SSR Markers for Trichoderma virens: Their Evaluation and Application to Identify and Quantify Root-Endophytic Strains
Abstract
:1. Introduction
2. Experimental Section
2.1. Screening of Databases and Primer Design
2.2. Fungal Strains
2.3. Culture Conditions
2.4. Plant Material and Root Inoculation
| Primer Name | Primer Sequence (5′-3′) | Locus No. | Tm | Fragment Sizes in bp: Maize9B, (Range), Reference Sequence | NCBI Accession No. | Alleles | Scaffold No. | T. harzianum |
|---|---|---|---|---|---|---|---|---|
| TvCTT56f | CTTGATGACAAGCCAAAAGG | 58.4 | 289 (283–439) 456 | KM010303 | 8 | 64 | − | |
| TvCTT56r | GAAGAGAGGACATAGGGTCTGG | L1 | 59.2 | |||||
| TvCAT32f | GTGTAGCAGCCCAACAGTCC | 60.7 | 409(364–457) 481 | KM010304 | 8 | 89 | − | |
| TvCAT32r | CAGGTGTCGTGACAGATTCG | L2 | 60.3 | |||||
| TvCTTT29f | GGAAGATAGCACGATGAAGTCG | 61.1 | 350 (291–410) 402 | KM010305 | 9 | 81 | − | |
| TvCTTT29r | AACCGTGGAAGTAGGTGTCG | L3 | 60.0 | |||||
| TvCTTTT27f | TCATCCACCCTGCTAACTCG | 61.2 | 420 (378–563) 482 | KM010306 | 9 | 81 | − | |
| TvCTTTT27r | CGCTGCGTCATCCTAAACC | L4 | 61.7 | |||||
| TvAAC21f | CACCATTCCATTATTACGCGACG | 60.4 | 234 (210–269) 268 | KM010307 | 7 | 2 | + | |
| TvAAC21r | CTGCACTCCCTCCCAATGC | L5 | 60.8 | |||||
| TvCAG13f | CCCAGGAAACCCTCAGAACG | 60.3 | 180(161–180) 206 | KM010308 | 7 | 92 | − | |
| TvCAG13r | TCTTTGCAGTTTCCAAGTCGG | L6 | 59.1 | |||||
| TvGAAA34f | GGGGTGCTGAATAGCTAACG | 59.7 | 325 (315–491) 423 | KM010310 | 6 | 3 | + | |
| TvGAAA34r | TGCCGTCTTGTCTTATTTTCG | L7 | 60.3 | |||||
| TvTGTC18f | GTGGTGAGGACTTGCTTGG | 59.3 | 425 (393–469) 483 | KM010311 | 7 | 2 | + | |
| TvTGTC18r | TCTGCCTGTCAGTTGTTTGC | L8 | 60.0 | |||||
| TvGAT18f | GGGATCTGATTTGGCCTACC | 60.7 | 371 (333–423) 387 | KM010312 | 8 | 3 | + | |
| TvGAT18r | ACTTCCCCCATCCAATAACG | L9 | 60.9 | |||||
| TvCA39f | GCATCTGCACCTGATATATTCC | 58.6 | 256 (236–271) 306 | KM010313 | 8 | 6 | − | |
| TvCA39r | CCTTGTACGATCTCCAGAACC | L10 | 58.7 | |||||
| TvGTT23f | GCATCAAAGCGTGCTGTTGG | 60.8 | 216 (206–237) 279 | KM010309 | 4 | 87 | − | |
| TvGTT23r | GCAAACACAAGCTGACAATGC | L11 | 60.6 | |||||
| TvAG29f | TGTGCCCACTGAGATTTCG | 60.8 | 449 (423–462) 470 | KM010314 | 8 | 93 | − | |
| TvAG29r | TCAGCATGAGATTACACATACCG | L12 | 60.0 |
| Isolate Origin | CBS Number | Habitat | Year of Isolation |
|---|---|---|---|
| Ivory Coast | CBS 123790 | soil | Unknown |
| Italy | CBS 116947 | sandy soil, Pinus pinea | 1982 |
| Iran | CBS 111249 | soil | 2001 |
| Guadeloupe | CBS 100946 | rain forest soil | 1998 |
| Papua New Guinea | CBS 350.96 | coastal region soil | 1995 |
| The Netherlands | CBS 609.95 | compost | 1995 |
| Australia (Perth) | CBS 497.84 | sandy soil, lettuce | 1984 |
| Moldova | CBS 512.66 | soil | 1966 |
| USA (Maryland) | CBS 430.54 | soil | 1954 |
| Germany | - | maize roots | 2012 |
2.5. DNA Extraction
2.5.1. Mycelia
2.5.2. Roots
2.5.3. Leaves
2.6. PCR Conditions
2.6.1. Touchdown PCR
2.6.2. Multiplex PCR
2.6.3. In-Root Detection of Fungal DNA
2.6.4. Real-Time PCR
2.7. Electrophoresis
2.7.1. Agarose Gels
2.7.2. Polyacrylamide (PAA) Gels
2.7.3. Capillary Gel Electrophoresis
