Tropical Range Extension for the Temperate, Endemic South-Eastern Australian Nudibranch Goniobranchus splendidus (Angas, 1864)
Abstract
:1. Introduction
2. Materials and Methods
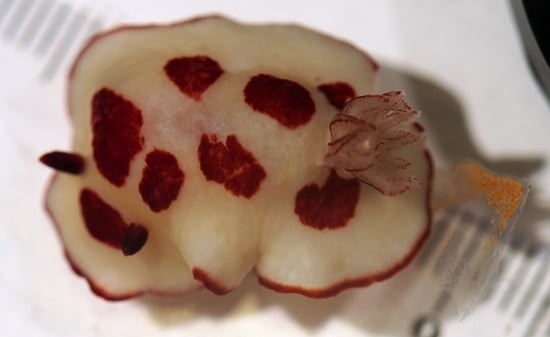
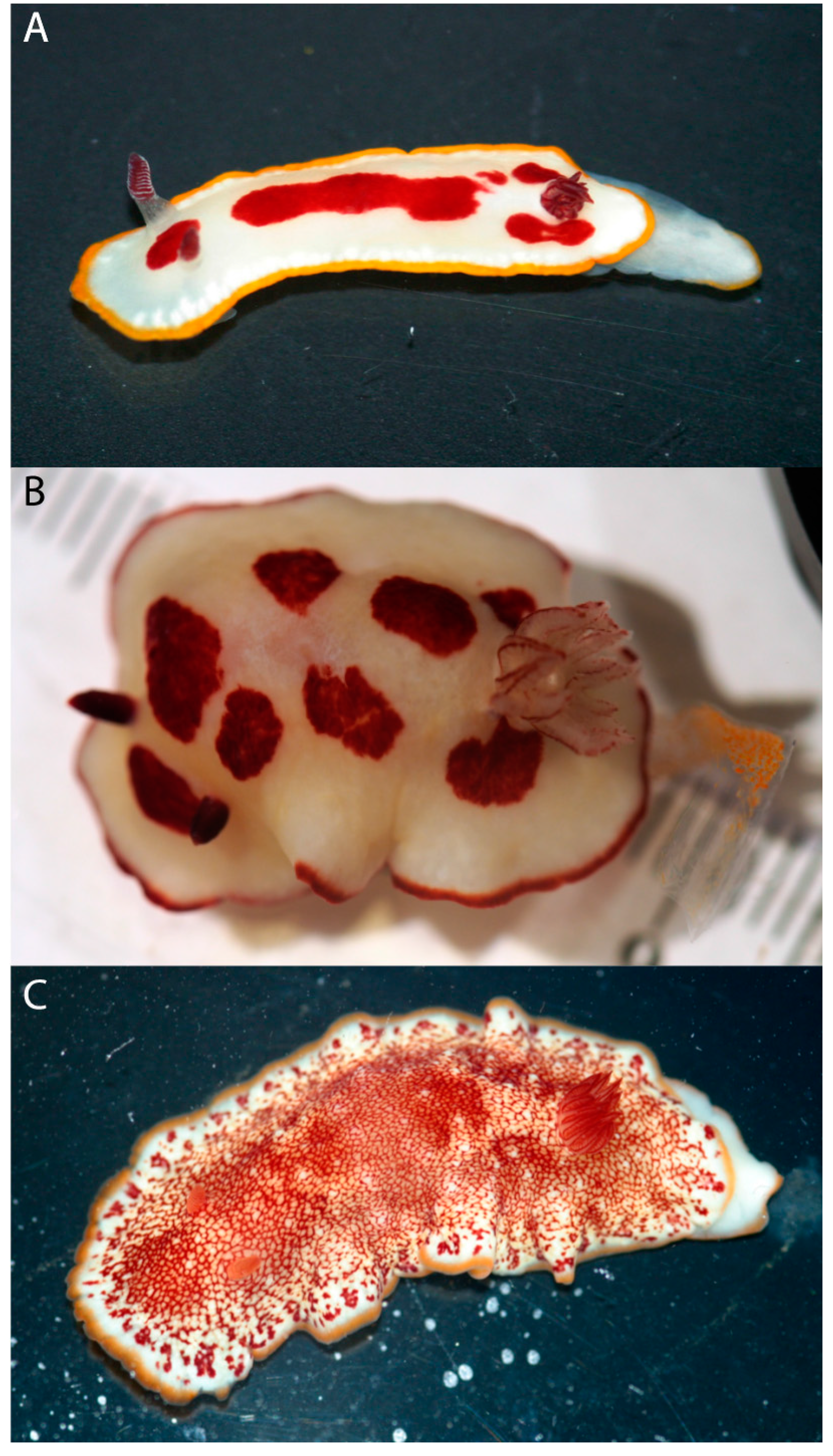
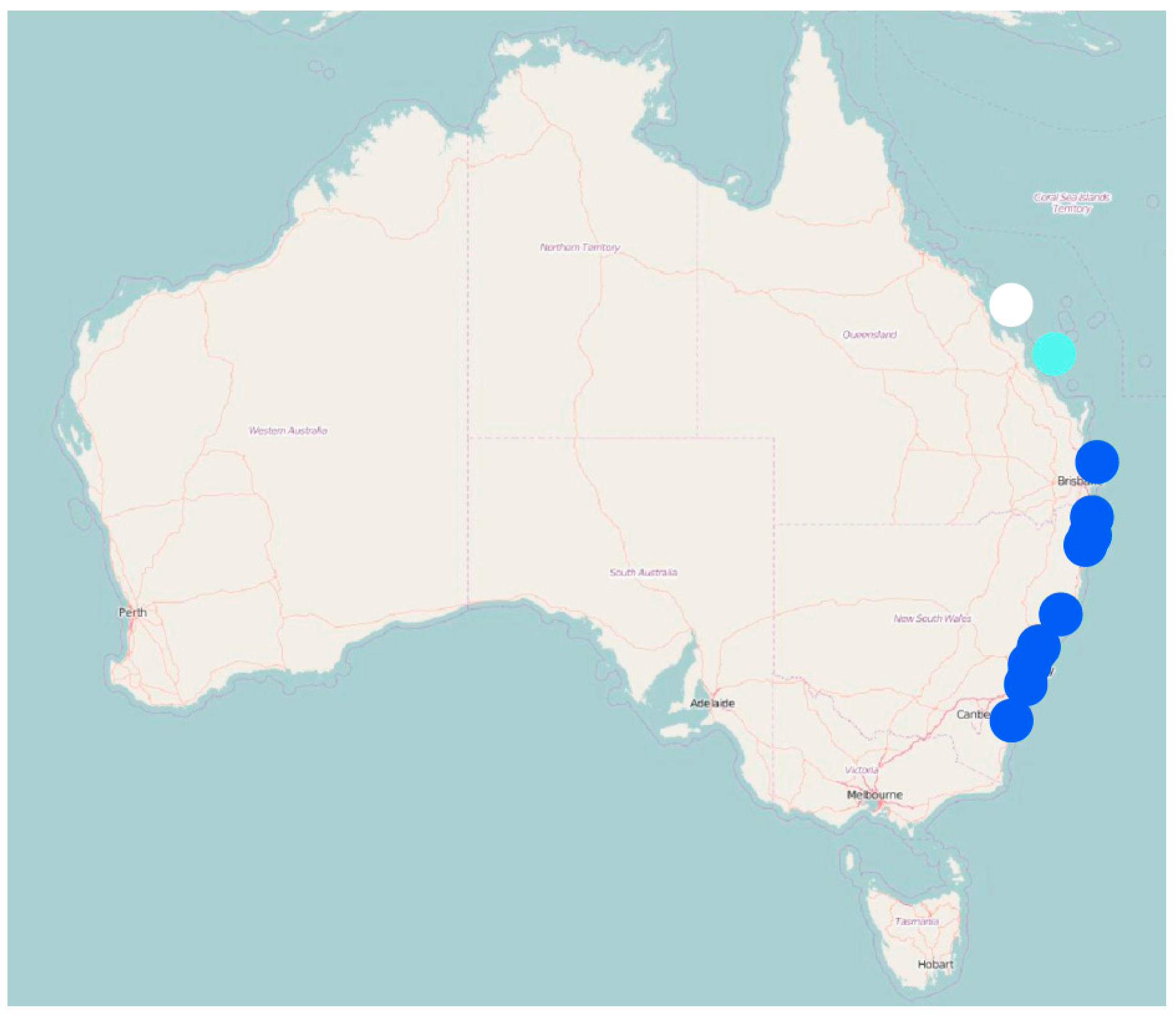
3. Results
4. Discussion
Acknowledgements
Author Contributions
Conflicts of Interest
Abbreviations
| AMS | Australian Museum, Sydney |
| CASIZ | California Academy of Sciences, Invertebrate Zoology |
| COI | Cytochrome Oxidase I |
| GBR | Great Barrier Reef |
| MV | Museum Victoria |
| NSW | New South Wales, Australia |
| QLD | Queensland Australia |
| SAM | South Australian Museum |
| UQ | University of Queensland |
| 16S rDNA | 16S ribosomal DNA |
References
- Rudman, W.B. The Chromodorididae (Opisthobranchia: Mollusca) of the Indo-West Pacific: Chromodoris splendida, C. aspersa and Hypselodoris placida colour groups. Zool. J. Linn. Soc. 1983, 78, 105–173. [Google Scholar] [CrossRef]
- Rudman, W.B. Purpose in pattern: The evolution of colour in chromodorid nudibranchs. J. Mollusc. Stud. 1991, 57, 5–21. [Google Scholar] [CrossRef]
