A Bacterial Biosensor for Oxidative Stress Using the Constitutively Expressed Redox-Sensitive Protein roGFP2
Abstract
:1. Introduction
2. Results
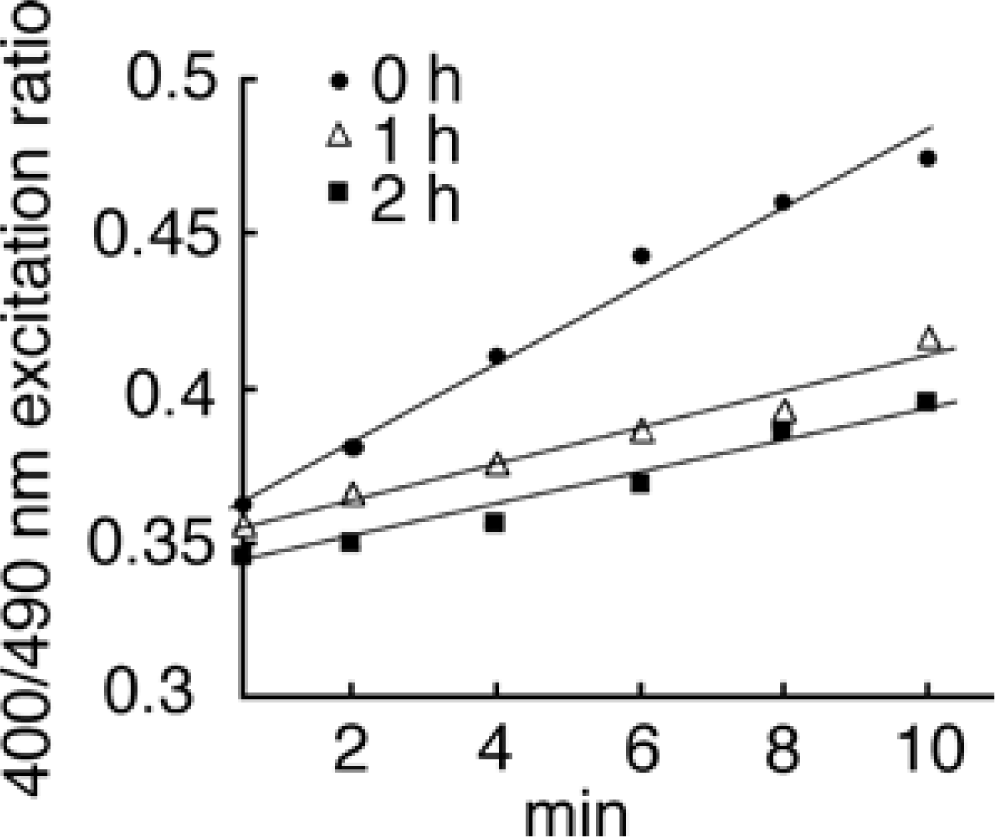
2.1. Autofluorescence normalization and background oxidation
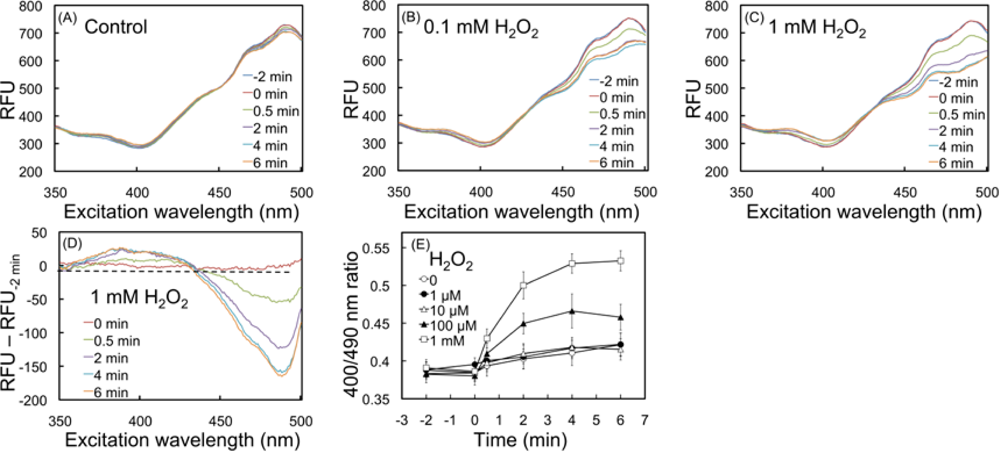
2.2. Response of the biosensor to H2O2
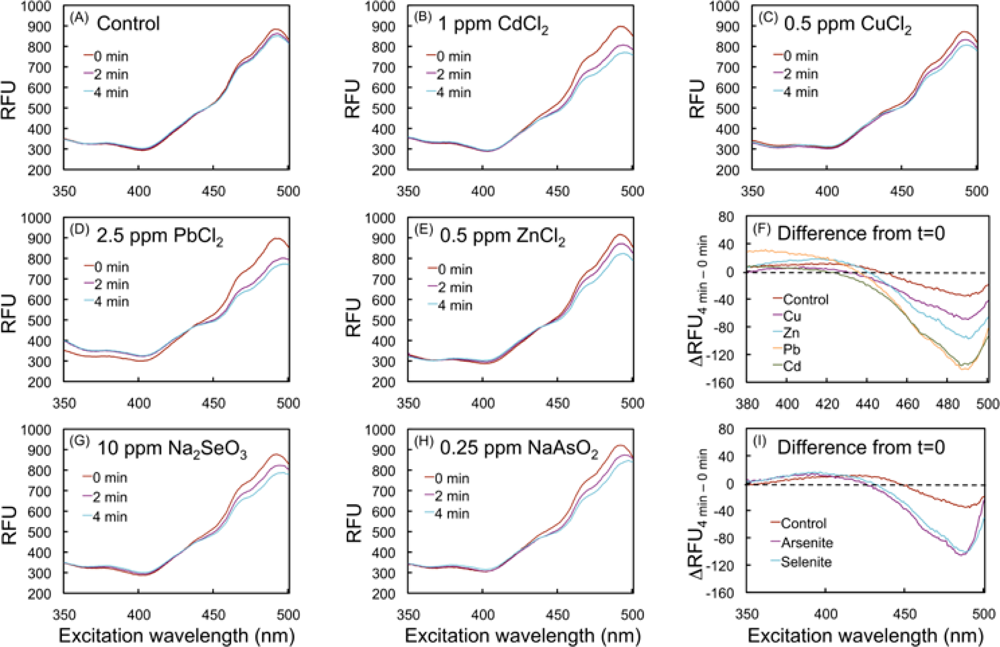
2.3. Spectral changes of the biosensor upon exposure to toxic metal compounds
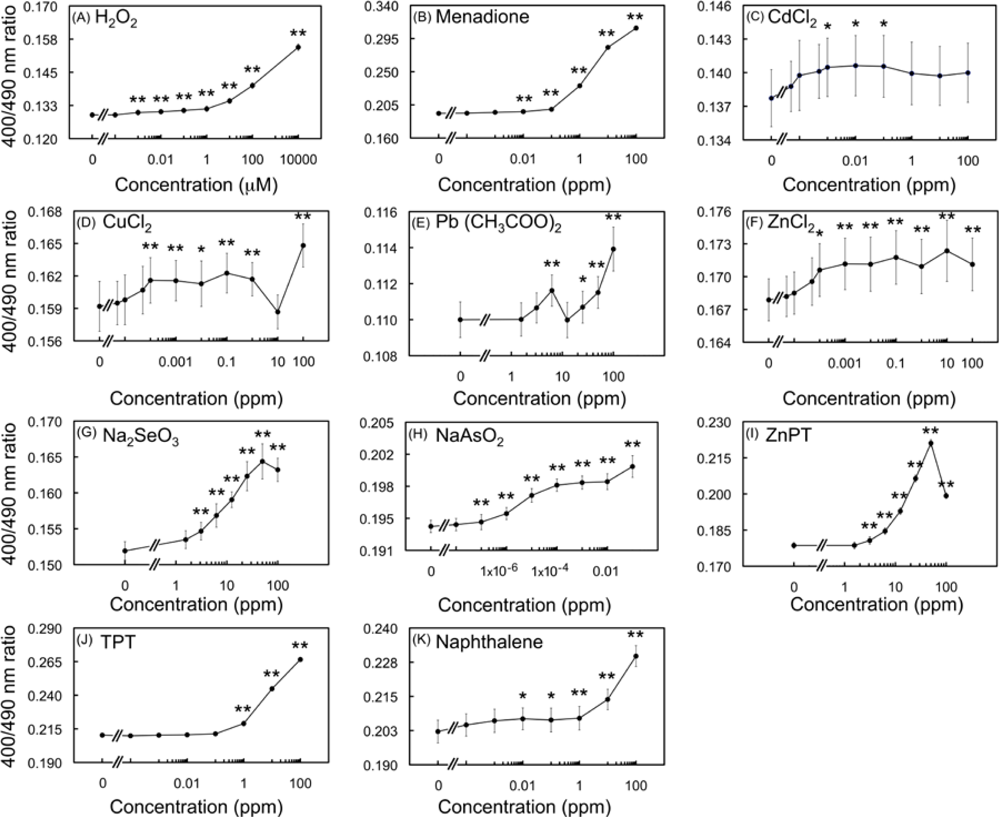
2.4. Ratiometric characterization of the biosensor
2.4.1. Detection of oxidation upon exposure to H2O2 and menadione
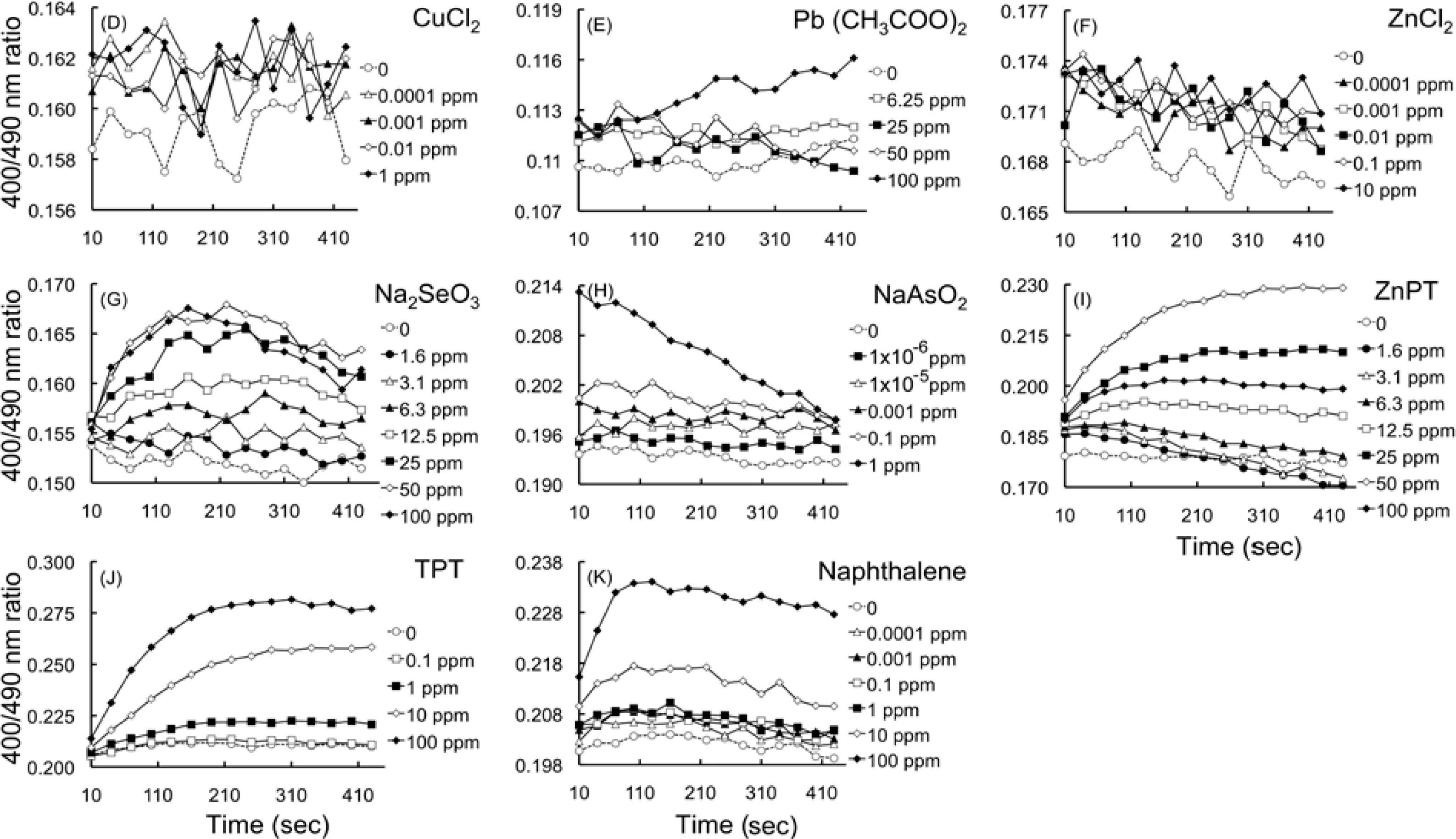
2.4.2. Effects of HM cations on E. coli-roGFP2
2.4.3. Oxidation detected upon exposure to selenite and arsenite
2.4.4. Detection of oxidation caused by organometallic compounds
2.4.5. Detection of oxidation after exposure to naphthalene
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Bacterial strain
4.3. Spectrophotometric measurement of roGFP2 fluorescence
4.4. Microplate-based measurement of roGFP2 fluorescence
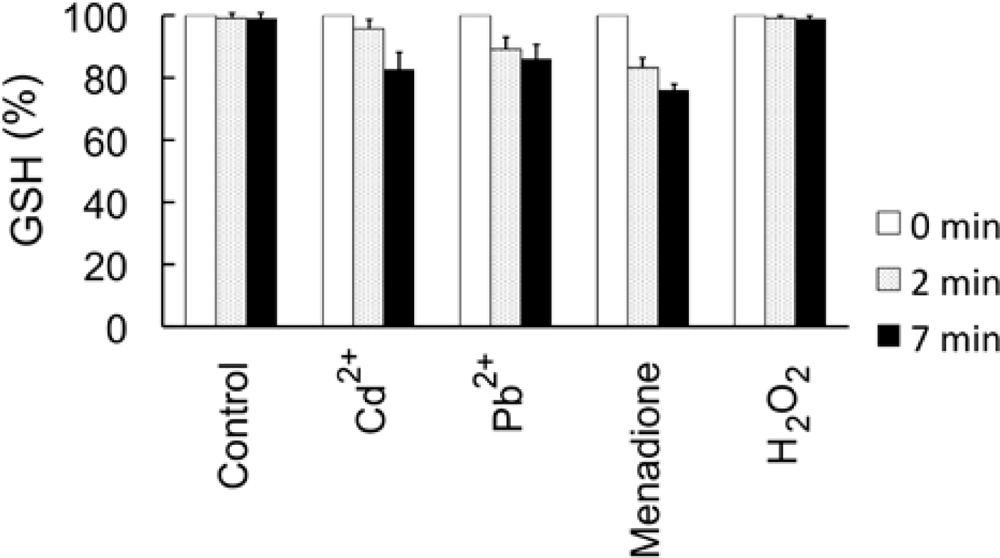
4.5. Measurement of cellular GSH
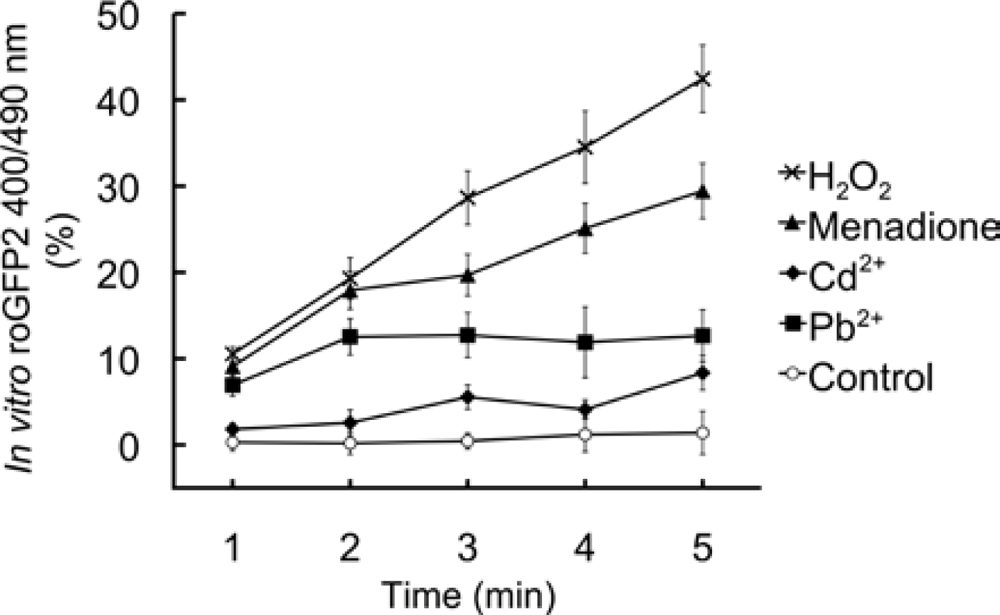
4.6. In vitro effects of chemicals on roGFP2
4.7. Data analysis
Acknowledgments
References
