Microfluidics Integrated Biosensors: A Leading Technology towards Lab-on-a-Chip and Sensing Applications
Abstract
:1. Introduction
2. Biosensors
2.1. Biosensors Categorized Based on the Type of Biological Recognition Element and Immobilization Techniques

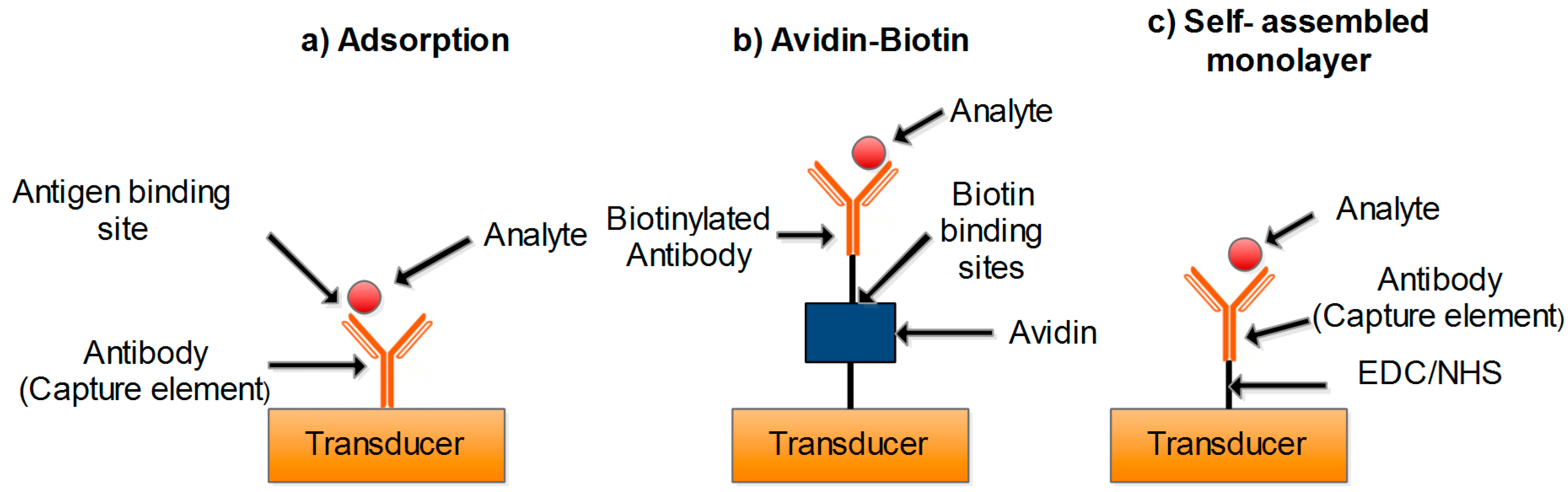
2.1.1. Enzyme-Based Biosensors
2.1.2. Antibody-Based Biosensor
2.1.3. Aptamer-Based Biosensor
2.2. Biosensors Categorized Based on the Type of Transducers
2.2.1. Electrochemical-Based Biosensors
| Transducer | Technique | Advantages | Disadvantages |
|---|---|---|---|
| Electrochemical | Amperometric [43,44] | Simplicity, miniaturization, low cost | Need redox elements to enhance the current production; time consuming; sensitive to the surrounding environment |
| Potentiometric [45,46] | Real-time detection; the possibility of continuous analysis on different analytes | Sensitive to the surrounding environment; time consuming; sensitive to temperature | |
| Impedimetric [47,48] | Simplicity and real-time detection | Sensitive to the surrounding environment; bulky devices required; require theoretical stimulation for data analysis | |
| Optical | Surface plasmon resonance (SPR) [49,50] | Real-time detection; reliable, high sensitivity | Sensitive to the surrounding environment; surface modification as one of the main challenges; bulky optical devices required |
| Mechanical | Cantilever [51,52] | Real-time detection; ability to detect more than one analyte with high sensitivity | Sensitive to the surrounding environment; sensitive to temperature; bulky devices required |