- Hirayama, Y.; Katavic, P.L.; White, A.M.; Pierens, G.K.; Lambert, L.K.; Winters, A.E.; Kigoshi, H.; Kita, M.; Garson, M.J. New cytotoxic norditerpenes from the Australian nudibranchs Goniobranchus splendidus and Goniobranchus daphne. Aust. J. Chem. 2015, 69, 136–144. [Google Scholar] [CrossRef]
- González, M.A. Spongiane diterpenoids. Curr. Bioact. Compd. 2007, 3, 1–36. [Google Scholar]
- Marshall, J.G.; Willan, R.C. Nudibranchs of Heron Island, Great Barrier Reef: A survey of the Opisthobranchia (sea slugs) of Heron and Wistari Reefs; Backhuys: Leiden, The Netherlands, 1999. [Google Scholar]
- Rudman, W.B. The Chromodorididae (Opisthobranchia: Mollusca) of the Indo-West Pacific: further species of Glossodoris, Thorunna and the Chromodoris aureomarginata colour group. Zool. J. Linn. Soc. 1990, 100, 263–326. [Google Scholar] [CrossRef]
- Debelius, H.; Kuiter, R.H. Nudibranchs of the World; Ikan-Unterwasserarchiv: Frankfurt, Germany, 2007. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- Palumbi, S; Martin, A; Romano, S; McMillan, W; Stice, L; Grabowski, G. The Simple Fool’s Guide to PCR; University of Hawaii: Honolulu, HI, USA, 1991; p. 361. [Google Scholar]