- Magrisso, S; Erel, Y; Belkin, S. Microbial reporters of metal bioavailability. Microb. Biotechnol 2008, 1, 320–330. [Google Scholar]
- Farré, M; Rodríguez-Mozaz, S; López de Alda, MJ; Barceló, D; Hansen, PD. The Handbook of Environmental Chemistry; Barceló, D, Hansen, PD, Eds.; Springer-Verlag: Berlin, Germany, 2009; Volume 5, Chapter 1; pp. 1–32. [Google Scholar]
- Franco, R; Sánchez-Olea, R; Reyes-Reyes, EM; Panayiotidis, MI. Environmental toxicity, oxidative stress and apoptosis; Ménage à trois. Mutat. Res. Genet. Toxicol. Environ. Mutagen 2009, 674, 3–22. [Google Scholar]
- Shen, H; Yang, C; Liu, J; Ong, C. Dual role of glutathione in selenite-induced oxidative stress and apoptosis in human hepatoma cells. Free Radic. Biol. Med 2000, 28, 1115–1124. [Google Scholar]
- Ercal, N; Gurer-Orhan, H; Aykin-Burns, N. Toxic metals and oxidative stress part I: Mechanisms involved in metal-induced oxidative damage. Curr. Top. Med. Chem 2001, 1, 529–539. [Google Scholar]
- Valko, M; Morris, H; Cronin, MT. Metals, toxicity and oxidative stress. Curr. Med. Chem 2005, 12, 1161–1208. [Google Scholar]
- Kumagai, Y; Sumi, D. Arsenic: Signal transduction, transcription factor, and biotransformation involved in cellular response and toxicity. Annu. Rev. Pharmacol. Toxicol 2007, 47, 243–262. [Google Scholar]