| Quartz crystal microbalance (QCM) [53,54] | Real-time detection; simplicity; high compatibility with point-of-care (POC) devices | Sensitive to the surrounding environment; sensitive to temperature and stress |
2.2.2. Optical-Based Biosensors
2.2.3. Colorimetric Biosensors
2.2.4. Mass Biosensors
2.2.5. Magnetic Sensors
3. Microfluidics
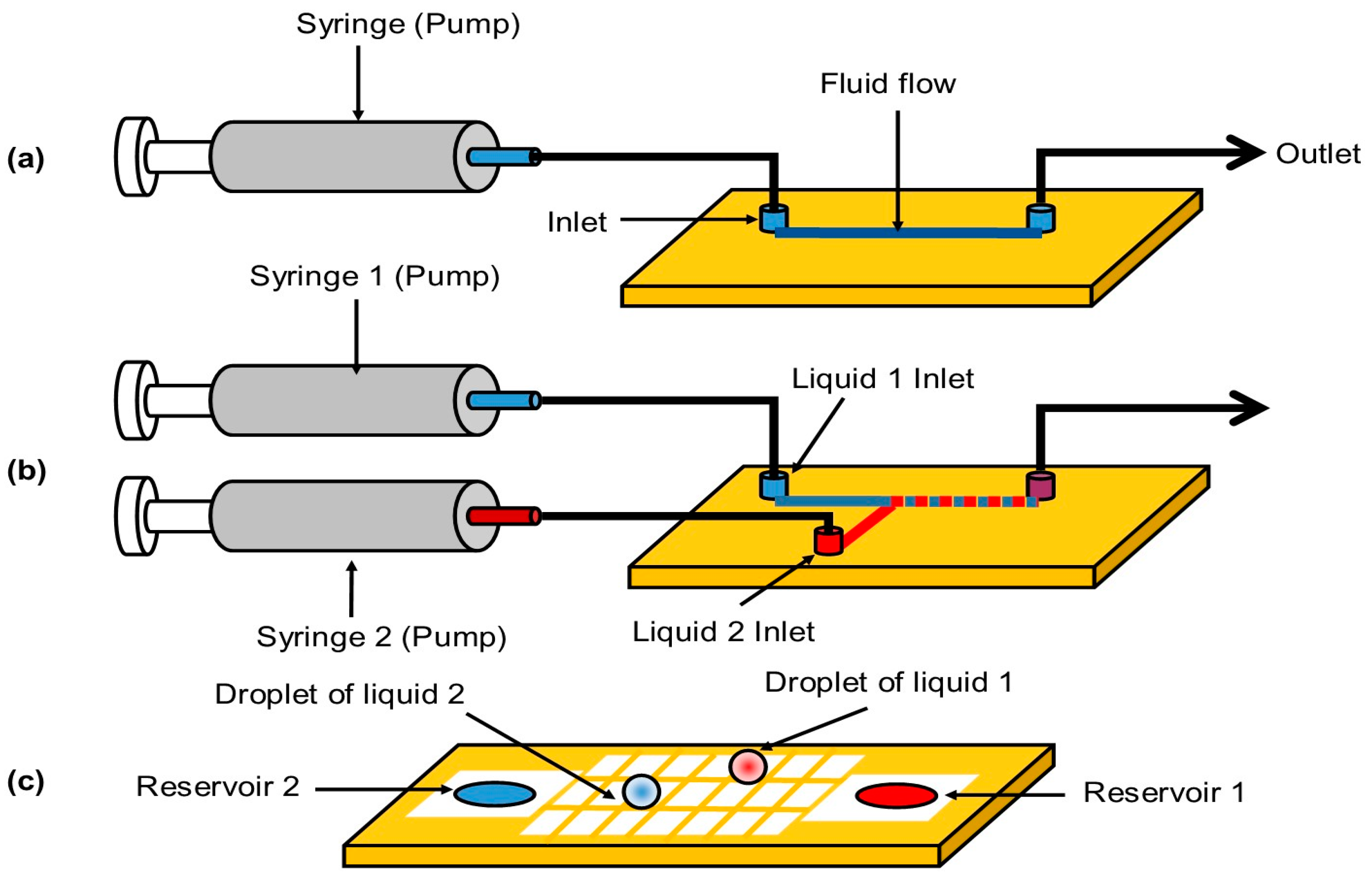
| Continuous-Flow Microfluidics | Droplet-Based Microfluidics | Digital Microfluidics | |
|---|---|---|---|
| Operating Method | Motion of continuous fluid in micro-channels | Motion of droplets in micro-channels using streams of immiscible fluids | Motion of discrete droplets on an array of planar electrodes |
| Flow Actuation | Mechanical (syringe) pumps, Pneumatic pressure, Electrokinetic | Mechanical (syringe) pumps, Pneumatic pressure | Electrowetting On Dielectric, Dielectrophoresis |
| Advantages | Ease of fabrication and operation, suitable for applications that require a continuous flow with relatively high sampling volume, and being compatible with most of current screening and sensing mechanisms | Ease of fabrication and operation, suitable for a applications that require isolated reaction sites to avoid cross contamination | Lower sample consumption, scalability, better localization, reconfigurability, and portability |
| Disadvantages | High sample volume consumption compared to other microfluidic systems, possible contamination, and not being scalable due to fabrication and physical limitations | No control over individual droplets, challenging to create droplets of different sizes using the same setup, and challenging to implement stable gas-liquid systems | Complicated fabrication procedure, and bio-adsorption and evaporation |