2.8. Re-Sequencing
2.9. Statistical Analysis
3. Results and Discussion
3.1. Primer Evaluation on a Global Set of Isolates

3.2. Data Analysis
| Italy | Austr | Iran | PapNG | IvoryC | USA | Germa | Netherl | Guade | Moldo | |
|---|---|---|---|---|---|---|---|---|---|---|
| Italy | 0.00 | 0.270 | 0.247 | 0.225 | 0.270 | 0.270 | 0.241 | 0.247 | 0.225 | 0.270 |
| Australia | 0.270 | 0.00 | 0.247 | 0.270 | 0.270 | 0.180 | 0.247 | 0.247 | 0.247 | 0.247 |
| Iran | 0.247 | 0.247 | 0.00 | 0.202 | 0.202 | 0.247 | 0.247 | 0.270 | 0.270 | 0.247 |
| Papua New Guinea | 0.225 | 0.270 | 0.202 | 0.00 | 0.247 | 0.270 | 0.270 | 0.270 | 0.225 | 0.270 |
| Ivory Coast | 0.270 | 0.270 | 0.202 | 0.247 | 0.00 | 0.270 | 0.247 | 0.270 | 0.247 | 0.270 |
| USA | 0.270 | 0.180 | 0.247 | 0.270 | 0.270 | 0.00 | 0.247 | 0.247 | 0.270 | 0.270 |
| Germany | 0.247 | 0.247 | 0.247 | 0.270 | 0.247 | 0.247 | 0.00 | 0.247 | 0.225 | 0.247 |
| The Netherlands | 0.247 | 0.247 | 0.270 | 0.270 | 0.270 | 0.247 | 0.247 | 0.00 | 0.247 | 0.247 |
| Guadeloupe | 0.225 | 0.247 | 0.270 | 0.225 | 0.247 | 0.270 | 0.225 | 0.247 | 0.00 | 0.247 |
| Moldova | 0.270 | 0.225 | 0.247 | 0,270 | 0.270 | 0.270 | 0.247 | 0.247 | 0.247 | 0.00 |
| L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | L9 | L10 | L11 | L12 | Total | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pop.1 | Na | 4 | 3 | 5 | 4 | 4 | 4 | 4 | 4 | 5 | 5 | 2 | 4 | 48 |
| Ne | 3.571 | 2.778 | 5.000 | 3.571 | 3.571 | 3.571 | 3.571 | 3.571 | 5.000 | 5.000 | 1.923 | 3.571 | ||
| I | 1.332 | 1.055 | 1.609 | 1.332 | 1.332 | 1.332 | 1.332 | 1.332 | 1.609 | 1.609 | 0.673 | 1.332 | ||
| h | 0.720 | 0.640 | 0.800 | 0.720 | 0.720 | 0.720 | 0.720 | 0.720 | 0.800 | 0.800 | 0.450 | 0.720 | ||
| uh | 0.900 | 0.800 | 1.000 | 0.900 | 0.900 | 0.900 | 0.900 | 0.900 | 1.000 | 1.000 | 0.600 | 0.900 | ||
| Pop.2 | Na | 4 | 5 | 4 | 5 | 4 | 2 | 3 | 4 | 3 | 3 | 3 | 4 | 44 |
| Ne | 3.571 | 5.000 | 3.571 | 5.000 | 3.571 | 1.923 | 2.273 | 3.571 | 2.273 | 2.273 | 2.273 | 3.571 | ||
| I | 1.332 | 1.609 | 1.332 | 1.609 | 1.332 | 0.673 | 0.950 | 1.332 | 1.055 | 0.950 | 1.055 | 1.332 | ||
| h | 0.720 | 0.800 | 0.720 | 0.800 | 0.720 | 0.480 | 0.560 | 0.720 | 0.640 | 0.560 | 0.640 | 0.720 | ||
| uh | 0.900 | 1.000 | 0.900 | 1.000 | 0.900 | 0.600 | 0.700 | 0.900 | 0.800 | 0.700 | 0.800 | 0.900 | ||
| Mean and SE of Loci for each Population and Grand Mean (Total) and SE of Loci and Populations | ||||||||||||||
| Mean | Na | 4.000 | 4.000 | 4.500 | 4.500 | 4.000 | 3.000 | 3.500 | 4.000 | 4.000 | 4.000 | 2.500 | 4.000 | 3.833 |
| SE | 0.000 | 1.000 | 0.500 | 0.500 | 0.000 | 1.000 | 0.500 | 0.000 | 1.000 | 1.000 | 0.500 | 0.000 | 0.177 | |
| Mean | Ne | 3.571 | 3.889 | 4.286 | 4.286 | 3.571 | 2.747 | 2.922 | 3.571 | 3.889 | 3.636 | 2.350 | 3.571 | 3.524 |
| SE | 0.000 | 1.111 | 0.714 | 0.714 | 0.000 | 0.824 | 0.649 | 0.000 | 1.111 | 1.364 | 0.427 | 0.000 | 0.194 | |
| Mean | I | 1.332 | 1.332 | 1.471 | 1.471 | 1.332 | 1.003 | 1.141 | 1.332 | 1.332 | 1.280 | 0.864 | 1.332 | 1.269 |
| SE | 0.000 | 0.277 | 0.139 | 0.139 | 0.000 | 0.330 | 0.191 | 0.000 | 0.277 | 0.330 | 0.191 | 0.000 | 0.055 | |