- Wilson, N.G.; Maschek, J.A.; Baker, B.J. A species flock driven by predation? Secondary metabolites support diversification of slugs in Antarctica. PLoS ONE 2013, 8, e80277. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M; Moir, R; Wilson, A; Stones-Havas, S; Cheung, M; Sturrock, S; Buxton, S; Cooper, A; Markowitz, S; Duran, C; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.F.; Gosliner, T.M. Traditional taxonomic groupings mask evolutionary history: A molecular phylogeny and new classification of the chromodorid nudibranchs. PLoS ONE 2012, 7, e33479. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.P. Chromodoris tumulifera from Singapore. Available online: http://www.seaslugforum.net/find/13223 (accessed on 15 July 2016).
- Hadfield, M.G.; Switzer-Dunlap, M. Opisthobranchs. In The Mollusca; Wilbur, K., Ed.; Academic Press: New York, NY, USA, 1984; pp. 209–350. [Google Scholar]
- Wilson, N.G. Egg masses of chromodorid nudibranchs (Mollusca: Gastropoda: Opisthobranchia). Malacologia 2002, 44, 289–306. [Google Scholar]
- Atkinson, L.; Atkinson, D. Re: Chromodoris splendida laying eggs. Available online: http://www.seaslugforum.net/find/21334 (accessed on 15 July 2016).