| Fabrication Method | Fabrication Material | Advantages | Disadvantages |
|---|---|---|---|
| Photolithography [89] | PDMS | Portability | Low throughput |
| Cost-effective and high automation | |||
| High sensitivity | |||
| Soft lithography [90] | PDMS | Real-time detection | Requiring high sample concentration |
| Portable | |||
| Disposable | |||
| Cost-effective | |||
| Nano-imprinting [91] | PMMA | Cost-effective | Expensive Low throughput |
| High sensitivity |
4. Integration of Microfluidics with Biosensor Technology
| Biological Recognition Element | Advantages | Disadvantages |
|---|---|---|
| Enzymes [95,97] | High sensitivity | Possibility of losing their activity upon immobilization |
| High selectivity towards their targets | ||
| Suitable for oxidation reduction reactions | Most suitable for small analytes, e.g., glucose, urea and lactate | |
| Antibodies [106,107] | Rapid analysis for direct immunoassays | Requiring labeling for indirect immune assays which can result in the increase cost and time required for analysis |
| Suitable for bioaffinity interaction e.g., antibody-antigen interaction | Not suitable for detection of small targets using direct and sandwich immunoassays | |
| Suitable for the detection of large targets e.g., bacteria and pathogens | Not suitable for oxidation reduction reactions | |
| Aptamers [108,109] | Highly sensitive and selective | Higher toxicity than antibodies |
| Suitable for the detection of a wide range of analytes | Faster excretion due to their small size | |