| Mean | h | 0.720 | 0.720 | 0.760 | 0.760 | 0.720 | 0.600 | 0.640 | 0.720 | 0.720 | 0.680 | 0.560 | 0.720 | 0.693 |
| SE | 0.000 | 0.080 | 0.040 | 0.040 | 0.000 | 0.120 | 0.080 | 0.000 | 0.080 | 0.120 | 0.080 | 0.000 | 0.019 | |
| Mean | uh | 0.900 | 0.900 | 0.950 | 0.950 | 0.900 | 0.750 | 0.800 | 0.900 | 0.900 | 0.850 | 0.700 | 0.900 | 0.867 |
| SE | 0.000 | 0.100 | 0.050 | 0.050 | 0.000 | 0.150 | 0.100 | 0.000 | 0.100 | 0.150 | 0.100 | 0.000 | 0.024 | |
| Pop. | Na | Ne | I | H | uh | |
|---|---|---|---|---|---|---|
| Pop. 1 | Mean | 4.000 | 3.725 | 1.323 | 0.713 | 0.892 |
| SE | 0.246 | 0.264 | 0.075 | 0.025 | 0.031 | |
| Pop. 2 | Mean | 3.667 | 3.323 | 1.214 | 0.673 | 0.842 |
| SE | 0.256 | 0.285 | 0.081 | 0.029 | 0.036 | |
| Total | Mean | 3.833 | 3.524 | 1.269 | 0.693 | 0.867 |
| SE | 0.177 | 0.194 | 0.055 | 0.019 | 0.024 |
| L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | L9 | L10 | L11 | L12 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PhiPT | −0.071 | −0.023 | −0.080 | −0.080 | 0.022 | 0.148 | 0.091 | −0.125 | −0.023 | 0.115 | −0.029 | −0.125 | −0.012 |
| PhiPT max | 0.100 | 0.100 | 0.050 | 0.050 | 0.100 | 0.250 | 0.200 | 0.100 | 0.100 | 0.150 | 0.300 | 0.100 | 0.133 |
| Phi’PT | −0.714 | −0.227 | −1.591 | −1.591 | 0.217 | 0.591 | 0.455 | −1.250 | −0.227 | 0.764 | −0.098 | −1.250 | −0.088 |
| Probability PhiPT | 1.000 | 0.701 | 1.000 | 1.000 | 0.624 | 0.129 | 0.352 | 1.000 | 0.719 | 0.155 | 0.501 | 1.000 | 0.556 |
3.3. Strain Discrimination Using Multiplex PCR
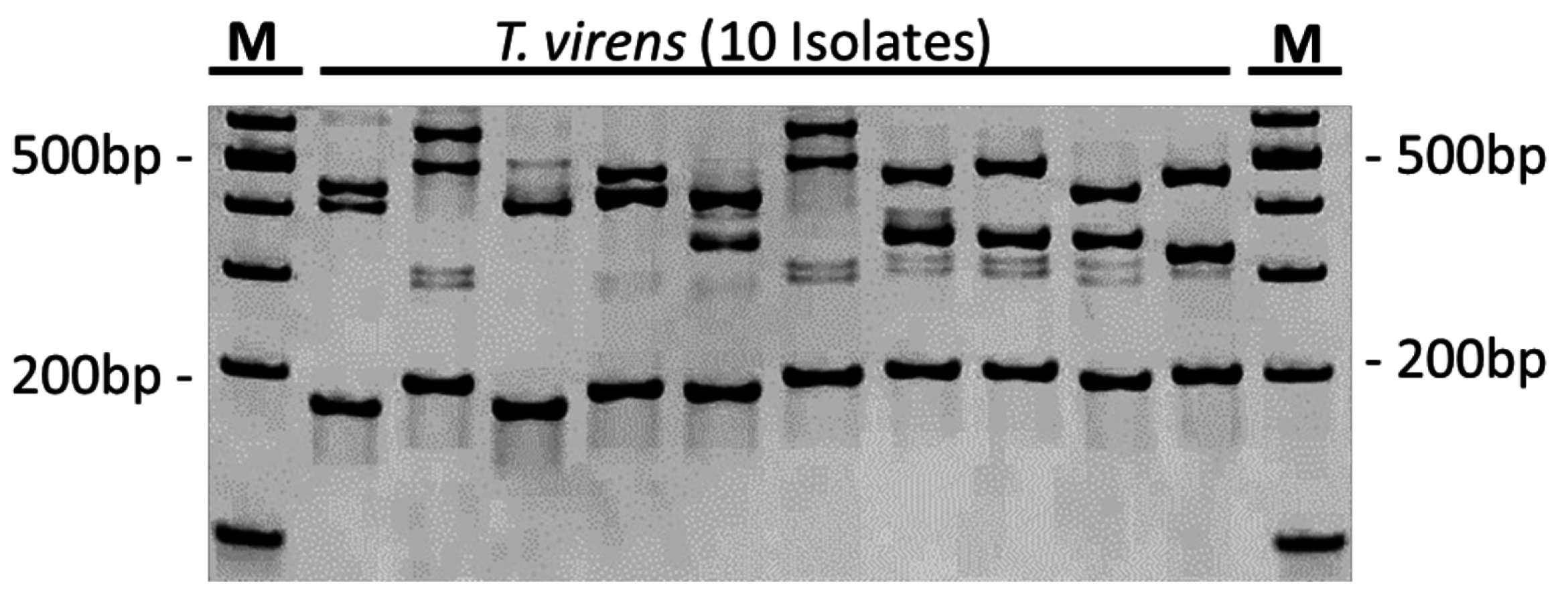
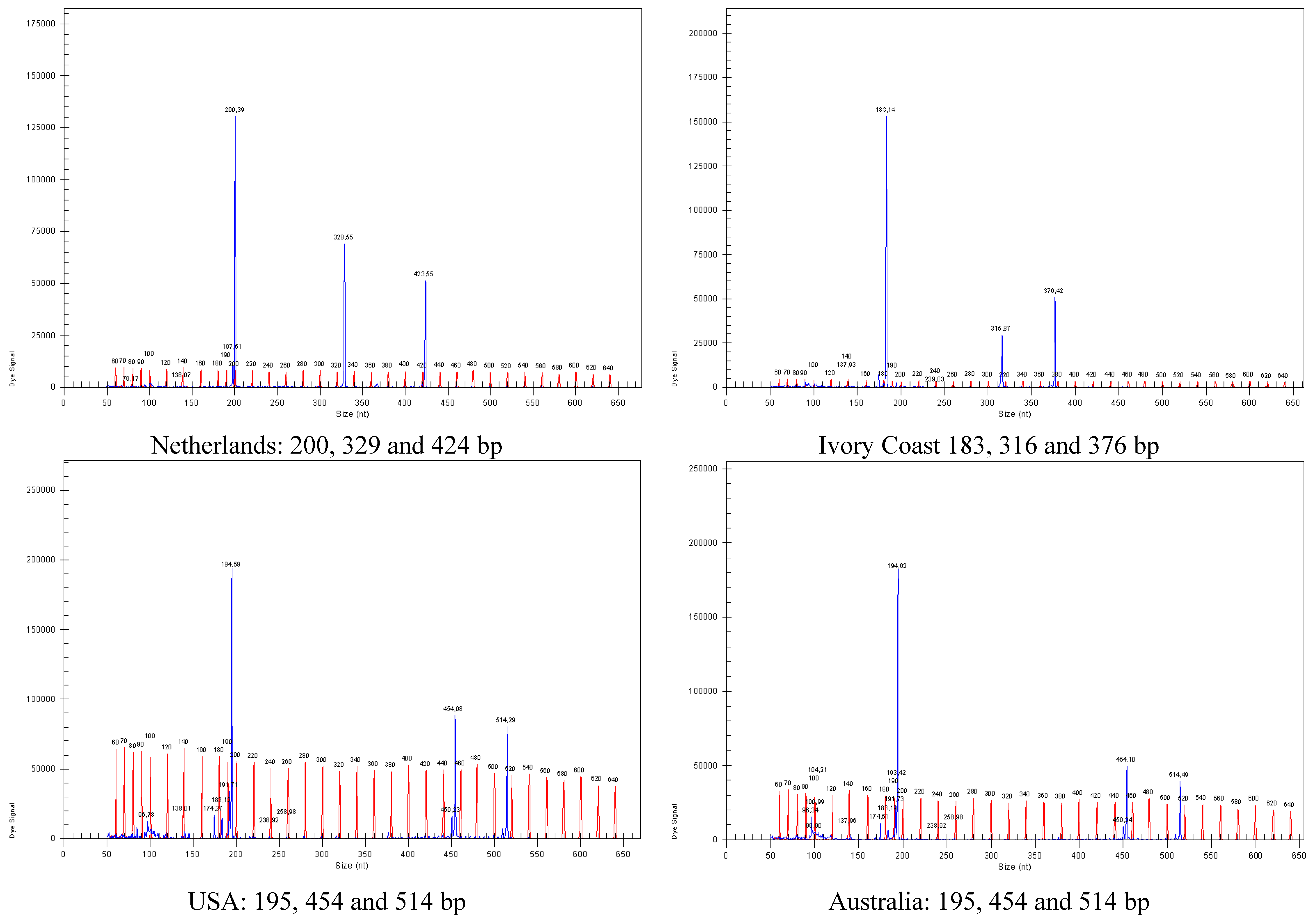
3.4. Parallel In-Root Detection of Endophytic T. virens Strains
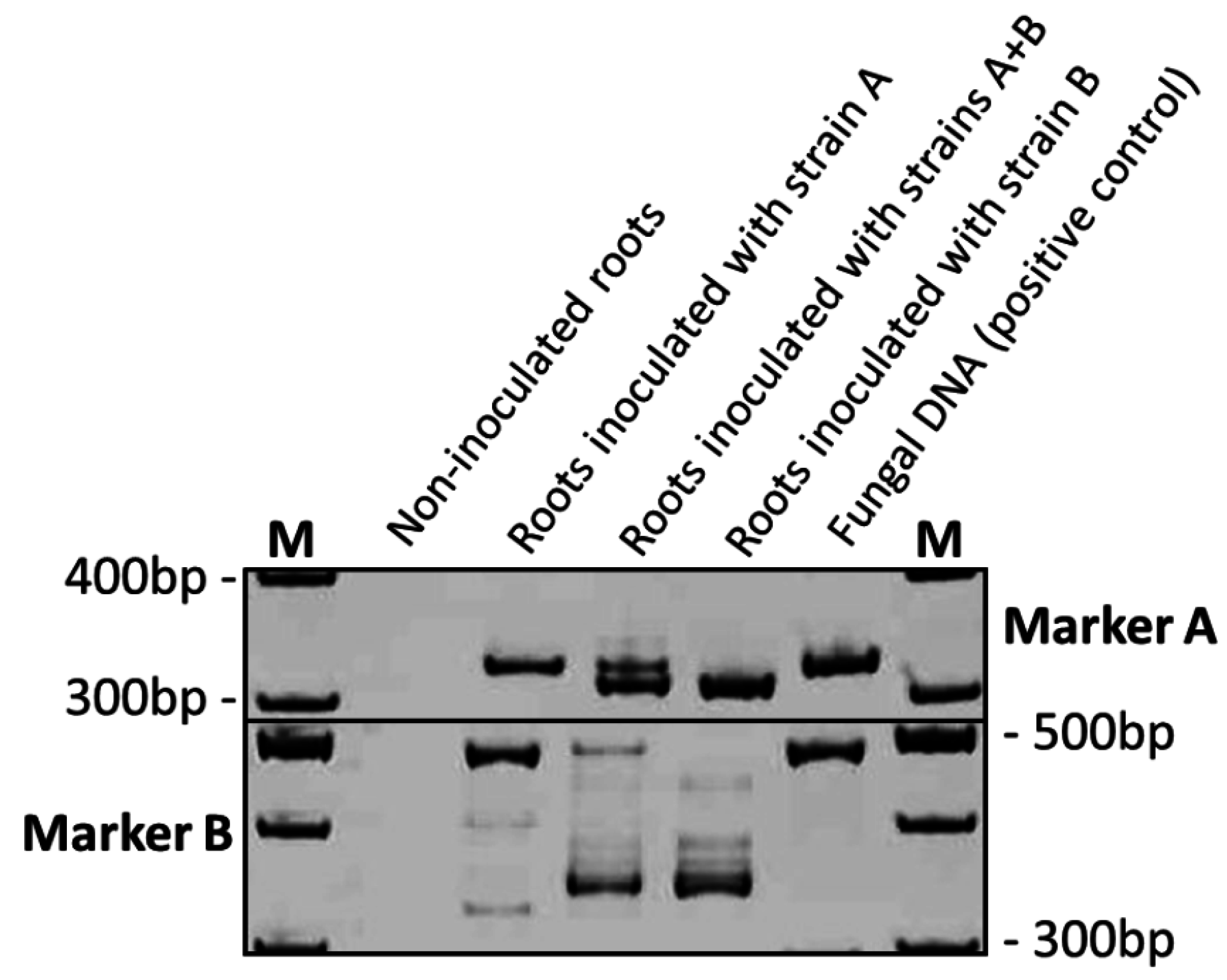
3.5. Determination of Strain-Specific Root Colonization Efficiencies by Quantitative PCR
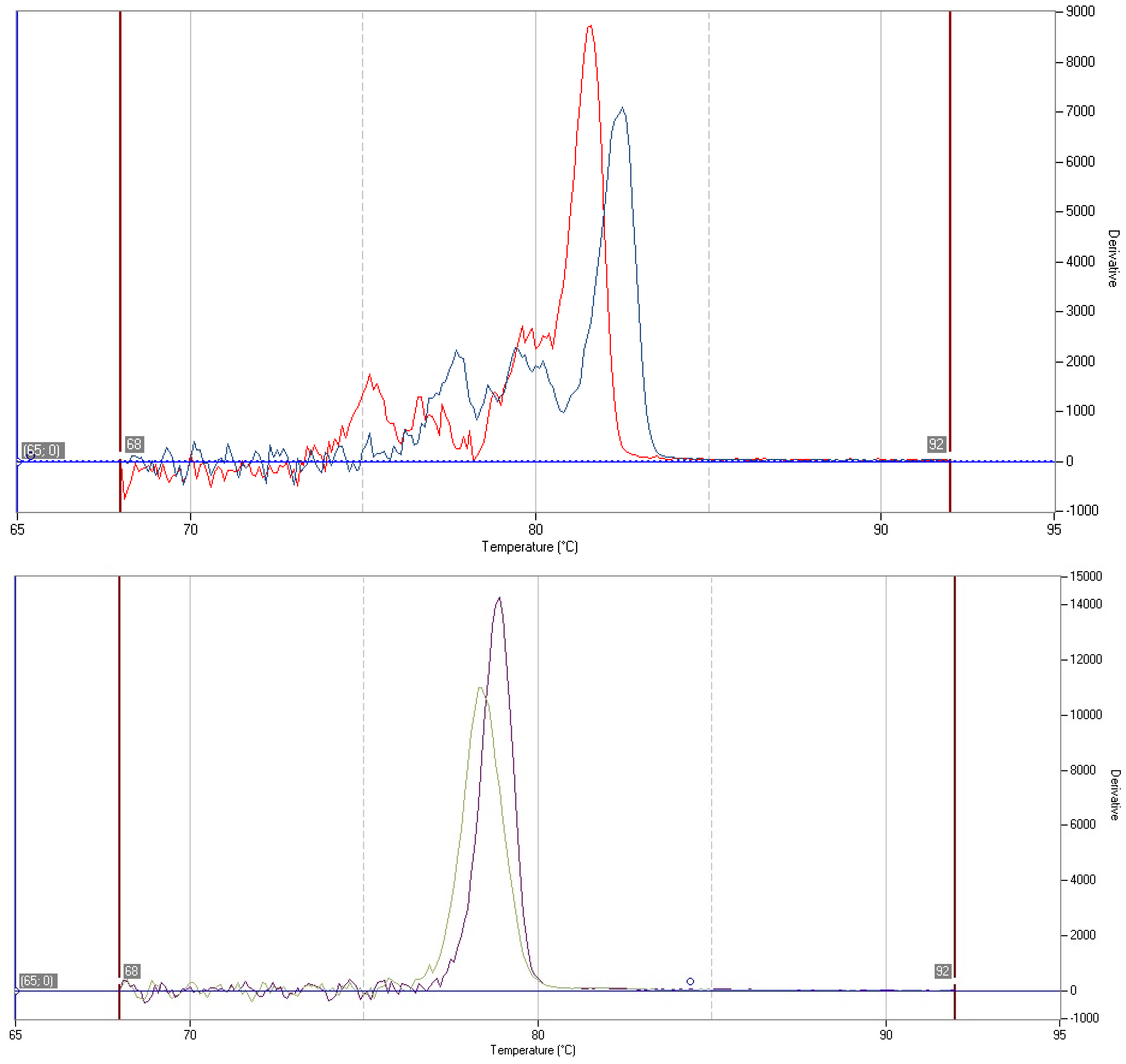
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chaverri, P.; Samuels, G.J.; Stewart, E.L. Hypocrea. virens sp. nov., the teleomorph of Trichoderma. virens. Mycologia 2001, 95, 1113–1124. [Google Scholar] [CrossRef]
- Miller, J.H.; Giddens, J.E.; Foster, A.A. A survey of the fungi of forest and cultivated soils in Georgia. Mycologia 1957, 49, 779–808. [Google Scholar] [CrossRef]
- Von Arx, J.A. Plant pathogenic fungi. Nova Hedwig. Beih. 1987, 87, 1–288. [Google Scholar]
- Atanasova, L.; Druzhinina, I.S.; Jaklitsch, W.M. Two hundred Trichoderma species recognized based on molecular phylogeny. In Trichoderma: Biology and Applications, 2nd ed.; Mukherjee, P.K., Sigh, U.S., Horwitz, B.A., Schmoll, M., Mukherjee, M., Eds.; CABI of Nosworthy Way: Wallingford, UK, 2013; pp. 10–26. [Google Scholar]
- Jaklitsch, W.M. European species of Hypocrea. Part 1. The green-spored species. Stud. Mycol. 2009, 63, 1–91. [Google Scholar] [CrossRef] [PubMed]