- Belkin, S; Smulski, DR; Vollmer, AC; Van Dyk, TK; LaRossa, RA. Oxidative stress detection with Escherichia coli harboring a katG′::lux fusion. Appl. Environ. Microbiol 1996, 62, 2252–2256. [Google Scholar]
- Lee, HJ; Gu, MB. Construction of a sodA::luxCDABE fusion Escherichia coli: Comparison with a katG fusion strain through their responses to oxidative stresses. Appl. Microbiol. Biotechnol 2003, 60, 577–580. [Google Scholar]
- Mitchell, RJ; Gu, MB. Construction and characterization of novel dual stress-responsive bacterial biosensors. Biosens. Bioelectron 2004, 19, 977–985. [Google Scholar]
- Kim, BC; Youn, CH; Ahn, JM; Gu, MB. Screening of target-specific stress-responsive genes for the development of cell-based biosensors using a DNA microarray. Anal. Chem 2005, 77, 8020–8026. [Google Scholar]
- Niazi, JH; Kim, BC; Ahn, JM; Gu, MB. A novel bioluminescent bacterial biosensor using the highly specific oxidative stress-inducible pgi gene. Biosens. Bioelectron 2008, 24, 670–675. [Google Scholar]
- Hanson, GT; Aggeler, R; Oglesbee, D; Cannon, M; Capaldi, RA; Tsien, RY; Remington, SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem 2004, 279, 13044–13053. [Google Scholar]
- Yu, S; Qin, W; Zhuang, G; Zhang, X; Chen, G; Liu, W. Monitoring oxidative stress and DNA damage induced by heavy metals in yeast expressing a redox-sensitive green fluorescent protein. Curr. Microbiol 2009, 58, 504–510. [Google Scholar]