| Long-term stability, inexpensive and rapid synthesis | Weaker binding to analytes | |
| Flexibility to be modified with labels without losing their performance or binding properties |
4.1. Continuous Microfluidic-Based Biosensor
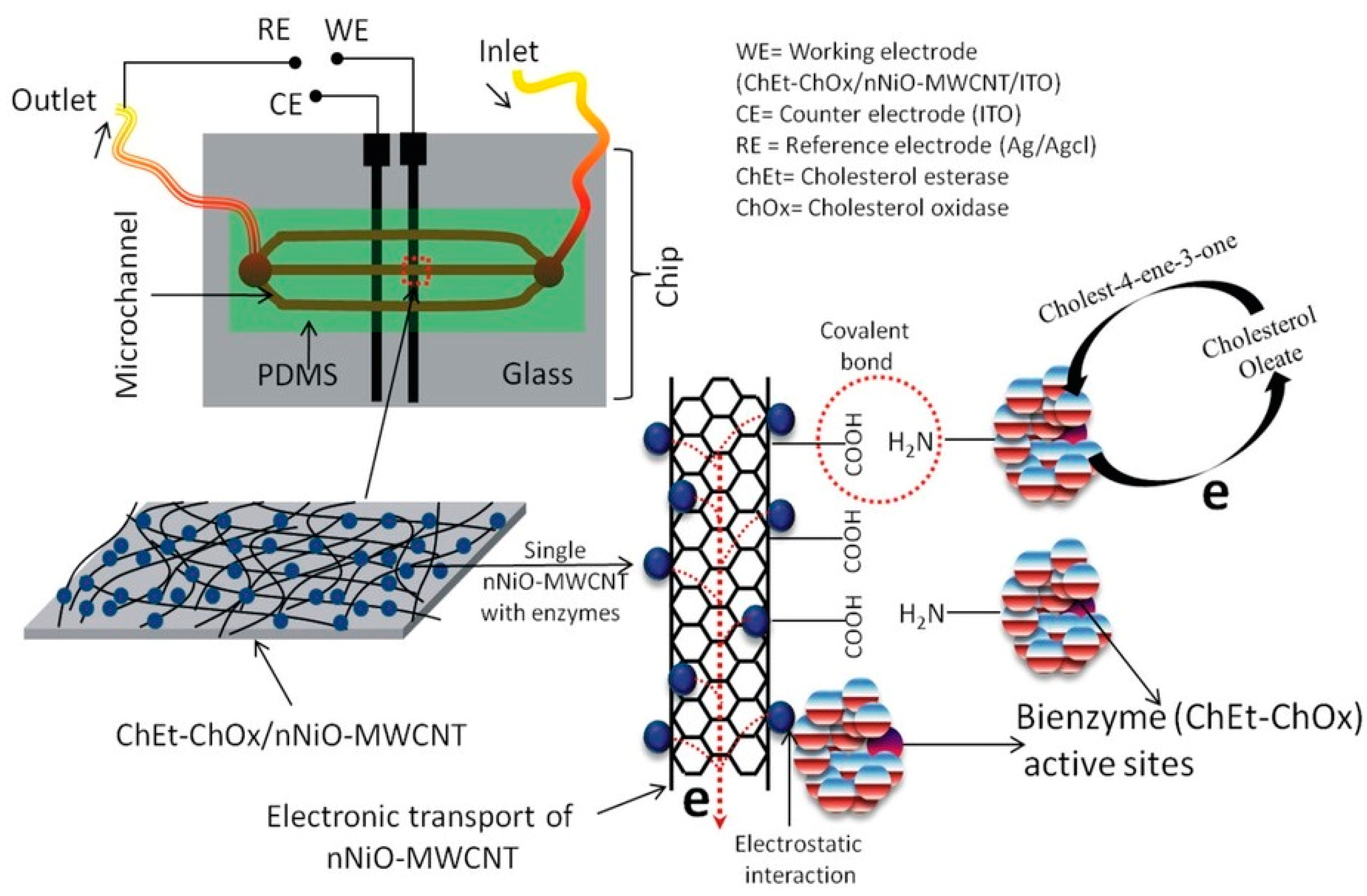
4.1.1. Enzyme-Based
4.1.2. Antibody-Based
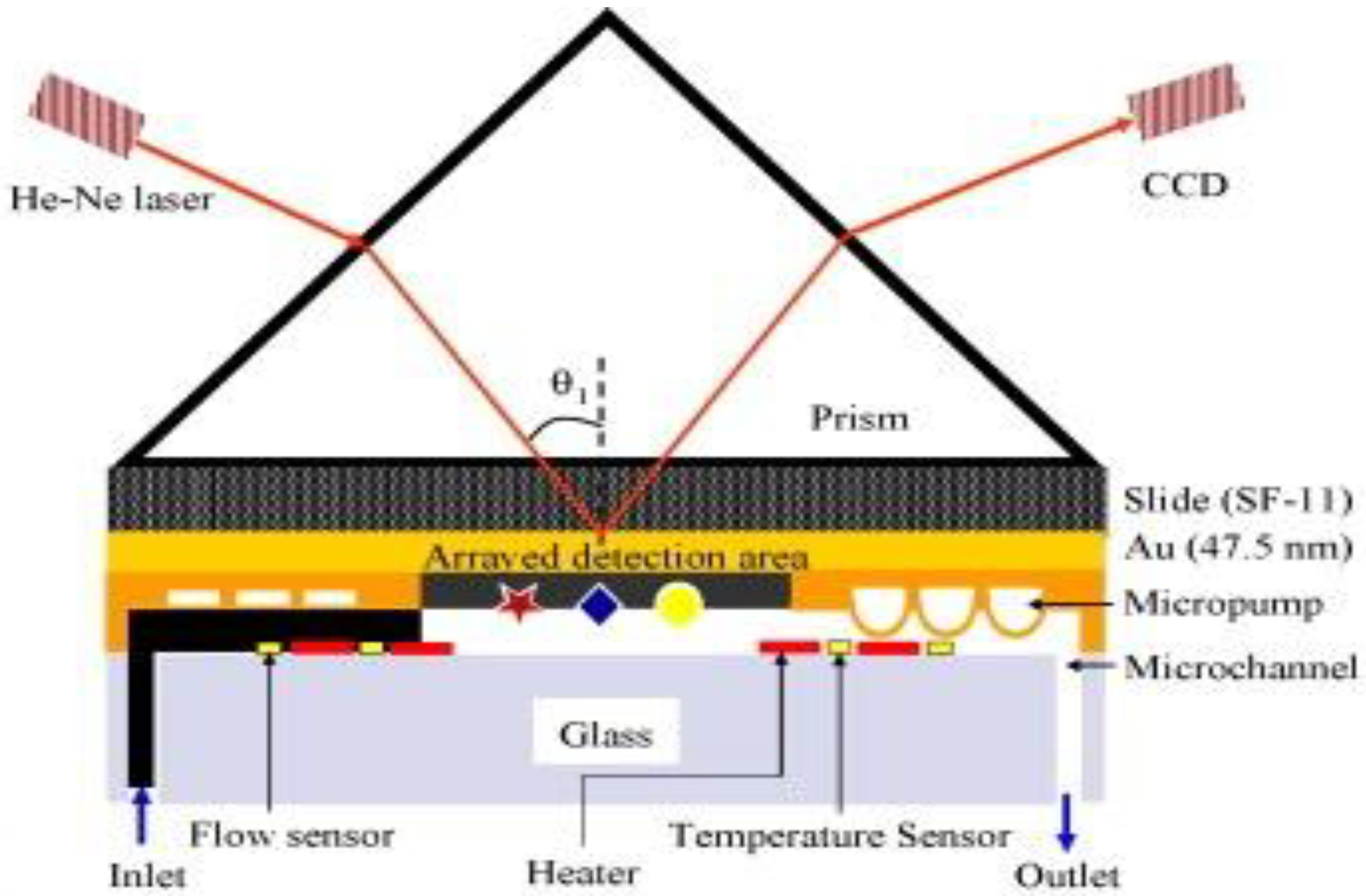
4.1.3. Aptamer-Based
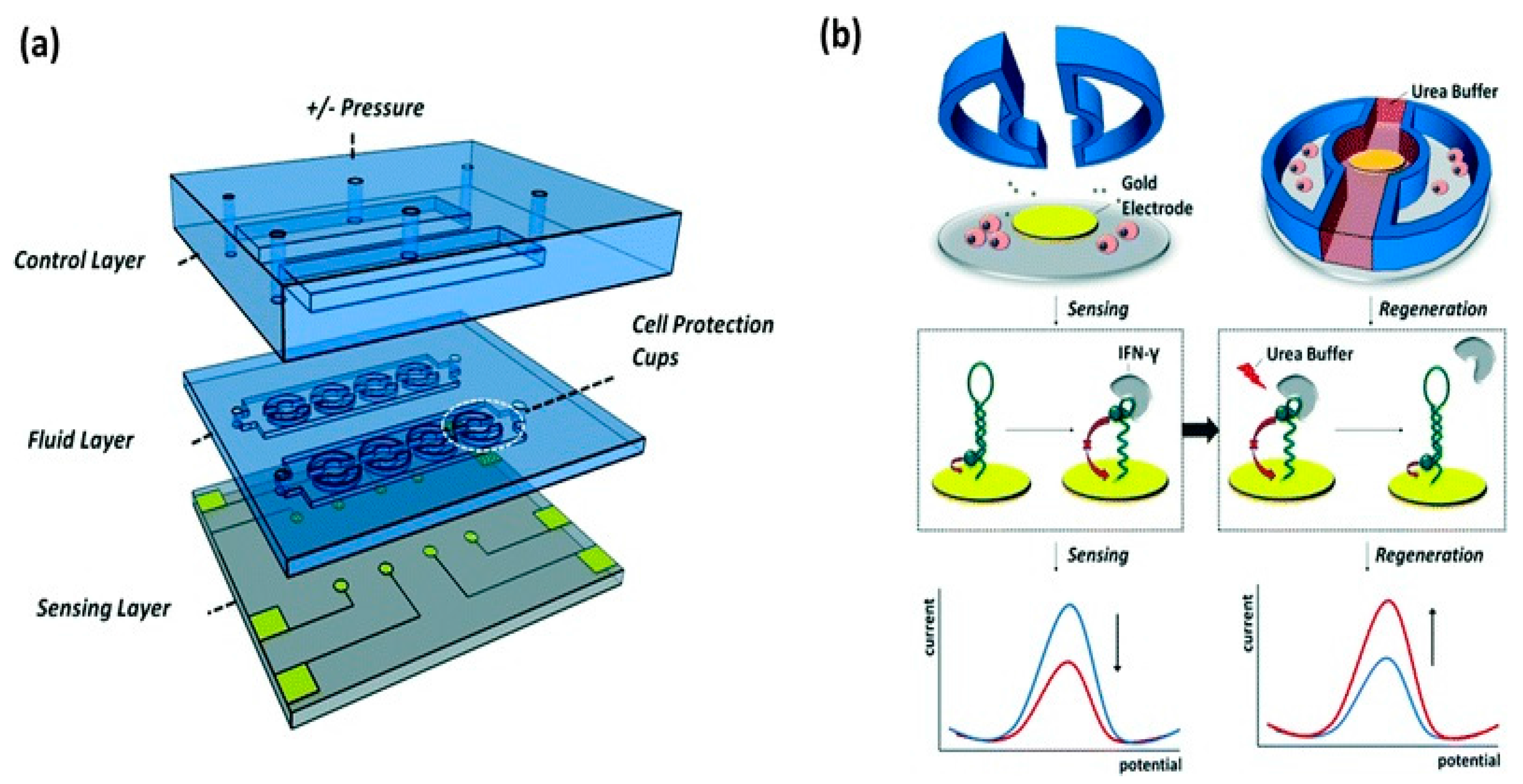
4.2. Droplet Microfluidic-Based Biosensor
4.2.1. Enzyme-Based
4.2.2. Antibody-Based
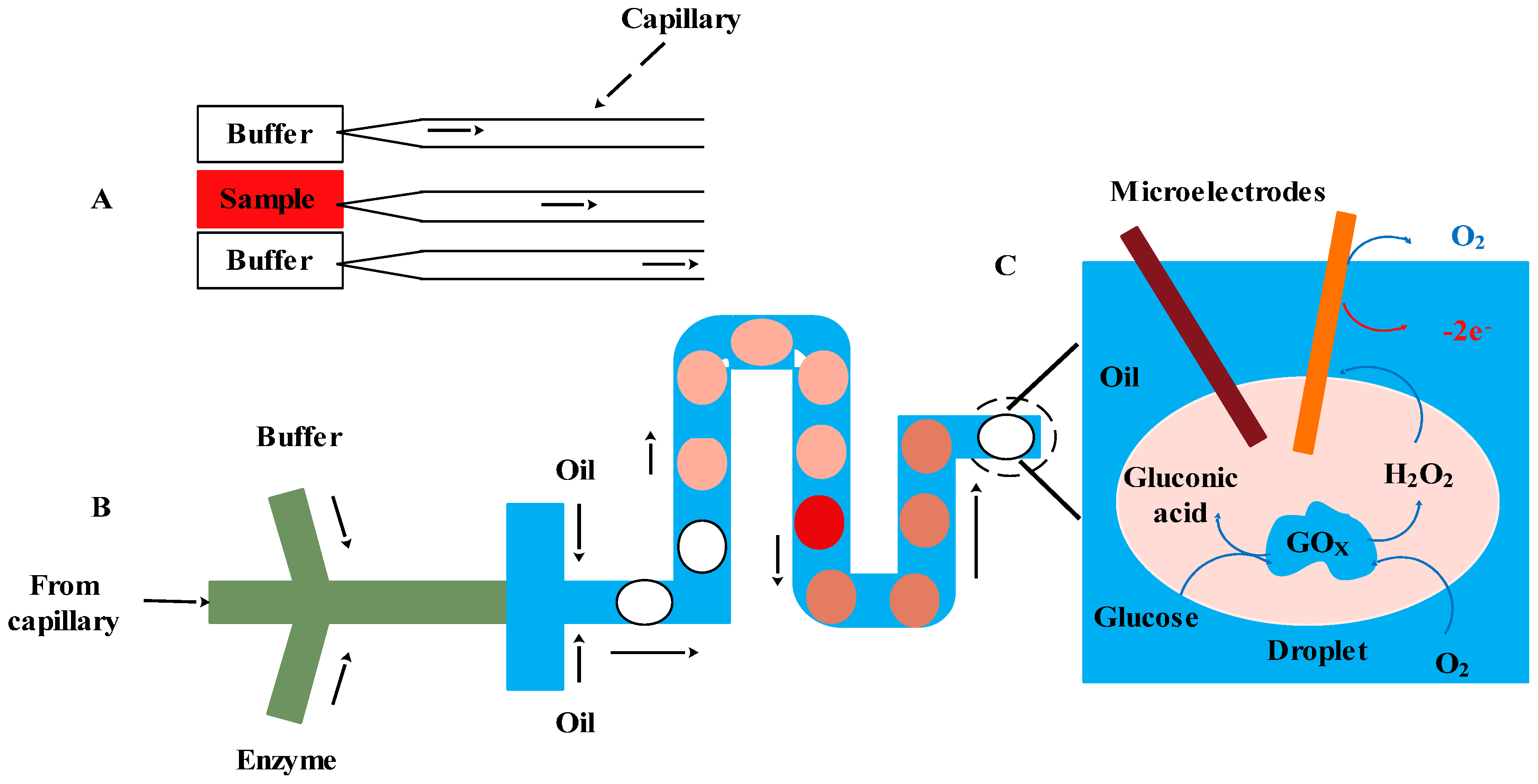
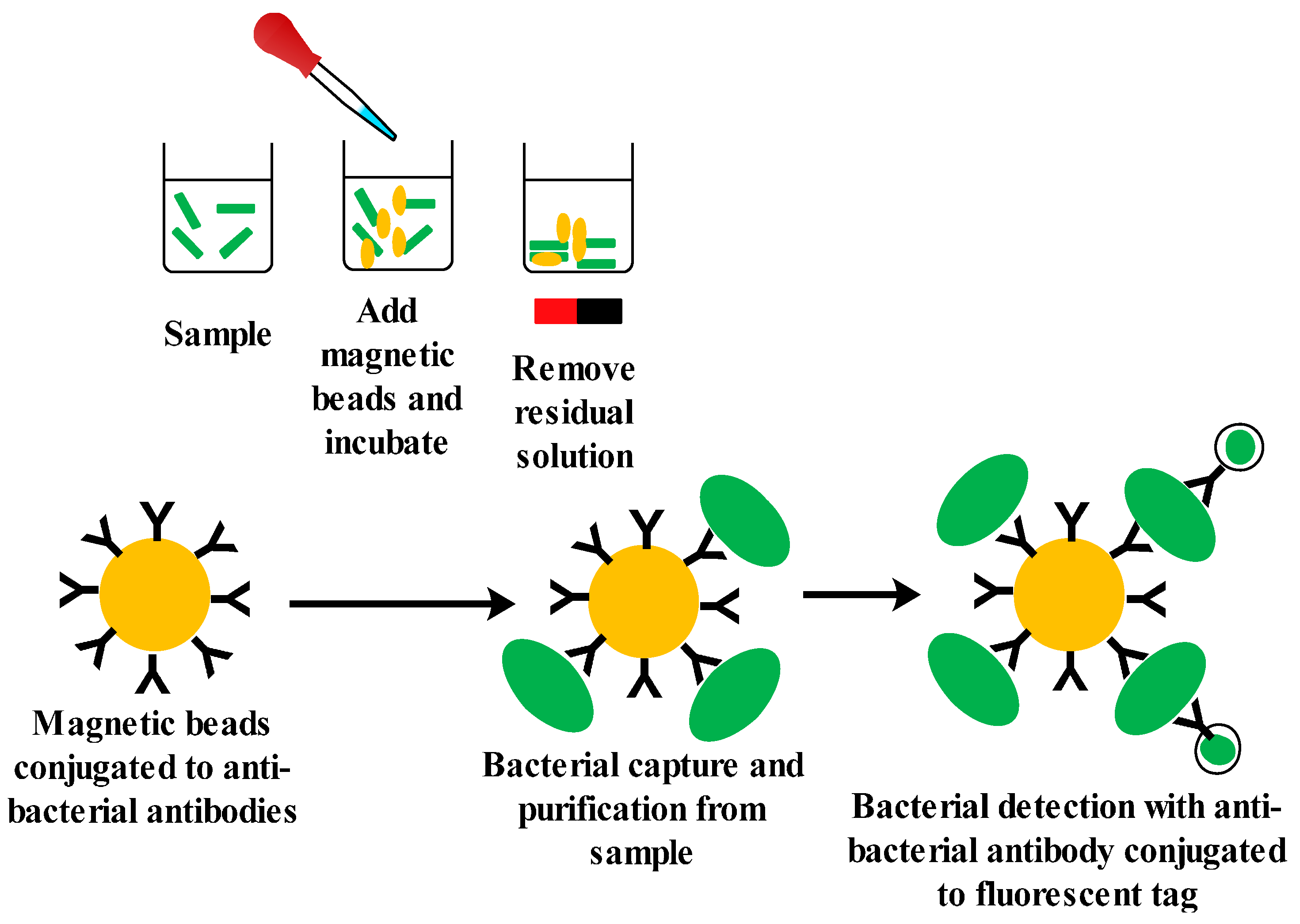
4.2.3. Aptamer-Based
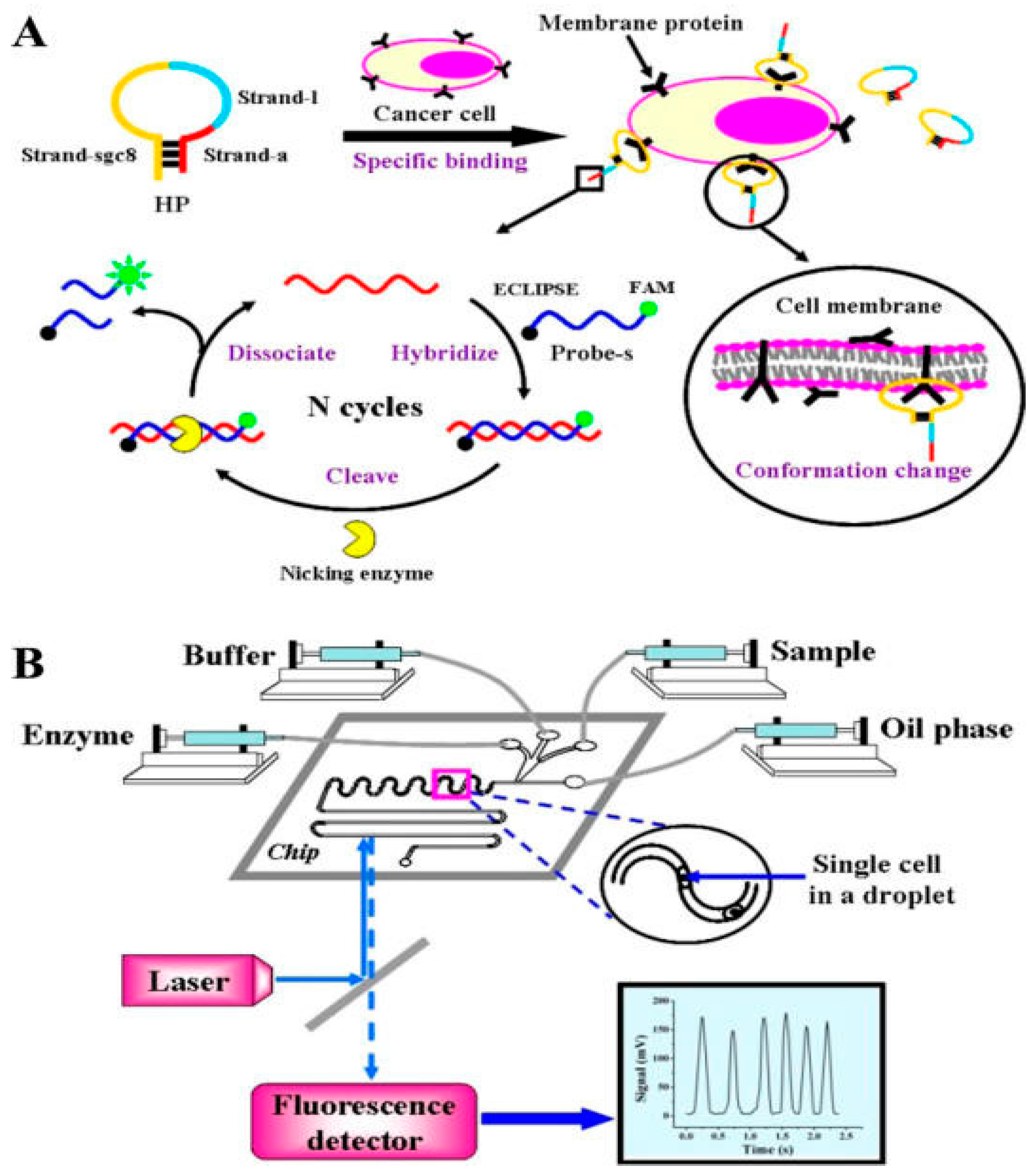
4.3. Digital Microfluidic-Based Biosensor
4.3.1. Enzyme-Based
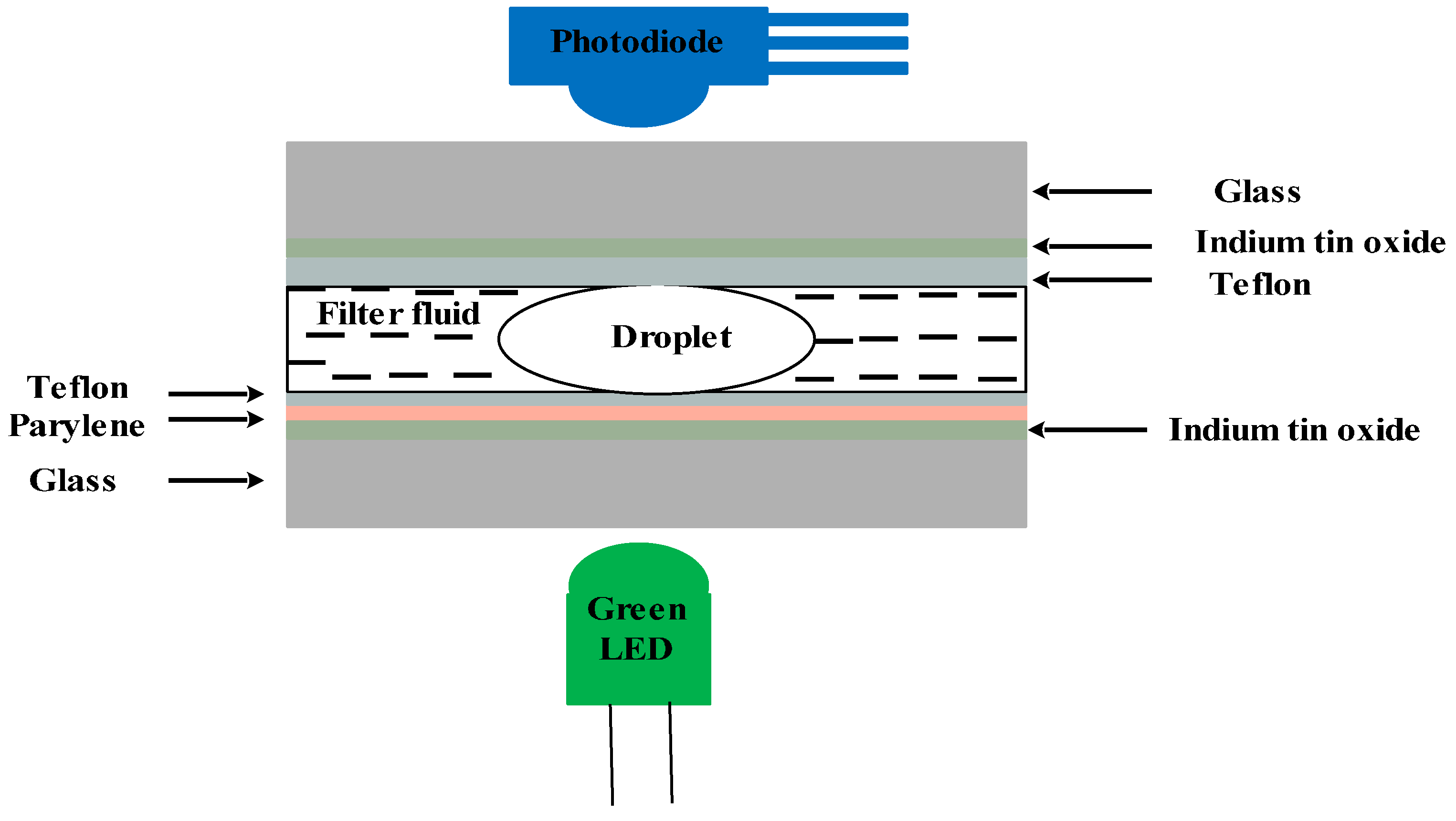
4.3.2. Antibody-Based
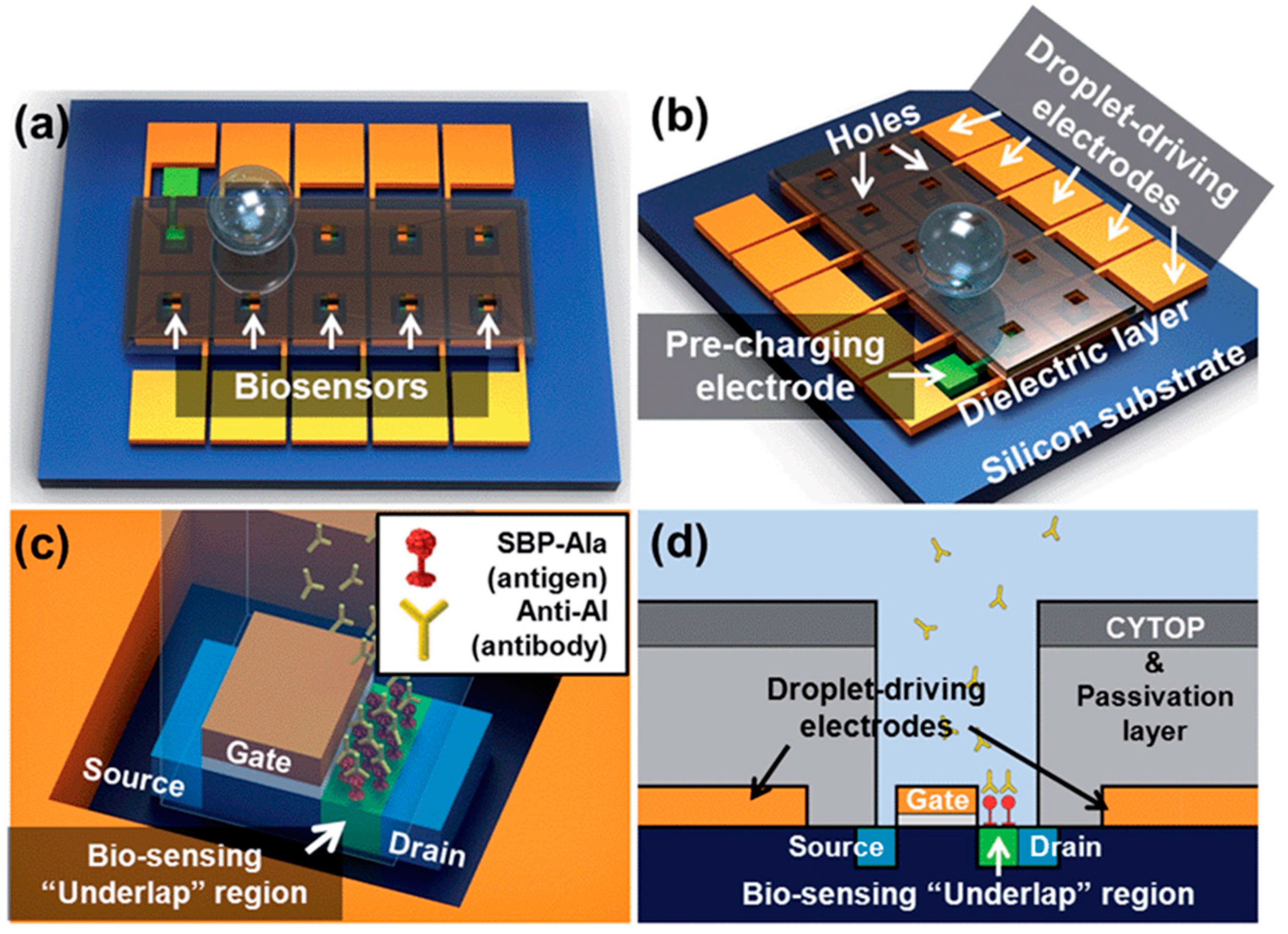
4.3.3. Aptamer-Based
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lindholm-Sethson, B.; Nyström, J.; Geladi, P.; Koeppe, R.; Nelson, A.; Whitehouse, C. Are biosensor arrays in one membrane possible? A combination of multifrequency impedance measurements and chemometrics. Anal. Bioanal. Chem. 2003, 377, 478–485. [Google Scholar] [PubMed]
- Subrahmanyam, S.; Piletsky, S.A.; Turner, A.P.F. Application of Natural Receptors in Sensors and Assays. Anal. Chem. 2002, 74, 3942–3951. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.P.; Kougianos, E. Biosensors: A tutorial review. IEEE Potentials 2006, 25, 35–40. [Google Scholar] [CrossRef]