- Druzhinina, I.S.; Kopchinskiy, A.G.; Komon, M.; Bissett, J.; Szakacs, G.; Kubicek, C.P. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 2005, 42, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Zafra, G.; Moreno-Montano, A.; Absalon, A.E.; Cortes-Espinosa, D.V. Degradation of polycyclic aromatic hydrocarbons in soil by a tolerant strain of Trichoderma asperellum. Environ. Sci. Pollut. Res. Int. 2015, 22, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Divya, L.M.; Prasanth, G.K.; Sadasivan, C. Potential of the salt-tolerant laccase-producing strain Trichoderma viride Pers. NFCCI-2745 from an estuary in the bioremediation of phenol-polluted environments. J. Basic Microbiol. 2014, 54, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Arfarita, N.; Imai, T.; Kanno, A.; Yarimizu, T.; Xiaofeng, S.; Jie, W.; Higuchi, T.; Akada, R. The potential use of Trichoderma viride strain FRP3 in biodegradation of the herbicide glyphosate. Biotechnol. Biotechnol. Equip. 2013, 27, 3518–3521. [Google Scholar] [CrossRef]
- Hoyos-Carvajal, L.; Orduz, S.; Bisset, J. Growth stimulation in bean (Phaseolus vulgaris L.) by Trichoderma. Biol. Control 2009, 51, 409–416. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma Species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Blaszczyk, L.; Popiel, D.; Chelkowski, J.; Koczyk, G.; Samuels, G.J.; Sobieralski, K.; Siwulski, M. Species diversity of Trichoderma in Poland. J. Appl. Genet. 2011, 52, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, N.W.; Maruthachalam, K.; Subbarao, K.V.; Brown, M.; Xiao, Y.; Robertson, C.L.; Schneider, R.W. Mycoparasitism of Phakopsora pachyrhizi, the soybean rust pathogen, by Simplicillium lanosoniveum. Biol. Control 2014, 76, 87–94. [Google Scholar] [CrossRef]
- Zeng, L.-M.; Zhang, J.; Han, Y.-C.; Wu, M.; Jiang, D.-H.; Chen, W.; Li, G.-Q. Degradation of oxalic acid by the mycoparasite Coniothyrium minitans plays an important role in interacting with Sclerotinia sclerotiorum. Environ. Microbiol. 2014, 16, 2591–2610. [Google Scholar] [CrossRef] [PubMed]
- Qualhato, T.F.; Lopes, F.A.C.; Steindorff, A.S.; Brandao, R.S.; Jesuino, R.S.A.; Ulhoa, C.J. Mycoparasitism study of Trichoderma species against three phytopathogenic fungi: Evaluation of antagonism and hydrolytic enzyme production. Biotechnol. Lett. 2013, 35, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Matarese, F.; Sarrocco, S.; Gruber, S.; Seidl-Seiboth, V.; Vannacci, G. Biocontrol of Fusarium head blight: Interactions between Trichoderma and mycotoxigenic Fusarium. Microbiology 2012, 158, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Mulaw, T.B.; Druzhinina, I.; Kubicek, C.P.; Atanasova, L. Novel endophytic Trichoderma spp. isolated from healthy Coffea arabica roots are capable of controlling coffee tracheomycosis. Diversity 2013, 5, 750–766. [Google Scholar] [CrossRef]
- Yang, P. Pathogenic fungi induce the expression of Trichoderma asperellum cell wall degrading enzymes in the process of mycoparasitism. Adv. Mater. Res. 2014, 937, 282–285. [Google Scholar] [CrossRef]
- Shi, M.; Chen, L.; Wang, X.-W.; Zhang, T.; Zhao, P.-B.; Song, X.-Y.; Sun, C.-Y.; Chen, X.-L.; Zhou, B.-C.; Zhang, Y.-Z. Antimicrobial peptaibols from Trichoderma pseudokoningii induce programmed cell death in plant fungal pathogens. Microbiology 2012, 158, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Mbarga, J.B.; Begoude, B.A.D.; Ambang, Z.; Meboma, M.; Kuate, J.; Schiffers, B.; Ewbank, W.; Dedieu, L.; Ten Hoopen, G.M. A new oil-based formulation of Trichoderma asperellum for the biological control of cacao black pot disease caused by Phytophthora megakarya. Biol. Control 2014, 77, 15–22. [Google Scholar] [CrossRef]
