; Atanasov, P.; Wilkins, E. Flow-through immunofiltration assay system for rapid detection of E. coli O157: H7. Biosens. Bioelectron. 1999, 14, 309–316. [Google Scholar] [CrossRef]
- Ertl, P.; Wagner, M.; Corton, E.; Mikkelsen, S.R. Rapid identification of viable Escherichia coli subspecies with an electrochemical screen-printed biosensor array. Biosens. Bioelectron. 2003, 18, 907–916. [Google Scholar] [CrossRef]
- McGraw, S.K.; Alocilja, E.; Senecal, K.; Senecal, A. A resistance based biosensor that utilizes conductive microfibers for microbial pathogen detection. Open J. Appl. Biosens. 2012, 1, 36–43. [Google Scholar] [CrossRef]
- Li, D.P.; Frey, M.W.; Baeumner, A.J. Electrospun polylactic acid nanofiber membranes as substrates for biosensor assemblies. J. Membr. Sci. 2006, 279, 354–363. [Google Scholar] [CrossRef]
- Jain, S.; Chattopadhyay, S.; Jackeray, R.; Abid, C.K.V.Z.; Kohli, G.S.; Singh, H. Highly sensitive detection of salmonella typhi using surface aminated polycarbonate membrane enhanced-ELISA. Biosens. Bioelectron. 2012, 31, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Connelly, J.T.; Kondapalli, S.; Skoupi, M.; Parker, J.S.L.; Kirby, B.J.; Baeumner, A.J. Micro-total analysis system for virus detection: Microfluidic pre-concentration coupled to liposome-based detection. Anal. Bioanal. Chem. 2012, 402, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.J.; Suk, H.J.; Sung, H.Y.; Li, T.; Poo, H.; Kim, M.G. Novel antibody/gold nanoparticle/magnetic nanoparticle nanocomposites for immunomagnetic separation and rapid colorimetric detection of staphylococcus aureus in milk. Biosens. Bioelectron. 2013, 43, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Gorey, C.; Zaky, A.; Escobar, I.; Gruden, C. Thermally responsive membrane-based microbiological sensing component for early detection of membrane biofouling. Desalination 2011, 270, 116–123. [Google Scholar] [CrossRef]
- Dadarwal, R.; Namvar, A.; Thomas, D.F.; Hall, J.C.; Warriner, K. Organic conducting polymer electrode based sensors for detection of salmonella infecting bacteriophages. Mater. Sci. Eng. C 2009, 29, 761–765. [Google Scholar] [CrossRef]
- Ertl, P.; Mikkelsen, S.R. Electrochemical biosensor array for the identification of microorganisms based on lectin-lipopolysaccharide recognition. Anal. Chem. 2001, 73, 4241–4248. [Google Scholar] [CrossRef] [PubMed]
- Senapati, S.; Slouka, Z.; Shah, S.S.; Behura, S.K.; Shi, Z.; Stack, M.S.; Severson, D.W.; Chang, H.C. An ion-exchange nanomembrane sensor for detection of nucleic acids using a surface charge inversion phenomenon. Biosens. Bioelectron. 2014, 60, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Gramberg, B.; Kintzios, S.; Schmidt, U.; Mewis, I.; Ulrichs, C. A basic approach towards the development of bioelectric bacterial biosensors for the detection of plant viruses. J. Phytopathol. 2012, 160, 106–111. [Google Scholar] [CrossRef]
- Perdikaris, A.; Vassilakos, N.; Yiakoumettis, I.; Kektsidou, O.; Kintzios, S. Development of a portable, high throughput biosensor system for rapid plant virus detection. J. Virol. Methods 2011, 177, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Moschopoulou, G.; Vitsa, K.; Bem, F.; Vassilakos, N.; Perdikaris, A.; Blouhos, P.; Yialouris, C.; Frosyniotis, D.; Anthopoulos, I.; Mangana, O.; et al. Engineering of the membrane of fibroblast cells with virus-specific antibodies: A novel biosensor tool for virus detection. Biosens. Bioelectron. 2008, 24, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Michalke, A.; Galla, H.J.; Steinem, C. Channel activity of a phytotoxin of clavibacter michiganense ssp. nebraskense in tethered membranes. Eur. Biophys. J. Biophys. Lett. 2001, 30, 421–429. [Google Scholar] [CrossRef]
- Chen, H.; Hu, Q.Y.; Yue, Z.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Construction of supported lipid membrane modified piezoelectric biosensor for sensitive assay of cholera toxin based on surface-agglutination of ganglioside-bearing liposomes. Anal. Chim. Acta 2010, 657, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Gao, Z.; Zhou, H.; Yue, M. Detection of SEB gene by bilayer lipid membranes nucleic acid biosensor supported by modified patch-clamp pipette electrode. Biosens. Bioelectron. 2007, 22, 2371–2376. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.J.; Charych, D. Molecular recognition and optical detection of biological pathogens at biomimetic membrane interfaces. Smart Struct. Mater. 1997, 3040, 211–217. [Google Scholar]
- Villalobos, P.; Chavez, M.I.; Olguin, Y.; Sanchez, E.; Valdes, E.; Galindo, R.; Young, M.E. The application of polymerized lipid vesicles as colorimetric biosensors for real-time detection of pathogens in drinking water. Electron. J. Biotechnol. 2012, 15. [Google Scholar] [CrossRef]
- Luna, D.M.N.; Oliveira, M.D.L.; Nogueira, M.L.; Andrade, C.A.S. Biosensor based on lectin and lipid membranes for detection of serum glycoproteins in infected patients with dengue. Chem. Phys. Lipids 2014, 180, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Alocilja, E.C.; Downes, F.P. Nanowire labeled direct-charge transfer biosensor for detecting bacillus species. Biosens. Bioelectron. 2007, 22, 2329–2336. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Gusarov, S.; Evoy, S.; Kovalenko, A. Electronic structure, binding energy, and solvation structure of the streptavidin-biotin supramolecular complex: ONIOM and 3d-RISM study. J. Phys. Chem. B 2009, 113, 9958–9967. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hurk, R.V.d.; Evoy, S. A Review of Membrane-Based Biosensors for Pathogen Detection. Sensors 2015, 15, 14045-14078. https://doi.org/10.3390/s150614045
Hurk RVd, Evoy S. A Review of Membrane-Based Biosensors for Pathogen Detection. Sensors. 2015; 15(6):14045-14078. https://doi.org/10.3390/s150614045
Chicago/Turabian StyleHurk, Remko Van den, and Stephane Evoy. 2015. "A Review of Membrane-Based Biosensors for Pathogen Detection" Sensors 15, no. 6: 14045-14078. https://doi.org/10.3390/s150614045
APA StyleHurk, R. V. d., & Evoy, S. (2015). A Review of Membrane-Based Biosensors for Pathogen Detection. Sensors, 15(6), 14045-14078. https://doi.org/10.3390/s150614045