- Rasooly, A.; Jacobson, J. Development of biosensors for cancer clinical testing. Biosens. Bioelectron. 2006, 21, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Holford, T.R.J.; Davis, F.; Higson, S.P.J. Recent trends in antibody based sensors. Biosens. Bioelectron. 2012, 34, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.P.F. Biosensors: Sense and sensibility. Chem. Soc. Rev. 2013, 42, 3184–3196. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Chakma, B.; Patra, S.; Goswami, P. Potential biomarkers and their applications for rapid and reliable detection of malaria. Biomed Res. Int. 2014, 2014, 852645. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, L.; Li, L.; Liu, W.; Zhou, J.; Yu, W.; Tong, D.; Chen, S. Microfluidics: A new cosset for neurobiology. Lab Chip 2009, 9, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Henares, T.G.; Mizutani, F.; Hisamoto, H. Current development in microfluidic immunosensing chip. Anal. Chim. Acta 2008, 611, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Paper, R. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar]
- The, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Technical report Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar]
- Ruan, C.; Zeng, K.; Varghese, O.K.; Grimes, C.A. A magnetoelastic bioaffinity-based sensor for avidin. Biosens. Bioelectron. 2004, 19, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Zhu, A.; Shi, H. Recent advances in optical biosensors for environmental monitoring and early warning. Sensors 2013, 13, 13928–13948. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhou, C. SAW based mass-loading biosensor for DNA detection. In Proceedings of the 2013 IEEE International Conference of Electron Devices and Solid-State Circuits (EDSSC), Hong Kong, China, 3–5 June 2013.