- Perazzolli, M.; Moretto, M.; Fontana, P.; Ferrarini, A.; Velasco, R.; Moser, C.; Delledonne, M.; Pertot, I. Downy mildew resistance induced by Trichoderma harzianum T39 in susceptible grapevines partially mimics transcriptional changes of resistant genotypes. BMC Genomics 2012, 13, 660–679. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.M.; Johnsson Holmberg, A.-I.; Sundh, I.; Ricard, T.; Melin, P. Specific SCAR markers and a multiplex real-time PCR for quantification of two Trichoderma biocontrol strains in environmental samples. BioControl 2011, 56, 903–913. [Google Scholar] [CrossRef]
- Naeimi, S.; Kocsube, S.; Antal, Z.; Okhovvat, S.M.; Javan-Nikkhah, M.; Vagvölgyi, C.; Kredics, L. Strain-specific SCAR markers for the detection of Trichoderma harzianum AS12–2, a biological control agent against Rhizoctonia solani, the causal agent of rice sheath blight. Acta Biol. Hung. 2011, 62, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, A.; Jimenez, M.; Arcia, A.; Ulacio, D.; Mendez, N. Molecular characterization of 12 isolates of Trichoderma spp. using RAPD and rDNA-ITS. Bioagro 2013, 25, 167–174. [Google Scholar]
- Devi, S.S.; Sreenivasulu, Y.; Saritha, S.; Kumar, M.R.; Kumar, K.P.; Sudhakar, P. Molecular diversity of native Trichoderma isolates against Fusarium oxysporum f. sp. lycopersici (Sacc.), a causal agent of Fusarium wilt in tomato (Lycopersicon esculentum Mill.). Arch. Phytopathol. Plant Prot. 2012, 45, 686–698. [Google Scholar] [CrossRef]
- Lubeck, M.; Bulat, S.; Alekhina, I.; Lieckfeldt, E. Delineation of species within the Trichoderma viride/atroviride/koningii complex by UP-PCR cross-blot hybridization. FEMS Biol. Lett. 2004, 237, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Srivastava, M.; Kumar, V.; Singh, A.; Pandey, S. Genetic determination of potential Trichoderma. species using ISSR (microsatellite) marker in Uttar Pradesh, India. J. Microb. Biochem. Technol. 2014, 6, 174–178. [Google Scholar] [CrossRef]
- Siddiquee, S.; Tan, S.G.; Yusuf, U.K.; Fatihah, N.H.N.; Hasan, M.M. Characterization of Malaysian Trichoderma isolates using random amplified microsatellites. Mol. Biol. Rep. 2012, 39, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Geistlinger, J.; Weising, K.; Winter, P.; Kahl, G. Locus-specific microsatellite markers for the fungal chickpea pathogen Didymella rabiei (anamorph Ascochyta rabiei). Mol. Ecol. 2000, 9, 1939–1941. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Bahkali, A.H.; Abd-Elsalam, K.A.; Guo, J.-R.; Khiyami, M.A.; Verreet, J.-A. Characterization of novel di-, tri-, and tetranucleotide microsatellite primers suitable for genotyping various plant pathogenic fungi with special emphasis on fusaria and Mycospherella graminicola. Int. J. Mol. Sci. 2012, 13, 2951–2964. [Google Scholar] [CrossRef] [PubMed]