- OZCAM. Available online: http://ozcam.ala.org.au/ (accessed on 16 March 2016).
- White, A.M.; Pierens, G.K.; Forster, L.C.; Winters, A.E.; Cheney, K.L.; Garson, M.J. Rearranged diterpenes and norditerpenes from three Australian Goniobranchus mollusks. J. Nat. Prod. 2015, 79, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Angas, G.F.; Crosse, J.C.H. Description d’espèces nouvelles appartenant à plusieurs genres de mollusques nudibranches des environs de Port-Jackson (Nouvelle-Galles du Sud): Accompagnée de dessins faits d’après nature. J. Conchyliol. 1864, 12, 43–70. [Google Scholar]
- Burn, R. A centennial commentary and zoogeographical remarks on Angas’ Sydney nudibranchs (Molluscs, Gastropoda). J. Conchyliol. 1965, 104, 85–93. [Google Scholar]
- Abraham, P.S. Revision of the anthobranchiate nudibranchiate Mollusca, with descriptions or notices of forty-one hitherto undescribed species. Proc. Zoo. Soc. Lond. 1877, 1877, 196–269. [Google Scholar]
- Bergh, L.S.R. Malacologische Untersuchungen. System der Nudibranchiaten Gasteropoden. In Reisen im Archipel der Philippinen von Dr. Carl Gottfried Semper; Zweiter, T., Ed.; C.W. Kreidel’s Verlag: Wiesbaden, Germany, 1892; Wissenschaftliche Resultate Band 2, Theil 3 (Heft 18); pp. 995–1168. [Google Scholar]
- Eliot, C.N. On some nudibranchs from East Africa and Zanzibar. Part IV. Proc. Zool. Soc. Lond. 1904, 74, 380–406. [Google Scholar] [CrossRef]
- Rudman, W.B. Chromodorid opisthobranch Mollusca from the Indo-West Pacific. Zool. J. Linn. Soc. 1973, 52, 175–199. [Google Scholar] [CrossRef]
- Thompson, T.E. Chromodorid nudibranchs from eastern Australia (Gastropoda, Opisthobranchia). J. Zool. 1972, 166, 391–409. [Google Scholar] [CrossRef]
- Rudman, W.B. The Chromodorididae (Opisthobranchia: Mollusca) of the Indo-West Pacific: A review of the genera. Zool. J. Linn. Soc. 1984, 81, 115–273. [Google Scholar] [CrossRef]
- Basedow, H.; Hedley, C. South Australian nudibranchs, and an enumeration of the known Australian species. Roy. Soc. S. Aust. 1905, 29, 134–160. [Google Scholar]
- Allan, J. Nudibranchiata from Clarence River Heads, North Coast, New South Wales. Rec. Aust. Mus. 1947, 21, 433–463. [Google Scholar] [CrossRef]
- Winters, A.E.; Wilson, N.G.; White, A.M.; How, M.; Garson, M.J.; Cheney, K.L. Geographic divergence of warning signals in nudibranch mollusc. In preparation.