- Kissinger, P.T. Biosensors-a perspective. Biosens. Bioelectron. 2005, 20, 2512–2516. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2012, 3, 1–9. [Google Scholar] [CrossRef]
- Santano, E.; Pinto, C.; Macías, P. Xenobiotic oxidation by hydroperoxidase activity of lipoxygenase immobilized by adsorption on controlled pore glass. Enzyme Microb. Technol. 2002, 30, 639–646. [Google Scholar] [CrossRef]
- Hilvert, D. Critical anallysis of antibody catalysis. Annu. Rev. Biochem. 2000, 69, 751–793. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.; Lyons, C. Electrode systems for continous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Saerens, D.; Huang, L.; Bonroy, K.; Muyldermans, S. Antibody Fragments as Probe in Biosensor Development. Sensors 2008, 8, 4669–4686. [Google Scholar] [CrossRef]
- Yalow, R. Assay of plasma insulin in human subjects by immunological methods. Nature 1959, 185, 1648–1649. [Google Scholar] [CrossRef]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 496–497. [Google Scholar] [CrossRef]
- Cheng, A.K.H.; Ge, B.; Yu, H.-Z. Aptamer-based biosensors for label-free voltammetric detection of lysozyme. Anal. Chem. 2007, 79, 5158–5164. [Google Scholar] [CrossRef] [PubMed]
- Emanuel, P.A.; Dang, J.; Gebhardt, J.S.; Aldrich, J.; Garber, E.A.E.; Kulaga, H.; Stopa, P.; Valdes, J.J.; Dion-schultz, A. Recombinant antibodies: A new reagent for biological agent detection. Biosens. Bioelectron. 2000, 14, 751–759. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Velu, R.; Hill, K.; Bardóczy, V.; Mészáros, T.; DeRosa, M.C. Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins 2014, 6, 2435–2452. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.; Szostak, J. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Toledo, J.; McKeague, M.; Zhang, X.; Giamberardino, A.; McConnell, E.; Francis, T.; DeRosa, M.C.; Dumontier, M. Aptamer Base: A collaborative knowledge base to describe aptamers and SELEX experiments. Database 2012, 2012, bas006. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Bradley, C.R.; De Girolamo, A.; Visconti, A.; Miller, J.D.; Derosa, M.C. Screening and initial binding assessment of fumonisin b(1) aptamers. Int. J. Mol. Sci. 2010, 11, 4864–4881. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhang, W.; Jia, S.; Guan, Z.; Yang, C.J.; Zhu, Z. Microfluidic approaches to rapid and efficient aptamer selection. Biomicrofluidics 2014, 8, 041501. [Google Scholar] [CrossRef] [PubMed]
- Bayrac, A.T.; Sefah, K.; Parekh, P.; Bayrac, C.; Gulbakan, B.; Oktem, H.A.; Tan, W. In vitro Selection of DNA Aptamers to Glioblastoma Multiforme. ACS Chem. Neurosci. 2011, 2, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ciobanu, M.; Cliffel, D. Glucose and Lactate Biosensors for Scanning Electrochemical Microscopy Imaging of Single Live Cells. Anal. Chem. 2008, 80, 2717–2727. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, D.; Duclair, S.; Datta, S.A.K.; Ellington, A.; Rein, A.; Prasad, V.R. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011, 85, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Campanella, L.; Favero, G.; Persi, L.; Sammartino, M.; Tomassetti, M.; Visco, G. Organic phase enzyme electrodes: Applications and theoretical studies. Anal. Chim. Acta 2001, 426, 235–247. [Google Scholar] [CrossRef]
- Singh, M.; Kathuroju, P.K.; Jampana, N. Polypyrrole based amperometric glucose biosensors. Sens. Actuators B Chem. 2009, 143, 430–443. [Google Scholar] [CrossRef]
- Search, H.; Journals, C.; Contact, A.; Iopscience, M.; Address, I.P. Biosensors: Recent advances. Reports Prog. Phys. 1997, 60, 1397–1445. [Google Scholar]
- Bo, S.; Pijanowska, D.; Olthuis, W.; Bergveld, P. A flow-through amperometric sensor based on dialysis tubing and free enzyme reactors. Biosens. Bioelectron. Bioelectron. 2001, 16, 391–397. [Google Scholar]
- Radoi, A.; Compagnone, D. Recent advances in NADH electrochemical sensing design. Bioelectrochemistry 2009, 76, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Electrochemical nucleic acid biosensors. Anal. Chim. Acta 2002, 469, 63–71. [Google Scholar] [CrossRef]
- D’Orazio, P. Biosensors in clinical chemistry. Clin. Chim. Acta 2003, 334, 41–69. [Google Scholar] [CrossRef]
- Warsinke, A.; Benkert, A.; Scheller, F.W. Electrochemical immunoassays. Fresenius. J. Anal. Chem. 2000, 366, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Ma, Z. A novel label-free amperometric immunosensor for carcinoembryonic antigen based on redox membrane. Biosens. Bioelectron. 2011, 26, 3068–3071. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.D.; Huang, H.; Liang, R.P. Biocompatible and label-free amperometric immunosensor for hepatitis B surface antigen using a sensing film composed of poly(allylamine)-branched ferrocene and gold nanoparticles. Microchim. Acta 2011, 174, 97–105. [Google Scholar] [CrossRef]
- Zelada-Guillén, G.A.; Tweed-Kent, A.; Niemann, M.; Göringer, H.U.; Riu, J.; Rius, F.X. Ultrasensitive and real-time detection of proteins in blood using a potentiometric carbon-nanotube aptasensor. Biosens. Bioelectron. 2013, 41, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Koncki, R. Recent developments in potentiometric biosensors for biomedical analysis. Anal. Chim. Acta 2007, 599, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Danielsa, J.S. Label-Free Impedance Biosensors: Opportunities and Challenges. Electroanalysis 2008, 19, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Kulkarni, A.; Doepke, A.; Halsall, H.B.; Iyer, S.; Heineman, W.R. Carbohydrate-Based Label-Free Detection of Escherichia coli ORN 178 Using Electrochemical Impedance Spectroscopy. Anal. Chem. 2012, 84, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Guo, X. Surface plasmon resonance based biosensor technique: A review. J. Biophotonics 2012, 5, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, J.; Kosaka, P.M.; Ruz, J.J.; San Paulo, Á.; Calleja, M. Biosensors based on nanomechanical systems. Chem. Soc. Rev. 2013, 42, 1287–1311. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schweizer, L.M.; Wang, W.; Reuben, R.L.; Schweizer, M.; Shu, W. Label-free and real-time monitoring of yeast cell growth by the bending of polymer microcantilever biosensors. Sens. Actuators B Chem. 2013, 178, 621–626. [Google Scholar] [CrossRef]
- Lu, C.H.; Zhang, Y.; Tang, S.F.; Fang, Z.B.; Yang, H.H.; Chen, X.; Chen, G.N. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012, 31, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.I.; Chang, Y.-P.; Chu, Y.-H. Biomolecular interactions and tools for their recognition: Focus on the quartz crystal microbalance and its diverse surface chemistries and applications. Chem. Soc. Rev. 2012, 41, 1947–1971. [Google Scholar] [CrossRef] [PubMed]
- Bottazzi, B.; Fornasari, L.; Frangolho, A.; Giudicatti, S.; Mantovani, A.; Marabelli, F.; Marchesini, G.; Pellacani, P.; Therisod, R.; Valsesia, A. Multiplexed label-free optical biosensor for medical diagnostics. J. Biomed. Opt. 2014, 19, 017006. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Yadav, A.R.; Lifson, M.A.; Baker, J.E.; Fauchet, P.M.; Miller, B.L. Selective virus detection in complex sample matrices with photonic crystal optical cavities. Biosens. Bioelectron. 2013, 44, 229–234. [Google Scholar] [CrossRef] [PubMed]