- Consolo, V.F.; Monaco, C.I.; Cordo, C.A.; Salerno, G.L. Characterization of novel Trichoderma spp. isolates as a search for effective biocontrollers of fungal diseases of economically important crops in Argentina. World J. Microbiol. Biotechnol. 2012, 28, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.M.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA for phylogenetics. In PCR Protocols: A Guide to Methods and Applications, 1st ed.; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–321. [Google Scholar]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–291. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.C.; Wilken, M.; Coetzee, M.P.A.; Wingfield, M.J.; Wingfield, B.D. Analysis of microsatellite markers in the genome of the plant pathogen Ceratocystis fimbriata. Fungal Biol. 2013, 117, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, G.S.; Bassil, N.V.; Barney, D.L.; Knaus, B.J.; Hummer, K.E. Short-read DNA sequencing yields microsatellite markers for Rheum. J. Am. Soc. Hortic. Sci. 2014, 139, 22–29. [Google Scholar]
- Anderson, S.J.; Stone, C.L.; Posada-Buitrago, M.L.; Boore, J.L.; Neelam, B.A.; Stephens, R.M.; Luster, D.G.; Frederick, R.D.; Pedley, K.F. Development of simple sequence repeat markers for the soybean rust fungus, Phakopsora pachyrhizi. Mol. Ecol. Resour. 2008, 8, 1310–1312. [Google Scholar] [CrossRef] [PubMed]
- Bucheli, E.; Gautschi, B.; Shykoff, J.A. Host-specific differentiation in the anther smut fungus Microbotryum violaceum as revealed by microsatellites. J. Evolut. Biol. 2000, 13, 188–198. [Google Scholar] [CrossRef]
- Reineke, A.; Bischoff-Schaefer, M.; Rondot, Y.; Galidevara, S.; Hirsch, J.; Devi, K.U. Microsatellite markers to monitor a commercialized isolate of the entomopathogenic fungus Beauveria bassiana in different environments: Technical validation and first applications. Biol. Control 2014, 70, 1–8. [Google Scholar] [CrossRef]
- Cipriani, G.; Marrazzo, M.T.; di Gaspero, G.; Pfeiffer, A.; Morgante, M.; Testolin, R. A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biol. 2008, 8, 127. [Google Scholar] [CrossRef] [PubMed]
- Schoebel, C.N.; Jung, E.; Prospero, S. Development of new polymorphic microsatellite markers for three closely related Phytophthora species using 454-pyrosequencing and their potential applications. Phytopathology 2013, 103, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Prospero, S.; Jung, E.; Tsykun, T.; Rigling, D. Eight microsatellite markers for Armillaria cepistipes and their transferability to other Armillaria species. Eur. J. Plant Pathol. 2010, 127, 165–170. [Google Scholar] [CrossRef]
- Dillon, N.L.; Innes, D.J.; Bally, I.S.E.; Wright, C.L.; Devitt, L.C.; Dietzgen, R.G. Expressed sequence tag-simple sequence repeat (EST-SSR) marker resources for diversity analysis of mango (Magnifera indica L.). Diversity 2014, 6, 72–87. [Google Scholar] [CrossRef]
- Sehic, J.; Garkava-Gustavsson, L.; Fernandez-Fernandez, F.; Nyboom, H. Genetic diversity in a collection of European pears (Pyrus communis) cultivars determined with SSR markers chosen by ECPGR. Sci. Hortic. 2012, 145, 39–45. [Google Scholar] [CrossRef]