- Delic, M; Mattanovich, D; Gasser, B. Monitoring intracellular redox conditions in the endoplasmic reticulum of living yeasts. FEMS Microbiol. Lett 2010, 306, 61–66. [Google Scholar]
- Meyer, AJ; Brach, T; Marty, L; Kreye, S; Rouhier, N; Jacquot, JP; Hell, R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 2007, 52, 973–986. [Google Scholar]
- Jubany-Mari, T; Alegre-Battle, L; Jiang, K; Feldman, LJ. Use of a redox-sensing GFP (c-roGFP1) for real-time monitoring of cytosol redox status in Arabidopsis thaliana water-stressed plants. FEBS Lett 2010, 584, 889–897. [Google Scholar]
- Dooley, CT; Dore, TM; Hanson, GT; Jackson, WC; Remington, SJ; Tsien, RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem 2004, 279, 22284–22293. [Google Scholar]
- Gutscher, M; Pauleau, AL; Marty, L; Brach, T; Wabnitz, GH; Samstag, Y; Meyer, AJ; Dick, TP. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 2008, 5, 553–559. [Google Scholar]
- Hobman, JL; Yamamoto, K; Oshima, T. Molecular Microbiology of Heavy Metals; Nies, DH, Silver, S, Eds.; Springer-Verlag, Berlin, Germany, 2007; Volume 1, Chapter 3; pp. 73–115. [Google Scholar]
- Smirnova, GV; Muzyka, NG; Glukhovchenko, MN; Oktyabrsky, ON. Effects of menadione and hydrogen peroxide on glutathione status in growing Escherichia coli. Free Radic. Biol. Med 2000, 28, 1009–1016. [Google Scholar]