- McGrath, T.F.; Andersson, K.; Campbell, K.; Fodey, T.L.; Elliott, C.T. Development of a rapid low cost fluorescent biosensor for the detection of food contaminants. Biosens. Bioelectron. 2013, 41, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, X.; Li, Y.; Ying, Y. A simple and rapid optical biosensor for detection of aflatoxin B1 based on competitive dispersion of gold nanorods. Biosens. Bioelectron. 2013, 47, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, Z.; Li, X. Advances in pesticide biosensors: Current status, challenges, and future perspectives. Anal. Bioanal. Chem. 2013, 405, 63–90. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Fritzsche, M.; Pieper, S.B.; Wood, T.K.; Lear, K.L.; Dandy, D.S.; Reardon, K.F. Fiber optic monooxygenase biosensor for toluene concentration measurement in aqueous samples. Biosens. Bioelectron. 2011, 26, 2407–2412. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Wartchow, C.A.; Podlaski, F.; Li, S.; Rowan, K.; Zhang, X.; Mark, D.; Huang, K.-S. Biosensor-based small molecule fragment screening with biolayer interferometry. J. Comput. Aided. Mol. Des. 2011, 25, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Gooding, J.J. Biosensor technology for detecting biological warfare agents: Recent progress and future trends. Anal. Chim. Acta 2006, 559, 137–151. [Google Scholar] [CrossRef]
- Lim, D.V.; Simpson, J.M.; Kearns, E.A.; Kramer, M.F. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin. Microbiol. Rev. 2005, 18, 583–607. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Ibrahim, F.; Djordjevic, I.; Koole, L.H. Recent advances in surface functionalization techniques on polymethacrylate materials for optical biosensor applications. Analyst 2014, 139, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Brogan, K.L.; Walt, D.R. Optical fiber-based sensors: Application to chemical biology. Curr. Opin. Chem. Biol. 2005, 9, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, P.D.; Ordeig, O.; Mogensen, K.B.; Kutter, J.P. Electrophoresis microchip with integrated waveguides for simultaneous native UV fluorescence and absorbance detection. Electrophoresis 2009, 30, 4172–4178. [Google Scholar] [CrossRef] [PubMed]
- Kozma, P.; Kehl, F.; Ehrentreich-Förster, E.; Stamm, C.; Bier, F.F. Integrated planar optical waveguide interferometer biosensors: A comparative review. Biosens. Bioelectron. 2014, 58, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Sciacca, B.; Monro, T.M. Dip biosensor based on localized surface plasmon resonance at the tip of an optical fiber. Langmuir 2014, 30, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, A.J.; Harpster, M.H.; Johnson, P.A. The development of surface-enhanced Raman scattering as a detection modality for portable in vitro diagnostics: Progress and challenges. Phys. Chem. Chem. Phys. 2013, 15, 20415–20433. [Google Scholar] [CrossRef] [PubMed]
- Online, V.A.; Citartan, M.; Gopinath, S.C.B.; Tominaga, J.; Tang, T. Label-free methods of reporting biomolecular interactions by optical biosensors. Analyst 2013, 138, 3576–3592. [Google Scholar]
- Gabig-Ciminska, M. Developing nucleic acid-based electrical detection systems. Microb. Cell Fact. 2006, 5. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wei, W.; Qu, X. Colorimetric biosensing using smart materials. Adv. Mater. 2011, 23, 4215–4236. [Google Scholar] [CrossRef] [PubMed]
- Leonard, P.; Hearty, S.; Brennan, J.; Dunne, L.; Quinn, J.; Chakraborty, T.; O’Kennedy, R. Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Technol. 2003, 32, 3–13. [Google Scholar] [CrossRef]
- Lucklum, R.; Hauptmann, P. Acoustic microsensors—The challenge behind microgravimetry. Anal. Bioanal. Chem. 2005, 384, 667–682. [Google Scholar] [CrossRef]
- Haun, J.B.; Yoon, T.-J.; Lee, H.; Weissleder, R. Magnetic nanoparticle biosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Kosel, J. A Magnetic Method to Concentrate and Trap Biological Targets. IEEE Trans. Magn. 2012, 48, 2854–2856. [Google Scholar] [CrossRef]
- Bi, S.; Cui, Y.; Dong, Y.; Zhang, N. Target-induced self-assembly of DNA nanomachine on magnetic particle for multi-amplified biosensing of nucleic acid, protein, and cancer cell. Biosens. Bioelectron. 2014, 53, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Annu. Rev. Mater. Sci. 1998, 28, 153–184. [Google Scholar] [CrossRef]
- Brouzes, E.; Medkova, M.; Savenelli, N.; Marran, D.; Twardowski, M.; Hutchison, J.B.; Rothberg, J.M.; Link, D.R.; Perrimon, N.; Samuels, M.L. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar] [CrossRef] [PubMed]
- Garstecki, P.; Fuerstman, M.J.; Stone, H.A.; Whitesides, G.M. Formation of droplets and bubbles in a microfluidic T-junction-scaling and mechanism of break-up. Lab Chip 2006, 6, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Pollack, M.G.; Shenderov, A.D.; Fair, R.B. Electrowetting-based actuation of droplets for integrated microfluidics. Lab Chip 2002, 2, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Berge, B.; Peseux, J. Variable focal lens controlled by an external voltage: An application of electrowetting. Eur. Phys. J. E 2000, 3, 159–163. [Google Scholar] [CrossRef]
- Ahmadi, A.; Devlin, K.D.; Najjaran, H.; Holzman, J.F.; Hoorfar, M. In situ characterization of microdroplet interfacial properties in digital microfluidic systems. Lab Chip 2010, 10, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.M.K.; Gitlin, I.; Stroock, A.D.; Whitesides, G.M. Components for integrated poly(dimethylsiloxane) microfluidic systems. Electrophoresis 2002, 23, 3461–3473. [Google Scholar] [CrossRef]
- Becker, H.; Locascio, L.E. Polymer microfluidic devices. Talanta 2002, 56, 267–287. [Google Scholar] [CrossRef]
- Sia, S.K.; Whitesides, G.M. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef] [PubMed]
- Feltis, B.N.; Sexton, B.A.; Glenn, F.L.; Best, M.J.; Wilkins, M.; Davis, T.J. A hand-held surface plasmon resonance biosensor for the detection of ricin and other biological agents. Biosens. Bioelectron. 2008, 23, 1131–1136. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Nijhuis, C.A.; Gong, J.; Chen, X.; Kumachev, A.; Martinez, A.W.; Narovlyansky, M.; Whitesides, G.M. Electrochemical sensing in paper-based microfluidic devices. Lab Chip 2010, 10, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Jing, G.; Polaczyk, A.; Oerther, D.B.; Papautsky, I. Development of a microfluidic biosensor for detection of environmental mycobacteria. Sens. Actuators B Chem. 2007, 123, 614–621. [Google Scholar] [CrossRef]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Bashir, R. Microfuidic Biochip for Impedance Spectroscopy of Biological Species. Biomed. Microdevices 2001, 3, 201–209. [Google Scholar]
- Weibel, D.B.; Whitesides, G.M. Applications of microfluidics in chemical biology. Curr. Opin. Chem. Biol. 2006, 10, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Edel, J.B.; deMello, A.J. Micro- and nanofluidic systems for high-throughput biological screening. Drug Discov. Today 2009, 14, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Bringer, M.R.; Gerdts, C.J.; Song, H.; Tice, J.D.; Ismagilov, R.F. Microfluidic systems for chemical kinetics that rely on chaotic mixing in droplets. Philos. Trans. A. Math. Phys. Eng. Sci. 2004, 362, 1087–1104. [Google Scholar] [CrossRef] [PubMed]