- Bouajila, A.; Zoghlami, N.; Murad, S.; Baum, M.; Ghorbel, A.; Nazari, K. Genetic differentiation in Pyrenophora teres f. teres populations from Syria and Tunisia as assessed by AFLP markers. Lett. Appl. Microbiol. 2013, 56, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Urbanelli, S.; della Rosa, V.; Fanelli, C.; Fabbri, A.A.; Reverberi, M. Genetic diversity and population structure of the Italian fungi belonging to the taxa Pleurotus eryngii and P. ferulae. Heredity 2013, 90, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wadl, P.A.; Wood-Jones, A.; Windham, G.; Trigiano, R.N.; Scruggs, M.; Pilgrim, C.; Baird, R. Characterization of expressed sequence tag-derived simple sequence repeat markers for Aspergillus flavus: Emphasis on variability of isolates from the southern United States. Mycopathologia 2012, 174, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lascoux, M.; Overall, A.D.J.; Waxman, D. The characteristic trajectory of a fixing allele: A consequence of fictitious selection that arises from conditioning. Genetics 2013, 195, 993–1006. [Google Scholar] [CrossRef] [PubMed]
- Aguilée, R.; Claessen, D.; Lambert, A. Allele fixation in a dynamic meta-population: Founder effects vs. refuge effects. Theor. Popul. Biol. 2009, 76, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Boose, D.; Harrison, S.; Clement, S.; Meyer, S. Population genetic structure of the seed pathogen Pyrenophora semeniperda on Bromus tectorum in western North America. Mycologia 2011, 103, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Saldamondo, C.I.; Velez-Arango, A.M. Host plant association and genetic differentiation of corn and rice strains of Spodoptera. frugiperda in Colombia. Neotropical. Entomology 2010, 39, 921–929. [Google Scholar] [CrossRef]
- Dong, Z.; Lui, Z.; Lui, D. Genetic characterization of the scyphozoan jellyfish Aurelia spp. in Chinese coastal waters using mitochondrial markers. Biochem. Syst. Ecol. 2015, 60, 15–23. [Google Scholar] [CrossRef]
- Gordon, M.; van Norman, K. Molecular monitoring of protected fungi: Mycelium persistence in soil after timber harvest. Fungal Ecol. 2014, 9, 34–42. [Google Scholar] [CrossRef]
- Prabhakaran, N.; Promeeladevi, T.; Sathiyabama, M.; Kamil, D. Multiplex PCR for detection and differentiation of diverse Trichoderma species. Ann. Microbiol. 2015, 65, 1591–1595. [Google Scholar] [CrossRef]
- Haasbrock, M.P.; Craven, M.; Barnes, I.; Crampton, B.G. Microsatellite and mating type primers for the maize and sorghum pathogen Exserohilum turcicum. Aust. Plant Pathol. 2014, 43, 577–581. [Google Scholar] [CrossRef]
- Ohsumi, K.; Watanabe, M.; Fujie, A. AS2077715 is a selective inhibitor of fungal mitochondrial cytochrome BC 1 complex. J. Antibiot. 2014, 67, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.-Z.; Mao, L.-J.; Li, N.; Feng, X.-X.; Yuan, Z.-L.; Wang, L.-W.; Lin, F.-C.; Zhang, C.-L. Evidence for biotrophic lifestyle and biocontrol potential of dark septate endophyte Harpophora oryzae to rice blast disease. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed]