- Hambley, T.W.; Poiner, A.; Taylor, W.C. The constituents of marine sponges. V. The isolation from Chelonaplysilla violacea (Dendroceratida) of Aplyviolene and other diterpenes, and the determination of the crystal structure of Aplyviolene. Aust. J. Chem. 1990, 43, 1861–1870. [Google Scholar] [CrossRef]
- Poiner, A.; Taylor, W.C. The constituents of marine sponges. IV. The isolation of degraded Spongian diterpenoids from Aplysilla tango (sp. nov.) (Dendroceratida). Aust. J. Chem. 1990, 43, 1713–1727. [Google Scholar] [CrossRef]
- Nelson, G.; Platnick, N.I. Systematics and Biogeography; Columbia University Press: New York, NY, USA, 1981; p. 567. [Google Scholar]
- Hardin, G. The competitive exclusion principle. Science 1960, 131, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, J. Interspecific competition increases local extinction rate in a metapopulation system. Nature 1989, 340, 713–715. [Google Scholar] [CrossRef]
- Rudman, W.B.; Bergquist, P.R. A review of feeding specificity in the sponge-feeding Chromodorididae (Nudibranchia: Mollusca). Mollus. Res. 2007, 27, 60–88. [Google Scholar]
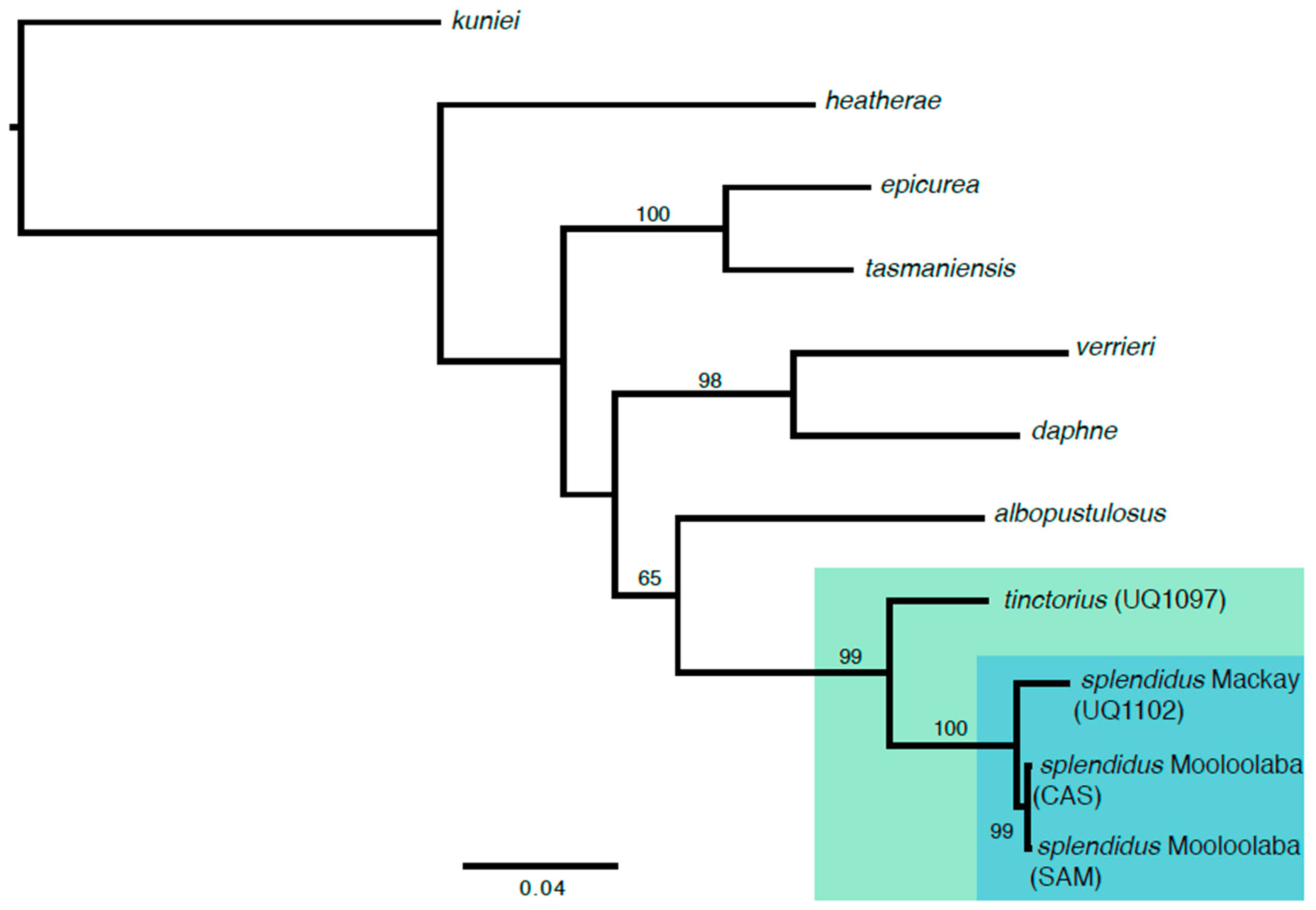
| Species | Voucher | Locality | COI | 16 S |
|---|---|---|---|---|
| G. albopustulosus | CASIZ 142953 | Maui, Hawaii | JQ 727828 | JQ 727701 |
| G. daphne | UQ collection | Gold Coast Seaway, QLD | KJ 001297 | KJ 018921 |
| G. epicurea | SAM D 19285 | Tasmania, Australia | EF 535114 | AY 458804 |
| G. heatherae | CASIZ 175546 | Cape Peninsula, South Africa | JQ 727844 | JQ 727720 |
| G. kuniei | SAM D 19261 | Heron Is., GBR, QLD | EF 535112 | AY 458807 |
| G. splendidus | SAM D 19292 | Mooloolaba, QLD | EF 535115 | AY 458815 |
| G. splendidus | CASIZ 146039 | Mooloolaba, QLD | EU 982738 | EU 982789 |
| G. cf. splendidus | UQ 1102 | Mackay, QLD | MH018011 | MH017998 |
| G. tasmaniensis | SAM 19295 | Triabunna, Tasmania | EF 535113 | AY 458817 |
| G. tinctorius | UQ 1097 | Mackay, QLD | MH018010 | MH01797 |
| G. verrieri | CASIZ 158796 | Batangas, Philippines | JQ 727858 | JQ 727740 |