- Squires, T.M. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977. [Google Scholar] [CrossRef]
- Choi, S.; Chae, J. A regenerative biosensing surface in microfluidics using electrochemical desorption of short-chain self-assembled monolayer. Microfluid. Nanofluidics 2009, 7, 819–827. [Google Scholar] [CrossRef]
- Auroux, P.-A.; Iossifidis, D.; Reyes, D.R.; Manz, A. Micro Total Analysis Systems. 2. Analytical Standard Operations and Applications. Anal. Chem. 2002, 74, 2637–2652. [Google Scholar] [CrossRef] [PubMed]
- Manz, A.; Graber, N.; Widmer, H.M. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Stone, H.A.; Kim, S. Microfluidics: Basic issues, applications, and challenges. AIChE J. 2001, 47, 1250–1254. [Google Scholar] [CrossRef]
- Anwar, K.; Han, T.; Yu, S.; Kim, S. An Integrated Micro-Nanofluidic system for sample preparation and preconcentration of proteins. R. Soc. Chem. 2010, 443–445. [Google Scholar]
- Liu, K.-K.; Wu, R.-G.; Chuang, Y.-J.; Khoo, H.S.; Huang, S.-H.; Tseng, F.-G. Microfluidic systems for biosensing. Sensors 2010, 10, 6623–6661. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zheng, G.; Lee, L.M. Optical imaging techniques in microfluidics and their applications. Lab Chip 2012, 12, 3566–3575. [Google Scholar] [CrossRef] [PubMed]
- Breslauer, D.N.; Lee, P.J.; Lee, L.P. Microfluidics-based systems biology. Mol. Biosyst. 2006, 2, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Mawatari, K.; Kitamori, T. Microchip-based cell analysis and clinical diagnosis system. Lab Chip 2008, 8, 1992–1998. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-H.; Sung, H.J.; Kim, S.; Kim, E.-O.; Lee, J.W.; Moon, J.Y.; Choi, K.; Jung, J.-E.; Lee, Y.; Koh, S.S.; et al. An RNA aptamer that specifically binds pancreatic adenocarcinoma up-regulated factor inhibits migration and growth of pancreatic cancer cells. Cancer Lett. 2011, 313, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Gardeniers, J.G.E.; van den Berg, A. Lab-on-a-chip systems for biomedical and environmental monitoring. Anal. Bioanal. Chem. 2004, 378, 1700–1703. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Srivastava, S.; Solanki, P.R.; Reddy, V.; Agrawal, V.V.; Kim, C.; John, R.; Malhotra, B.D. Highly efficient bienzyme functionalized nanocomposite-based microfluidics biosensor platform for biomedical application. Sci. Rep. 2013, 3, 2661. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Su, Y.-D.; Chen, S.-J.; Tseng, F.-G.; Lee, G.-B. Microfluidic systems integrated with two-dimensional surface plasmon resonance phase imaging systems for microarray immunoassay. Biosens. Bioelectron. 2007, 23, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, X.; Wang, E. Review: Aptamers in microfluidic chips. Anal. Chim. Acta 2010, 683, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Kwa, T.; Gao, Y.; Liu, Y.; Rahimian, A.; Revzin, A. On-chip regeneration of aptasensors for monitoring cell secretion. Lab Chip 2014, 14, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Ali-Cherif, A.; Begolo, S.; Descroix, S.; Viovy, J.L.; Malaquin, L. Programmable magnetic tweezers and droplet microfluidic device for high-throughput nanoliter multi-step assays. Angew. Chemie - Int. Ed. 2012, 51, 10765–10769. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Lu, Y.; Ding, Y.; Li, L.; Song, H.; Wang, J.; Wu, Q. A droplet-based microfluidic electrochemical sensor using platinum-black microelectrode and its application in high sensitive glucose sensing. Biosens. Bioelectron. 2014, 55, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Golberg, A.; Linshiz, G.; Kravets, I.; Stawski, N.; Hillson, N.J.; Yarmush, M.L.; Marks, R.S.; Konry, T. Cloud-enabled microscopy and droplet microfluidic platform for specific detection of Escherichia coli in water. PLoS ONE 2014, 9, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Iwai, K.; Sochol, R.D.; Lee, L.P.; Lin, L. Finger-powered bead-in-droplet microfluidic system for point-of-care diagnostics. In Proceedings of the 2012 IEEE 25th International Conference on Micro Electro Mechanical Systems (MEMS), Paris, France, 29 January 2015–2 February 2015; pp. 949–952.
- Li, L.; Wang, Q.; Feng, J.; Tong, L.; Tang, B. Highly sensitive and homogeneous detection of membrane protein on a single living cell by aptamer and nicking enzyme assisted signal amplification based on microfluidic droplets. Anal. Chem. 2014, 86, 5101–5107. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, J.; Zhou, J. Parallel-plate lab-on-a-chip based on digital microfluidics for on-chip electrochemical analysis. J. Micromech. Microeng. 2014, 24, 015020. [Google Scholar] [CrossRef]
- Cho, S.K.; Fan, S.; Moon, H.; Angeles, L. Towards digital microfluidic circuits creating, transporting, cutting and merging liquid droplets by electrowetting-based actuation. In Proceedings of the Fifteenth IEEE International Conference on Micro Electro Mechanical Systems, Las Vegas, NV, USA, 24 January 2002; pp. 32–35.
- Vergauwe, N.; Witters, D.; Ceyssens, F.; Vermeir, S.; Verbruggen, B.; Puers, R.; Lammertyn, J. A versatile electrowetting-based digital microfluidic platform for quantitative homogeneous and heterogeneous bio-assays. J. Micromech. Microeng. 2011, 21, 5. [Google Scholar] [CrossRef]
- Srinivasan, V.; Pamula, V.; Fair, R. A Digital microfluidic biosensor for multianalyte detection. In Proceedings of the 2003 IEEE The Sixteenth Annual International Conference on Micro Electro Mechanical Systems, Kyoto, Japan, 19–23 January 2003; pp. 327–330.
- Choi, K.; Kim, J.-Y.; Ahn, J.-H.; Choi, J.-M.; Im, M.; Choi, Y.-K. Integration of field effect transistor-based biosensors with a digital microfluidic device for a lab-on-a-chip application. Lab Chip 2012, 12, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- García-Alonso, J.; Greenway, G.M.; Hardege, J.D.; Haswell, S.J. A prototype microfluidic chip using fluorescent yeast for detection of toxic compounds. Biosens. Bioelectron. 2009, 24, 1508–1511. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luka, G.; Ahmadi, A.; Najjaran, H.; Alocilja, E.; DeRosa, M.; Wolthers, K.; Malki, A.; Aziz, H.; Althani, A.; Hoorfar, M. Microfluidics Integrated Biosensors: A Leading Technology towards Lab-on-a-Chip and Sensing Applications. Sensors 2015, 15, 30011-30031. https://doi.org/10.3390/s151229783
Luka G, Ahmadi A, Najjaran H, Alocilja E, DeRosa M, Wolthers K, Malki A, Aziz H, Althani A, Hoorfar M. Microfluidics Integrated Biosensors: A Leading Technology towards Lab-on-a-Chip and Sensing Applications. Sensors. 2015; 15(12):30011-30031. https://doi.org/10.3390/s151229783
Chicago/Turabian StyleLuka, George, Ali Ahmadi, Homayoun Najjaran, Evangelyn Alocilja, Maria DeRosa, Kirsten Wolthers, Ahmed Malki, Hassan Aziz, Asmaa Althani, and Mina Hoorfar. 2015. "Microfluidics Integrated Biosensors: A Leading Technology towards Lab-on-a-Chip and Sensing Applications" Sensors 15, no. 12: 30011-30031. https://doi.org/10.3390/s151229783
APA StyleLuka, G., Ahmadi, A., Najjaran, H., Alocilja, E., DeRosa, M., Wolthers, K., Malki, A., Aziz, H., Althani, A., & Hoorfar, M. (2015). Microfluidics Integrated Biosensors: A Leading Technology towards Lab-on-a-Chip and Sensing Applications. Sensors, 15(12), 30011-30031. https://doi.org/10.3390/s151229783