- Helbig, K; Bleuel, C; Krauss, GJ; Nies, DH. Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol 2008, 190, 5431–5438. [Google Scholar]
- Quig, D. Cysteine metabolism and metal toxicity. Alter. Med. Rev 1998, 3, 262–270. [Google Scholar]
- Nies, DH. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol 1999, 51, 730–750. [Google Scholar]
- Turner, RJ; Weiner, JH; Taylor, DE. Selenium metabolism in Escherichia coli. Biometals 1998, 11, 223–227. [Google Scholar]
- Stocker, J; Balluch, D; Gsell, M; Harms, H; Feliciano, J; Daunert, S; Malik, KA; van der Meer, JR. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol 2003, 37, 4743–4750. [Google Scholar]
- Wright, DA; Dawson, R; Cutler, SJ; Cutler, HG; Orano-Dawson, CE; Graneli, E. Naphthoquinones as broad spectrum biocides for treatment of ship's ballast water: Toxicity to phytoplankton and bacteria. Water Res 2007, 41, 1294–1302. [Google Scholar]
- Stohs, SJ; Ohia, S; Bagchi, D. Naphthalene toxicity and antioxidant nutrients. Toxicology 2002, 180, 97–105. [Google Scholar]
- Wilson, AS; Davis, CD; Williams, DP; Buckpitt, AR; Pirmohamed, M; Park, BK. Characterisation of the toxic metabolite(s) of naphthalene. Toxicology 1996, 114, 233–242. [Google Scholar]