- Reininger, V.; Sieber, T.N. Mitigation of antagonistic effects on plant growth due to root co-colonization by dark septate endophytes and ectomycorrhiza. Environ. Microbiol. Rep. 2013, 5, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Saravesi, K.; Ruotsalainen, A.L.; Cahill, J.F. Contrasting impacts of defoliation on root colonization by arbuscular mycorrhizal fungi and dark septate endophyte fungi of Medicago sativa. Mycorrhiza 2014, 24, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Anith, K.N.; Faseela, K.M.; Archana, P.A.; Prathapan, K.D. Compatibility of Piriformospora indica and Trichoderma harzianum as dual inoculants in black pepper (Piper nigrum L.). Symbiosis 2011, 55, 11–17. [Google Scholar] [CrossRef]
- Sathiyadash, K.; Muthukumar, T.; Uma, E. Arbuscular mycorrhizal and dark septate endophyte fungal associations in South Indian grasses. Symbiosis 2010, 52, 21–32. [Google Scholar] [CrossRef]
- Wang, X.-M.; Yang, B.; Wang, H.-W.; Yang, T.; Ren, C.-G.; Zheng, H.-L.; Dai, C.-C. Consequences of antagonistic interactions between endophytic fungus and bacterium on plant growth and defense responses in Atractylodes lancea. J. Basic Microbiol. 2015, 55, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kumar, S.; Kashyap, P.L.; Srivastava, A.K.; Mishra, S.; Sharma, A.K. Identification and characterization of microsatellite from Alternaria brassicicola to access cross-species transferability and utility as a diagnostic marker. Mol. Biotechnol. 2014, 56, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Shi, S.; Bao, Y.; Yao, H.; Jing, Y.; Lin, Z. Development of 90 EST-SNP markers in blood clam (Tegillarca granosa) using high resolution melting (HRM). Conserv. Genet. Resour. 2015, 7, 309–314. [Google Scholar] [CrossRef]
- Capper, R.L.; Jin, Y.K.; Lundgren, P.B.; Peplow, L.M.; Matz, M.V.; van Oppen, M.J.H. Quantitative high resolution melting: Two methods to determine SNP allele frequencies from pooled samples. BMC Genet. 2015, 16, 62–75. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geistlinger, J.; Zwanzig, J.; Heckendorff, S.; Schellenberg, I. SSR Markers for Trichoderma virens: Their Evaluation and Application to Identify and Quantify Root-Endophytic Strains. Diversity 2015, 7, 360-384. https://doi.org/10.3390/d7040360
Geistlinger J, Zwanzig J, Heckendorff S, Schellenberg I. SSR Markers for Trichoderma virens: Their Evaluation and Application to Identify and Quantify Root-Endophytic Strains. Diversity. 2015; 7(4):360-384. https://doi.org/10.3390/d7040360
Chicago/Turabian StyleGeistlinger, Joerg, Jessica Zwanzig, Sophie Heckendorff, and Ingo Schellenberg. 2015. "SSR Markers for Trichoderma virens: Their Evaluation and Application to Identify and Quantify Root-Endophytic Strains" Diversity 7, no. 4: 360-384. https://doi.org/10.3390/d7040360
APA StyleGeistlinger, J., Zwanzig, J., Heckendorff, S., & Schellenberg, I. (2015). SSR Markers for Trichoderma virens: Their Evaluation and Application to Identify and Quantify Root-Endophytic Strains. Diversity, 7(4), 360-384. https://doi.org/10.3390/d7040360