| Compound | G. splendidus Mackay QLD (n = 1) [21] | G. splendidus Mooloolaba QLD (n = 3) [21] | G. splendidus Mooloolaba QLD (n = 2) [3] | G. splendidus Coffs Harbour, NSW (n = 1) [21] |
|---|---|---|---|---|
| aplysulphurin | 1102 | 931, 928 | yes | 1105 |
| tetrahydroaplysulphurin-1 | 931, 927, 928 | 1105 | ||
| 7α-acetoxyspongian-16-one | yes | 1105 | ||
| aplyroseol-2 | 1102 | yes | ||
| splendidalactone-2 | 931, 927 | |||
| cadlinolide A | 931, 927 | |||
| membranolide | 931, 927 | |||
| spongian-16-one | 928 | yes | ||
| tetrahydroaplysulphurin-4 | 1102 | |||
| dendrillolide A | 1102 | |||
| macfarlandin C, D, E | 1102 | |||
| norrisolide | 1102 | |||
| spongionellin | 1102 | |||
| chromodorolide A | 1102 | |||
| spongialactone | 1102 | |||
| aplyroseol-3 | 1102 | |||
| 15,16-Deacetoxy-15-hydroxy-9,11-dihydrogracilin A | 931 | |||
| 16-Deacetoxy-9,11-dihydrogracilin A | 927 | |||
| tyrinnal, tyrinnal B | 928 | |||
| aplytandiene-3 | yes | |||
| gracilin A, M, N, O, Q, C, G | yes | |||
| 6Z isomers of gracilin B, C | yes | |||
| splendidalactone-1 | 1105 | |||
| aplyviolene | 1105 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, N.G.; Winters, A.E.; Cheney, K.L. Tropical Range Extension for the Temperate, Endemic South-Eastern Australian Nudibranch Goniobranchus splendidus (Angas, 1864). Diversity 2016, 8, 16. https://doi.org/10.3390/d8030016
Wilson NG, Winters AE, Cheney KL. Tropical Range Extension for the Temperate, Endemic South-Eastern Australian Nudibranch Goniobranchus splendidus (Angas, 1864). Diversity. 2016; 8(3):16. https://doi.org/10.3390/d8030016
Chicago/Turabian StyleWilson, Nerida G., Anne E. Winters, and Karen L. Cheney. 2016. "Tropical Range Extension for the Temperate, Endemic South-Eastern Australian Nudibranch Goniobranchus splendidus (Angas, 1864)" Diversity 8, no. 3: 16. https://doi.org/10.3390/d8030016
APA StyleWilson, N. G., Winters, A. E., & Cheney, K. L. (2016). Tropical Range Extension for the Temperate, Endemic South-Eastern Australian Nudibranch Goniobranchus splendidus (Angas, 1864). Diversity, 8(3), 16. https://doi.org/10.3390/d8030016