- Koutsaftis, A; Aoyama, I. The interactive effects of binary mixtures of three antifouling biocides and three heavy metals against the marine algae Chaetoceros gracilis. Environ. Toxicol 2006, 21, 432–439. [Google Scholar]
- Mochida, K; Fujii, K. Toxicity in plankton and fish Kazuhiko Mochida and Kazunori Fujii. In Ecotoxicology of Antifouling Biocides; Arai, T, Harino, H, Ohji, M, Langston, WJ, Eds.; Springer, Tokyo, Japan, 2009; Volume 1, Chapter 21; pp. 365–382. [Google Scholar]
- Appel, KE. Organotin compounds: Toxicokinetic aspects. Drug. Metab. Rev 2004, 36, 763–786. [Google Scholar]
- Zazueta, C; Reyes-Rivas, H; Bravo, C; Pichardo, J; Corona, N; Chávez, E. Triphenyltin as inductor of mitochondrial membrane permeability transition. J. Bioenerg. Biomembr 1994, 26, 457–462. [Google Scholar]
- Robertson, JD; Orrenius, S. Molecular mechanisms of apoptosis induced by cytotoxic chemicals. Crit. Rev. Toxicol 2000, 30, 609–627. [Google Scholar]
- Ellman, GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys 1959, 82, 70–77. [Google Scholar]
- Inayat-Hussain, SH; Chan, KM; Rajab, NF; Din, LB; Chow, SC; Kizilors, A; Farzaneh, F; Williams, GT. Goniothalamin-induced oxidative stress, DNA damage and apoptosis via caspase-2 independent and Bcl-2 independent pathways in Jurkat T-cells. Toxicol. Lett 2010, 193, 108–114. [Google Scholar]


© 2010 by the authors licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Arias-Barreiro, C.R.; Okazaki, K.; Koutsaftis, A.; Inayat-Hussain, S.H.; Tani, A.; Katsuhara, M.; Kimbara, K.; Mori, I.C. A Bacterial Biosensor for Oxidative Stress Using the Constitutively Expressed Redox-Sensitive Protein roGFP2. Sensors 2010, 10, 6290-6306. https://doi.org/10.3390/s100706290
Arias-Barreiro CR, Okazaki K, Koutsaftis A, Inayat-Hussain SH, Tani A, Katsuhara M, Kimbara K, Mori IC. A Bacterial Biosensor for Oxidative Stress Using the Constitutively Expressed Redox-Sensitive Protein roGFP2. Sensors. 2010; 10(7):6290-6306. https://doi.org/10.3390/s100706290
Chicago/Turabian StyleArias-Barreiro, Carlos R., Keisuke Okazaki, Apostolos Koutsaftis, Salmaan H. Inayat-Hussain, Akio Tani, Maki Katsuhara, Kazuhide Kimbara, and Izumi C. Mori. 2010. "A Bacterial Biosensor for Oxidative Stress Using the Constitutively Expressed Redox-Sensitive Protein roGFP2" Sensors 10, no. 7: 6290-6306. https://doi.org/10.3390/s100706290
APA StyleArias-Barreiro, C. R., Okazaki, K., Koutsaftis, A., Inayat-Hussain, S. H., Tani, A., Katsuhara, M., Kimbara, K., & Mori, I. C. (2010). A Bacterial Biosensor for Oxidative Stress Using the Constitutively Expressed Redox-Sensitive Protein roGFP2. Sensors, 10(7), 6290-6306. https://doi.org/10.3390/s100706290



